Circulating Cell-Free Microbial DNA Signatures and Plasma Soluble CD14 Level Are Associated with Clinical Outcomes of Anti-PD-1 Therapy in Advanced Melanoma Patients
Abstract
1. Introduction
2. Results
2.1. Cohort Characteristics
2.2. Taxonomic Profile of the Circulating cfmDNA at the Phylum Level
2.3. Bacterial ASV Alpha Diversity of the Circulating cfmDNA
2.4. The Comparison of the Circulating cfmDNA Signatures Between Patients with Favorable and Unfavorable Clinical Outcomes at Baseline (at T0) and During the Anti-PD-1 Therapy (at T1 and Tn)
2.5. Changes in the Circulating cfmDNA Signatures During the Anti-PD-1 Therapy Within Subgroups of Patients with Favorable and Unfavorable Clinical Outcomes (T0 vs. T1, T0 vs. Tn, and T1 vs. Tn)
2.6. The Association Between the Baseline Circulating cfmDNA Signatures and Survival Outcomes
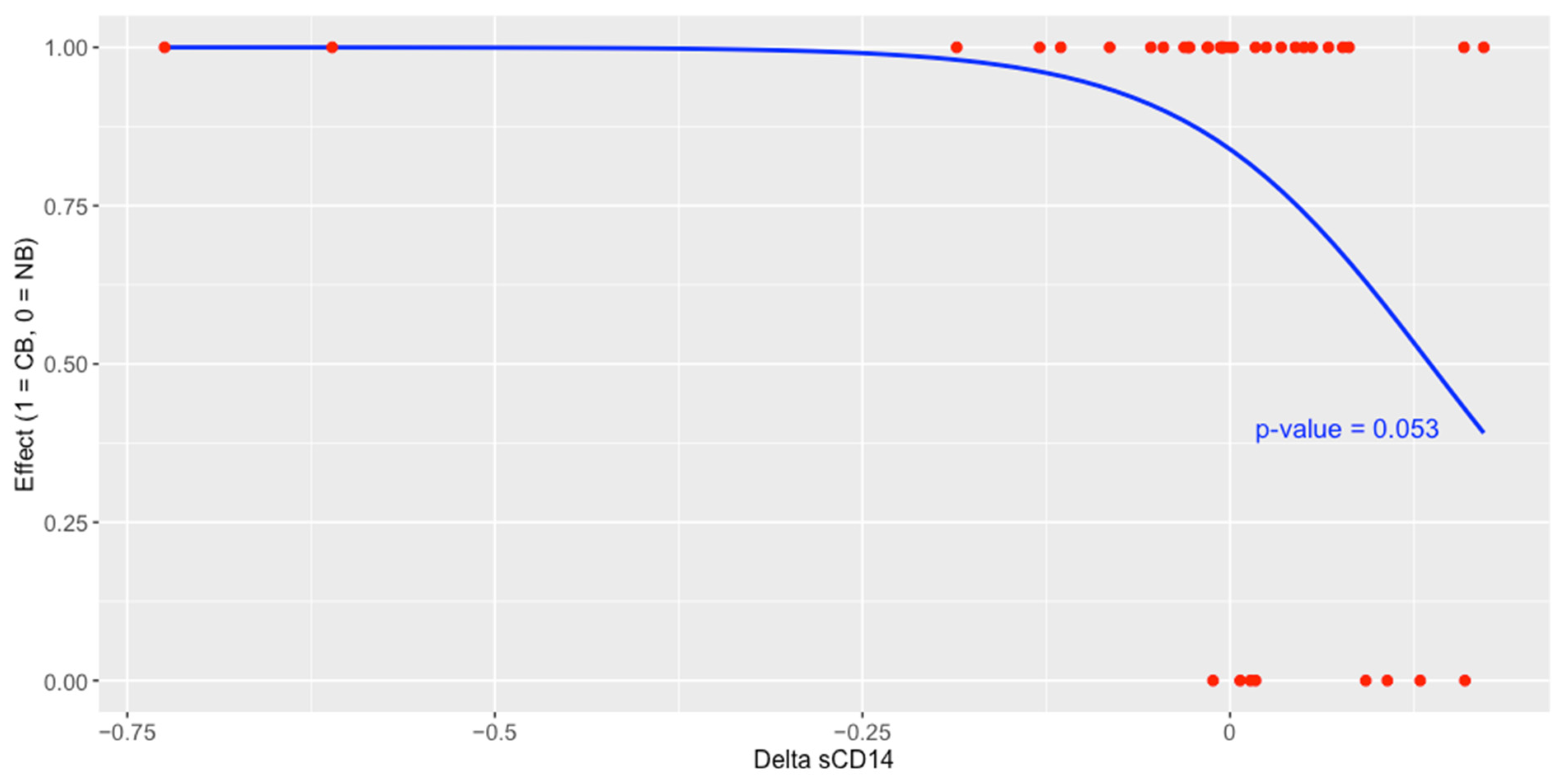
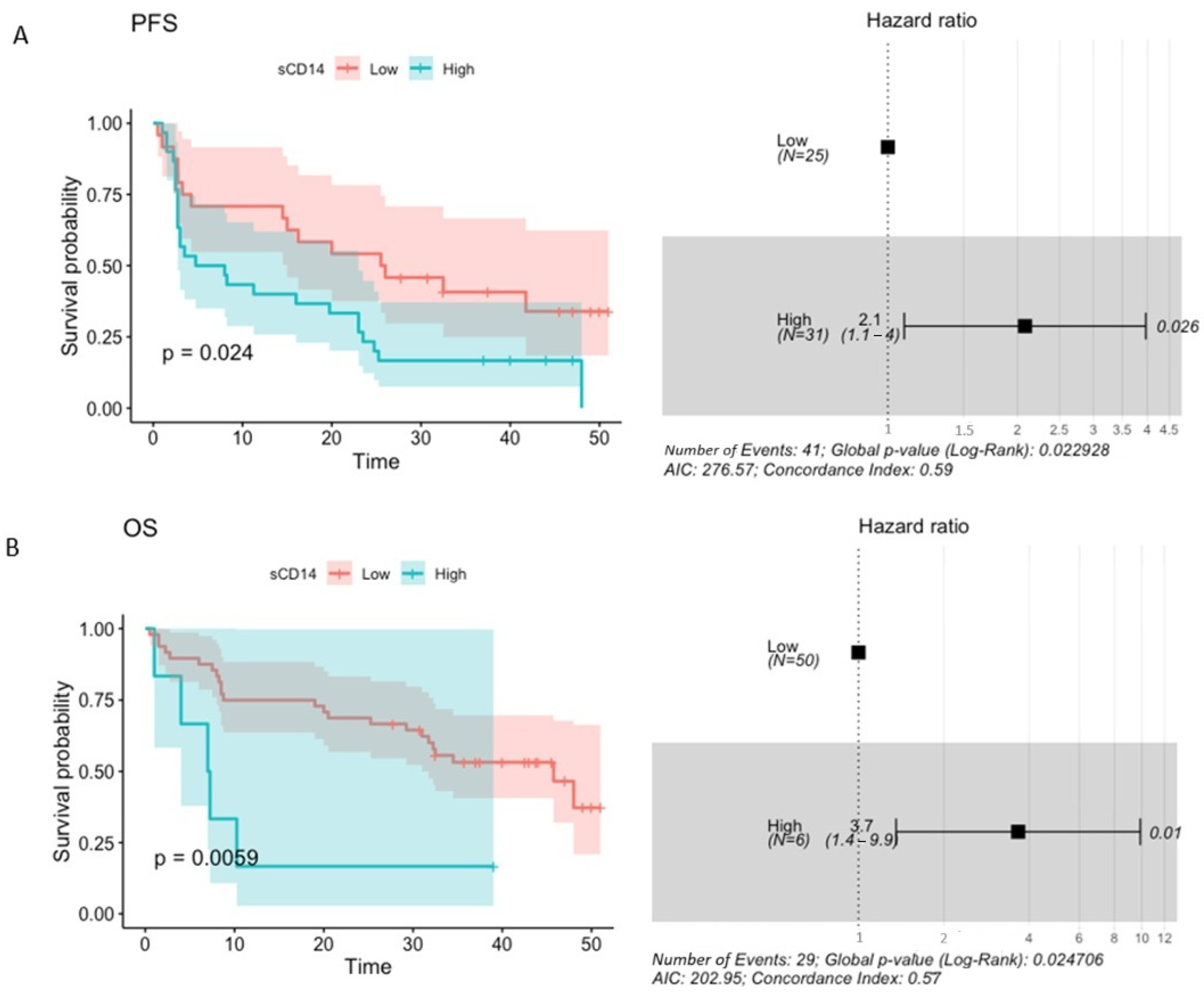
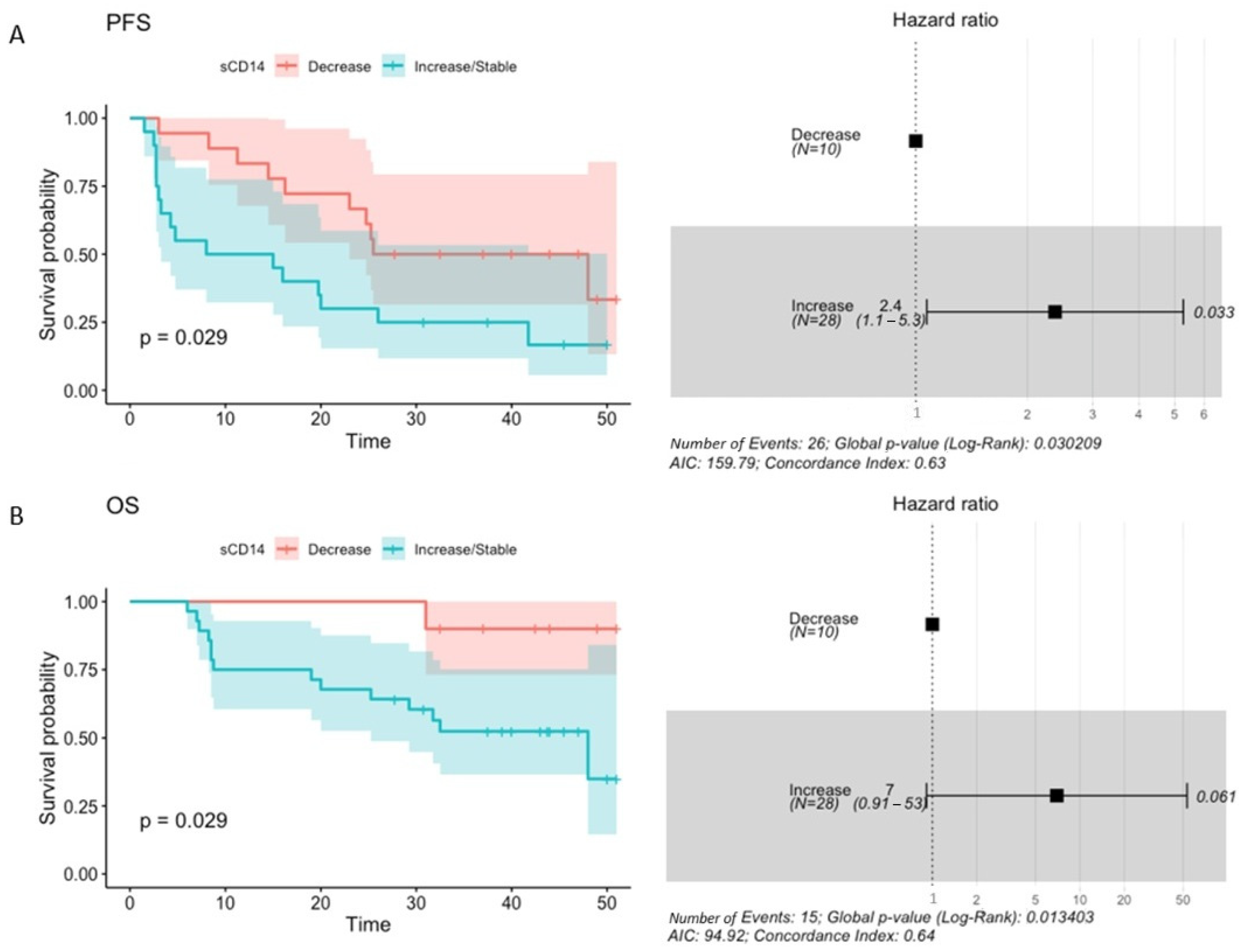
2.7. The Concentration of Plasma sCD14 and Its Association with Survival Outcomes
3. Discussion
4. Materials and Methods
4.1. Study Cohort and Clinical Data Collection
4.2. Blood Sample Collection and Measurement of Plasma sCD14 Concentration
4.3. Circulating cfDNA Extraction and 16S rRNA Gene Sequencing
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gosman, L.M.; Țăpoi, D.-A.; Costache, M. Cutaneous Melanoma: A Review of Multifactorial Pathogenesis, Immunohistochemistry, and Emerging Biomarkers for Early Detection and Management. Int. J. Mol. Sci. 2023, 24, 15881. [Google Scholar] [CrossRef]
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol. 2022, 158, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, R.W.; Barbie, D.A.; Flaherty, K.T. Mechanisms of Resistance to Immune Checkpoint Inhibitors. Br. J. Cancer 2018, 118, 9–16. [Google Scholar] [CrossRef]
- Tomela, K.; Pietrzak, B.; Schmidt, M.; Mackiewicz, A. The Tumor and Host Immune Signature, and the Gut Microbiota as Predictive Biomarkers for Immune Checkpoint Inhibitor Response in Melanoma Patients. Life 2020, 10, 219. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, Y.; Zhang, S.; Dong, L. Gut Microbiota-Mediated Immunomodulation in Tumor. J. Exp. Clin. Cancer Res. 2021, 40, 221. [Google Scholar] [CrossRef]
- Lee, K.A.; Thomas, A.M.; Bolte, L.A.; Björk, J.R.; de Ruijter, L.K.; Armanini, F.; Asnicar, F.; Blanco-Miguez, A.; Board, R.; Calbet-Llopart, N.; et al. Cross-Cohort Gut Microbiome Associations with Immune Checkpoint Inhibitor Response in Advanced Melanoma. Nat. Med. 2022, 28, 535–544. [Google Scholar] [CrossRef]
- McCulloch, J.A.; Davar, D.; Rodrigues, R.R.; Badger, J.H.; Fang, J.R.; Cole, A.M.; Balaji, A.K.; Vetizou, M.; Prescott, S.M.; Fernandes, M.R.; et al. Intestinal Microbiota Signatures of Clinical Response and Immune-Related Adverse Events in Melanoma Patients Treated with Anti-PD-1. Nat. Med. 2022, 28, 545–556. [Google Scholar] [CrossRef]
- Pietrzak, B.; Tomela, K.; Olejnik-Schmidt, A.; Galus, Ł.; Mackiewicz, J.; Kaczmarek, M.; Mackiewicz, A.; Schmidt, M. A Clinical Outcome of the Anti-PD-1 Therapy of Melanoma in Polish Patients Is Mediated by Population-Specific Gut Microbiome Composition. Cancers 2022, 14, 5369. [Google Scholar] [CrossRef]
- Björk, J.R.; Bolte, L.A.; Maltez Thomas, A.; Lee, K.A.; Rossi, N.; Wind, T.T.; Smit, L.M.; Armanini, F.; Asnicar, F.; Blanco-Miguez, A.; et al. Longitudinal Gut Microbiome Changes in Immune Checkpoint Blockade-Treated Advanced Melanoma. Nat. Med. 2024, 30, 785–796. [Google Scholar] [CrossRef]
- Worthley, D.L.; Cole, S.R.; Esterman, A.; Mehaffey, S.; Roosa, N.M.; Smith, A.; Turnbull, D.; Young, G.P. Screening for Colorectal Cancer by Faecal Occult Blood Test: Why People Choose to Refuse. Intern. Med. J. 2006, 36, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, B.; Kawacka, I.; Olejnik-Schmidt, A.; Schmidt, M. Circulating Microbial Cell-Free DNA in Health and Disease. Int. J. Mol. Sci. 2023, 24, 3051. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-F.; Chen, Y.-J.; Fan, T.-C.; Chang, N.-C.; Chen, Y.-J.; Midha, M.K.; Chen, T.-H.; Yang, H.-H.; Wang, Y.-T.; Yu, A.L.; et al. Analysis of Microbial Sequences in Plasma Cell-Free DNA for Early-Onset Breast Cancer Patients and Healthy Females. BMC Med. Genom. 2018, 11, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Zozaya-Valdés, E.; Wong, S.Q.; Raleigh, J.; Hatzimihalis, A.; Ftouni, S.; Papenfuss, A.T.; Sandhu, S.; Dawson, M.A.; Dawson, S.-J. Detection of Cell-Free Microbial DNA Using a Contaminant-Controlled Analysis Framework. Genome Biol. 2021, 22, 187. [Google Scholar] [CrossRef]
- Ouaknine Krief, J.; Helly de Tauriers, P.; Dumenil, C.; Neveux, N.; Dumoulin, J.; Giraud, V.; Labrune, S.; Tisserand, J.; Julie, C.; Emile, J.-F.; et al. Role of Antibiotic Use, Plasma Citrulline and Blood Microbiome in Advanced Non-Small Cell Lung Cancer Patients Treated with Nivolumab. J. Immunother. Cancer 2019, 7, 176. [Google Scholar] [CrossRef]
- Cauwels, A.; Frei, K.; Sansano, S.; Fearns, C.; Ulevitch, R.; Zimmerli, W.; Landmann, R. The Origin and Function of Soluble CD14 in Experimental Bacterial Meningitis1. J. Immunol. 1999, 162, 4762–4772. [Google Scholar] [CrossRef]
- Ichise, Y.; Saegusa, J.; Tanaka-Natsui, S.; Naka, I.; Hayashi, S.; Kuroda, R.; Morinobu, A. Soluble CD14 Induces Pro-Inflammatory Cytokines in Rheumatoid Arthritis Fibroblast-Like Synovial Cells via Toll-Like Receptor 4. Cells 2020, 9, 1689. [Google Scholar] [CrossRef]
- Sharygin, D.; Koniaris, L.G.; Wells, C.; Zimmers, T.A.; Hamidi, T. Role of CD14 in Human Disease. Immunology 2023, 169, 260–270. [Google Scholar] [CrossRef]
- Na, K.; Oh, B.-C.; Jung, Y. Multifaceted Role of CD14 in Innate Immunity and Tissue Homeostasis. Cytokine Growth Factor Rev. 2023, 74, 100–107. [Google Scholar] [CrossRef]
- Zaidi, A.H.; Pratama, M.Y.; Omstead, A.N.; Gorbonova, A.; Mansoor, R.; Melton-Kreft, R.; Jobe, B.A.; Wagner, P.L.; Kelly, R.J.; Goel, A. A Blood-Based Circulating Microbial Metagenomic Panel for Early Diagnosis and Prognosis of Oesophageal Adenocarcinoma. Br. J. Cancer 2022, 127, 2016–2024. [Google Scholar] [CrossRef]
- Glyn, T.; Purcell, R. Circulating Bacterial DNA: A New Paradigm for Cancer Diagnostics. Front. Med. 2022, 9, 831096. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Chen, B.; Pan, H.; Wang, D.; Liu, M.; Yang, Y.; Zou, M.; Yang, J.; Xiao, K.; Zhao, R.; et al. Detection of Microbial 16S rRNA Gene in the Serum of Patients With Gastric Cancer. Front. Oncol. 2019, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Lu, W.; Kong, X.; Shao, Y.W.; Hu, Y.; Wang, A.; Bao, H.; Cao, R.; Liu, K.; Wang, X.; et al. Alterations of Circulating Bacterial DNA in Colorectal Cancer and Adenoma: A Proof-of-Concept Study. Cancer Lett. 2021, 499, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, X.; Ho, C.L. Recent Development of Probiotic Bifidobacteria for Treating Human Diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef] [PubMed]
- Fidanza, M.; Panigrahi, P.; Kollmann, T.R. Lactiplantibacillus Plantarum–Nomad and Ideal Probiotic. Front. Microbiol. 2021, 12, 712236. [Google Scholar] [CrossRef]
- Martín, R.; Rios-Covian, D.; Huillet, E.; Auger, S.; Khazaal, S.; Bermúdez-Humarán, L.G.; Sokol, H.; Chatel, J.-M.; Langella, P. Faecalibacterium: A Bacterial Genus with Promising Human Health Applications. FEMS Microbiol. Rev. 2023, 47, fuad039. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The Commensal Microbiome Is Associated with Anti-PD-1 Efficacy in Metastatic Melanoma Patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Jin, Y.; Dong, H.; Xia, L.; Yang, Y.; Zhu, Y.; Shen, Y.; Zheng, H.; Yao, C.; Wang, Y.; Lu, S. The Diversity of Gut Microbiome Is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J. Thorac. Oncol. 2019, 14, 1378–1389. [Google Scholar] [CrossRef]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline Gut Microbiota Predicts Clinical Response and Colitis in Metastatic Melanoma Patients Treated with Ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef]
- Peters, B.A.; Wilson, M.; Moran, U.; Pavlick, A.; Izsak, A.; Wechter, T.; Weber, J.S.; Osman, I.; Ahn, J. Relating the Gut Metagenome and Metatranscriptome to Immunotherapy Responses in Melanoma Patients. Genome Med. 2019, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ye, J. Characterization of Gut Microbiota in Patients with Primary Hepatocellular Carcinoma Received Immune Checkpoint Inhibitors: A Chinese Population-Based Study. Medicine 2020, 99, e21788. [Google Scholar] [CrossRef]
- Coutzac, C.; Jouniaux, J.-M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic Short Chain Fatty Acids Limit Antitumor Effect of CTLA-4 Blockade in Hosts with Cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. All Disease Begins in the (Leaky) Gut: Role of Zonulin-Mediated Gut Permeability in the Pathogenesis of Some Chronic Inflammatory Diseases. F1000Research 2020, 9, 69. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The Human Tumor Microbiome Is Composed of Tumor Type-Specific Intracellular Bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Narunsky-Haziza, L.; Sepich-Poore, G.D.; Livyatan, I.; Asraf, O.; Martino, C.; Nejman, D.; Gavert, N.; Stajich, J.E.; Amit, G.; González, A.; et al. Pan-Cancer Analyses Reveal Cancer-Type-Specific Fungal Ecologies and Bacteriome Interactions. Cell 2022, 185, 3789–3806.e17. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wang, H. The Clinical and Immune Features of CD14 in Colorectal Cancer Identified via Large-Scale Analysis. Int. Immunopharmacol. 2020, 88, 106966. [Google Scholar] [CrossRef]
- Gustafson, M.P.; Lin, Y.; Bleeker, J.S.; Warad, D.; Tollefson, M.K.; Crispen, P.L.; Bulur, P.A.; Harrington, S.M.; Laborde, R.R.; Gastineau, D.A.; et al. Intratumoral CD14+ Cells and Circulating CD14+HLA-DRlo/Neg Monocytes Correlate with Decreased Survival in Patients with Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. 2015, 21, 4224–4233. [Google Scholar] [CrossRef]
- Kowarsky, M.; Camunas-Soler, J.; Kertesz, M.; De Vlaminck, I.; Koh, W.; Pan, W.; Martin, L.; Neff, N.F.; Okamoto, J.; Wong, R.J.; et al. Numerous Uncharacterized and Highly Divergent Microbes Which Colonize Humans Are Revealed by Circulating Cell-Free DNA. Proc. Natl. Acad. Sci. USA 2017, 114, 9623–9628. [Google Scholar] [CrossRef]
- Szmulski, Ł. Obwieszczenie Nr 65 Ministra Zdrowia z Dnia 30 Sierpnia 2019 r. w Sprawie Wykazu Refundowanych Leków, Środków Spożywczych Specjalnego Przeznaczenia Żywieniowego Oraz Wyrobów Medycznych. Available online: https://www.gov.pl/web/zdrowie/obwieszczenie-ministra-zdrowia-z-dnia-30-sierpnia-2019-r-w-sprawie-wykazu-refundowanych-lekow-srodkow-spozywczych-specjalnego-przeznaczenia-zywieniowego-oraz-wyrobow-medycznych-na-1-wrzesnia-2019-r (accessed on 23 October 2024).
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Schmidt, M. DADA2 Formatted Silva SSU Taxonomic Training Data (Silva Version 138.1) with Emended Description of the Genus Lactobacillus Beijerinck 1901. 2023. Available online: https://zenodo.org/records/8170966 (accessed on 23 October 2024).
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and Laboratory Contamination Can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Kers, J.G.; Saccenti, E. The Power of Microbiome Studies: Some Considerations on Which Alpha and Beta Metrics to Use and How to Report Results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef] [PubMed]
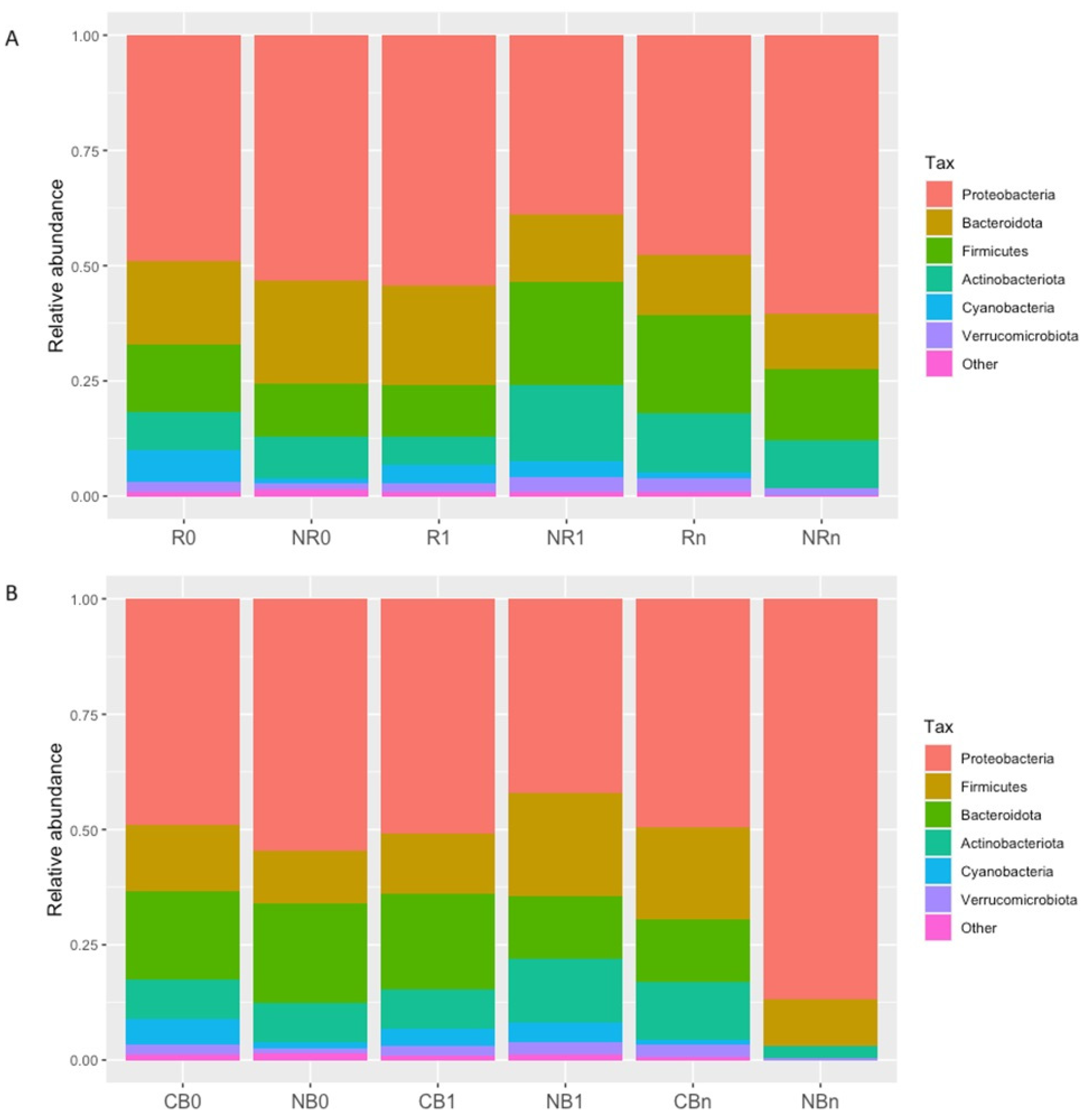
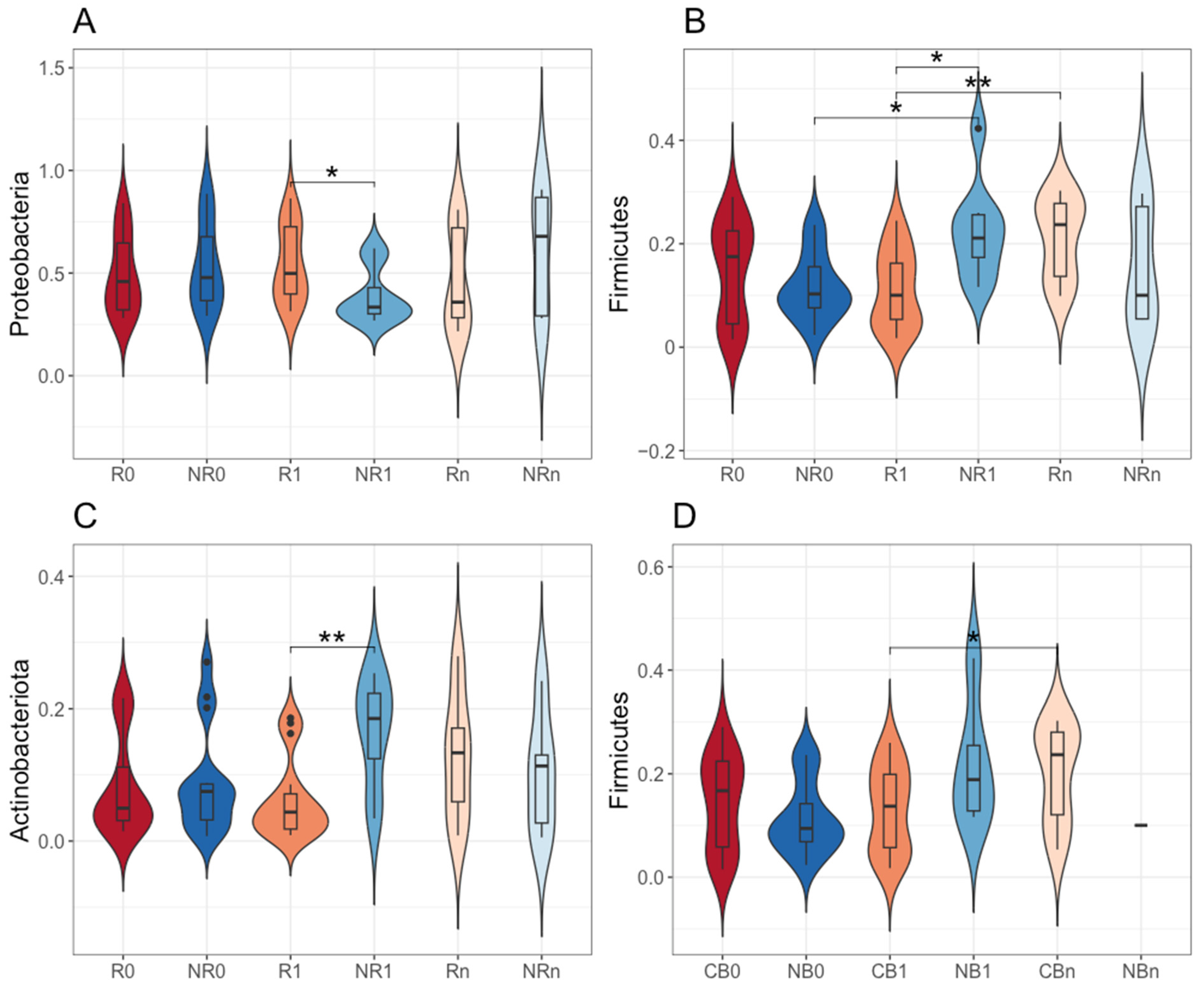

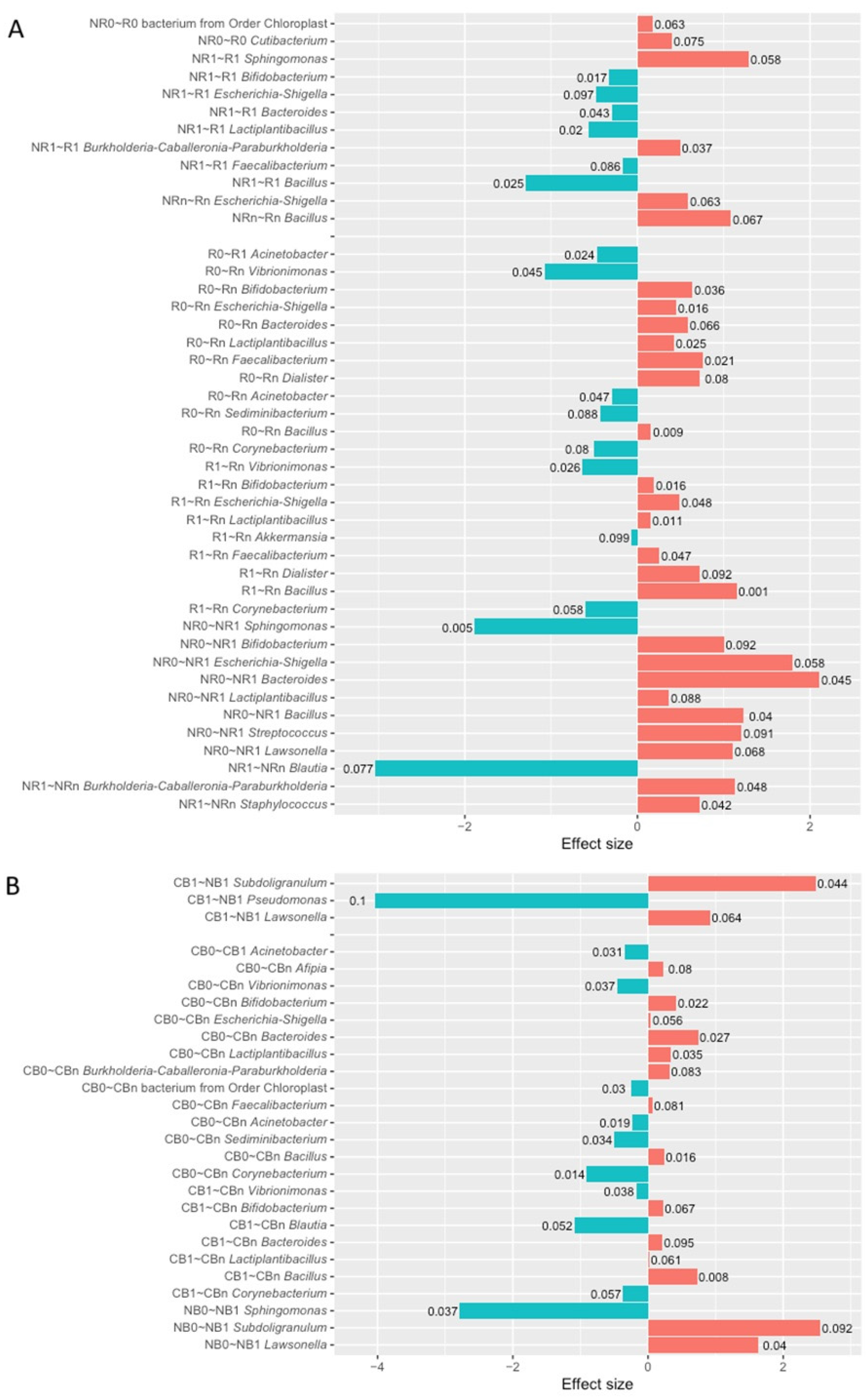

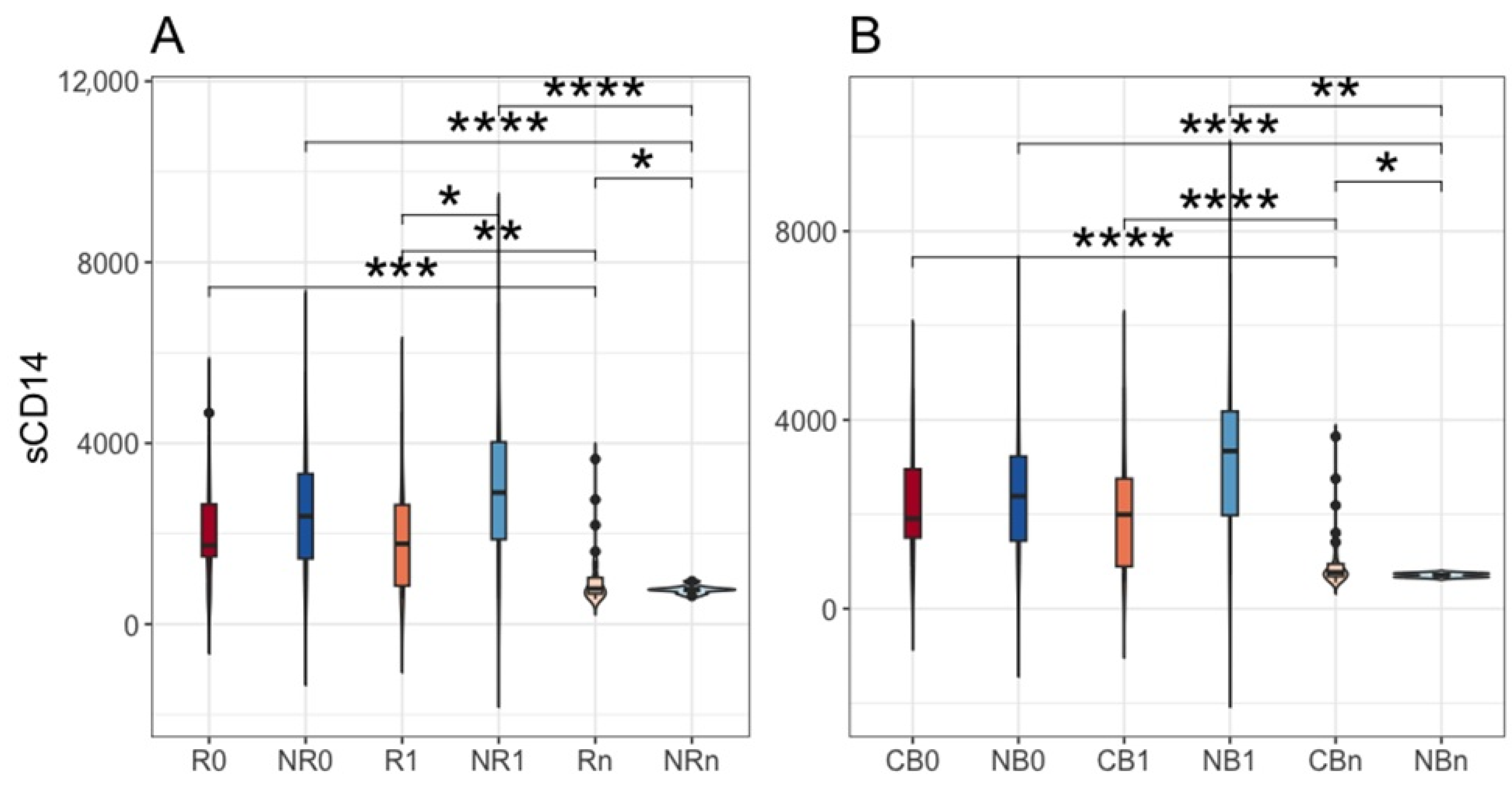
| Subject Characteristics | Responders (R; n = 28) | Non-Responders (NR; n = 38) | p-Value (R~NR) | Clinical Benefit (CB; n = 39) | No Benefit (NB; n= 27) | p-Value (CB~NB) |
|---|---|---|---|---|---|---|
| Sex, n (%) | ||||||
| Male | 15 (23) | 27 (41) | 0.197 a | 23 (35) | 19 (29) | 0.438 a |
| Female | 13 (20) | 11 (17) | 16 (24) | 8 (12) | ||
| Age (years), | ||||||
| median (range) | 64 (40–84) | 69 (32–92) | 0.215 b | 63 (32–85) | 70 (38–92) | 0.084 b |
| M-stage at diagnosis c, n (%) | ||||||
| IV M1a | 7 (11) | 9 (14) | 0.339 a | 13 (20) | 3 (5) | 0.067 a |
| IV M1b | 5 (8) | 5 (8) | 7 (11) | 3 (5) | ||
| IV M1c | 9 (14) | 13 (20) | 11 (17) | 11 (17) | ||
| IV M1d | 4 (6) | 9 (14) | 5 (8) | 8 (13) | ||
| IIIc | 3 (5) | 0 (0) | 3 (5) | 0 (0) | ||
| Serum LDH, n (%) | ||||||
| Normal (≤250 U/L) | 22 (34) | 21 (34) | 0.111 a | 31 (48) | 12 (19) | 0.014 a |
| Elevated (>250 U/L) | 6 (9) | 15 (23) | 8 (12) | 13 (20) | ||
| Serum LDH | ||||||
| median (range) | 197.5 (121–474) | 238 (141–1173) | 0.039 b | 195 (121–474) | 292 (141–1173) | 0.003 b |
| BRAF V600E/K mutation, n (%) | ||||||
| Present | 9 (14) | 16 (25) | 0.439 a | 13 (20) | 12 (19) | 0.298 a |
| Absent | 19 (30) | 20 (31) | 26 (41) | 13 (20) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drymel, B.; Tomela, K.; Galus, Ł.; Olejnik-Schmidt, A.; Mackiewicz, J.; Kaczmarek, M.; Mackiewicz, A.; Schmidt, M. Circulating Cell-Free Microbial DNA Signatures and Plasma Soluble CD14 Level Are Associated with Clinical Outcomes of Anti-PD-1 Therapy in Advanced Melanoma Patients. Int. J. Mol. Sci. 2024, 25, 12982. https://doi.org/10.3390/ijms252312982
Drymel B, Tomela K, Galus Ł, Olejnik-Schmidt A, Mackiewicz J, Kaczmarek M, Mackiewicz A, Schmidt M. Circulating Cell-Free Microbial DNA Signatures and Plasma Soluble CD14 Level Are Associated with Clinical Outcomes of Anti-PD-1 Therapy in Advanced Melanoma Patients. International Journal of Molecular Sciences. 2024; 25(23):12982. https://doi.org/10.3390/ijms252312982
Chicago/Turabian StyleDrymel, Bernadeta, Katarzyna Tomela, Łukasz Galus, Agnieszka Olejnik-Schmidt, Jacek Mackiewicz, Mariusz Kaczmarek, Andrzej Mackiewicz, and Marcin Schmidt. 2024. "Circulating Cell-Free Microbial DNA Signatures and Plasma Soluble CD14 Level Are Associated with Clinical Outcomes of Anti-PD-1 Therapy in Advanced Melanoma Patients" International Journal of Molecular Sciences 25, no. 23: 12982. https://doi.org/10.3390/ijms252312982
APA StyleDrymel, B., Tomela, K., Galus, Ł., Olejnik-Schmidt, A., Mackiewicz, J., Kaczmarek, M., Mackiewicz, A., & Schmidt, M. (2024). Circulating Cell-Free Microbial DNA Signatures and Plasma Soluble CD14 Level Are Associated with Clinical Outcomes of Anti-PD-1 Therapy in Advanced Melanoma Patients. International Journal of Molecular Sciences, 25(23), 12982. https://doi.org/10.3390/ijms252312982








