Stress Granules in Infectious Disease: Cellular Principles and Dynamic Roles in Immunity and Organelles
Abstract
1. Introduction
2. Principles and Properties of Stress Granules
2.1. Stress Granule Composition and Dynamics
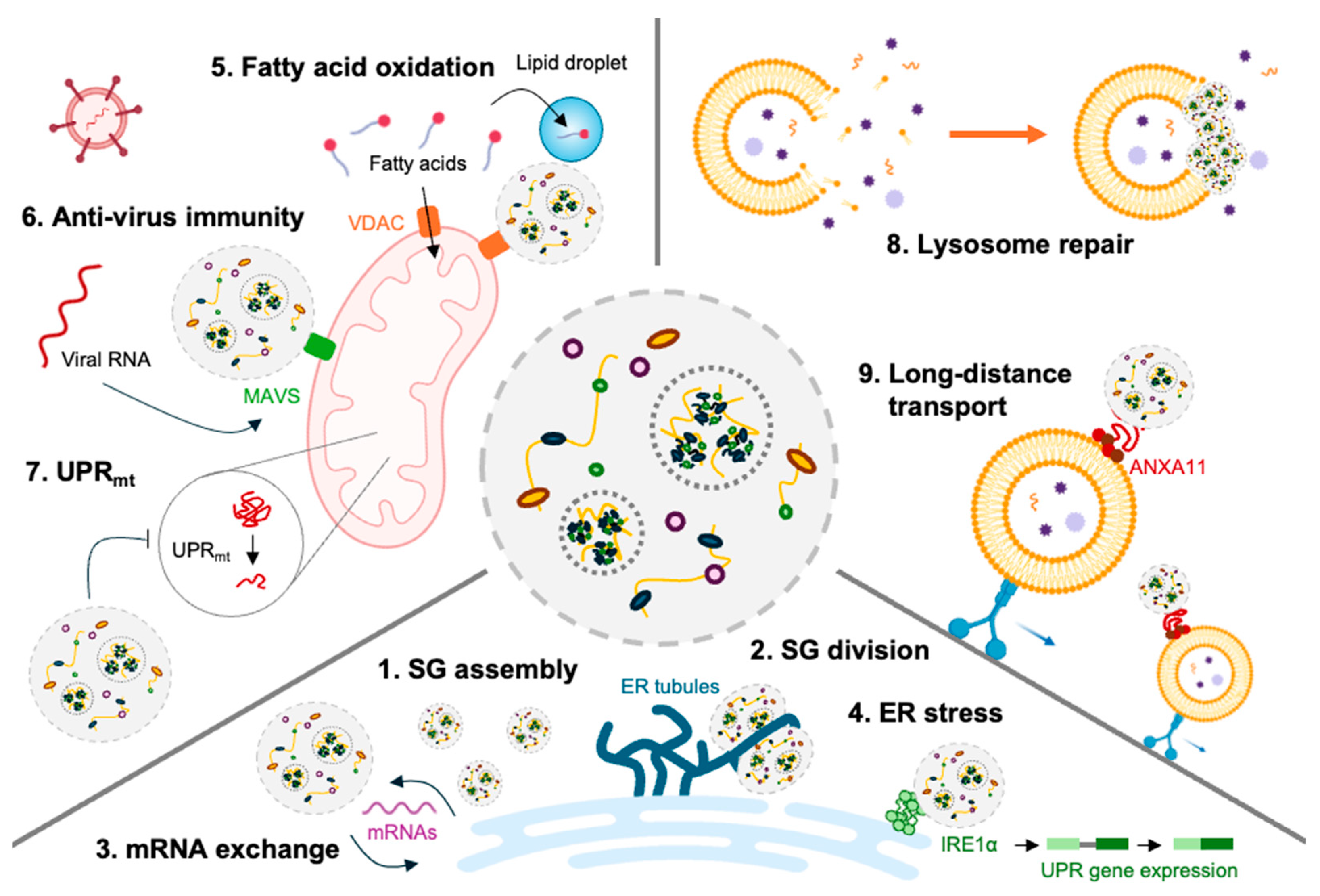
2.2. Stress Granules and Cellular Functions Beyond Translation
2.3. Stress Granules and Immune-Related Proteins
3. Stress Granules and Organelles in Infected Cells
3.1. Stress Granules and Endoplasmic Reticulum
3.2. Stress Granules and Mitochondria
3.3. Stress Granules and Lysosome
4. Stress Granules and Immunity
4.1. Stress Granules and Innate Immunity
4.1.1. Platforms for Immune Signaling Pathways
4.1.2. IFN Response
4.1.3. Dual Role of SGs in Immune Modulation and Pathogen Evasion
4.2. Stress Granules and Adaptive Immunity
4.2.1. Antigen Presenting Cells
4.2.2. T and B Cell Activation
5. Stress Granules and Infectious Diseases
5.1. Viral Infection and Diseases
5.2. Bacterial Infection and Diseases
5.3. Differences in Viral and Bacterial Stress Granules
| Pathogens | Effect on SGs | Key Viral Factors | Target | Mechanism | Outcome | Reference |
|---|---|---|---|---|---|---|
| Influenza A Virus | SG suppression | NS1 protein | PKR, G3BP1, eIF4G | Inhibits SG formation by preventing PKR activation and RNA sequestration | Enhances viral replication, evades immune responses | [122,123] |
| SARS-CoV-2 | SG suppression and remodeling | Nucleocapsid protein | G3BP1, G3BP2 | Remodels SGs, sequesters G3BP proteins | Suppresses innate immunity, enhances viral replication | [91,126,127] |
| Foot-and-Mouth Disease Virus | SG suppression | L and 3C proteases | G3BP1, G3BP2 | Degrades SG scaffolding proteins | Ensures viral mRNA translation, bypasses host defenses | [116] |
| Zika Virus | SG exploitation | Capsid protein | G3BP1, TIA-1 | Uses SG proteins to enhance replication | Facilitates viral replication within host cells | [21,117,118] |
| Ebola Virus | SG suppression | Viral protein (VP35) | SG-associated proteins | Blocks SG formation, sequesters SG proteins | Prevents immune detection, enhances viral replication | [119,120,121] |
| Poliovirus | SG disassembly | 3C protease | Cleaves G3BP1 and eIF4G | Disassembles existing SGs | Enhances viral RNA translation | [146] |
| Herpes Simplex Virus | SG inhibition | Endoribonuclease VHS | eIF2α dephospho-rylation, PKR | Prevents SG formation | Maintains host translation machinery for viral replication | [98,147] |
| Salmonella enterica | SG disassembly | Unknown | G3BP1, PKR | Inhibiting PKR | Evades host immune response, promotes intracellular survival | [87,131,133] |
| Mycobacterium tuberculosis | Persistent SG induction | Unknown | eIF2α phosphory-lation, lysosomal damage | Induces persistent SGs via ISR and endolysosomal damage | Repairs damaged lysosome, helps or suppresses bacterial survival within macrophsges | [32,67] |
| Listeria monocytogenes | SG induction and persistence | Unknown | eIF2α phosphory-lation | Induces SG formation during host invasion | Modulates host stress response to help survival | [136] |
| Shigella flexneri | Limited SG induction | Type III secretion system | eIF2α phosphory-lation | Induces transient SG formation, then inhibits assembly | Manipulates host immune response, facilitates bacterial spread | [22,23] |
| Escherichia coli | SG induction | Shiga toxins | Ribosome-inactivating proteins | Induces SG formation by halting translation | Sequesters host proteins, promotes bacterial survival | [134,135] |
| Helicobacter pylori | Unknown | Peptidoglycan | eIF2α phosphory-lation | Potentially induces SG formation | Unknown | [139,140] |
6. Prospects and Future Directions
6.1. Stress Granules in Host-Directed Therapy
6.2. Temporal Dynamics of Stress Granules During Infection
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef] [PubMed]
- Almanza, A.; Carlesso, A.; Chintha, C.; Creedican, S.; Doultsinos, D.; Leuzzi, B.; Luis, A.; McCarthy, N.; Montibeller, L.; More, S.; et al. Endoplasmic reticulum stress signalling—From basic mechanisms to clinical applications. FEBS J. 2019, 286, 241–278. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Shi, C.R.; He, M.H.; Xiong, S.Q.; Xia, X.B. Endoplasmic reticulum stress: Molecular mechanism and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 352. [Google Scholar] [CrossRef] [PubMed]
- Manoli, I.; Alesci, S.; Blackman, M.R.; Su, Y.A.; Rennert, O.M.; Chrousos, G.P. Mitochondria as key components of the stress response. Trends Endocrinol. Metab. 2007, 18, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Melber, A.; Haynes, C.M. UPR(mt) regulation and output: A stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018, 28, 281–295. [Google Scholar] [CrossRef]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef]
- van Leeuwen, W.; Rabouille, C. Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic 2019, 20, 623–638. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, Z.Y.; Lian, G.W.; Yang, P.G. Biomolecular phase separation in stress granule assembly and virus infection. Acta Biochim. Biophys. Sin. 2023, 55, 1099–1118. [Google Scholar] [CrossRef]
- Youn, J.Y.; Dyakov, B.J.A.; Zhang, J.P.; Knight, J.D.R.; Vernon, R.M.; Forman-Kay, J.D.; Gingras, A.C. Properties of Stress Granule and P-Body Proteomes. Mol. Cell 2019, 76, 286–294. [Google Scholar] [CrossRef]
- Tauber, D.; Tauber, G.; Parker, R. Mechanisms and Regulation of RNA Condensation in RNP Granule Formation. Trends Biochem. Sci. 2020, 45, 764–778. [Google Scholar] [CrossRef]
- Glauninger, H.; Hickernell, C.J.W.; Bard, J.A.M.; Drummond, D.A. Stressful steps: Progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol. Cell 2022, 82, 2544–2556. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Bio 2009, 10, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Horvathova, I.; Voigt, F.; Kotrys, A.V.; Zhan, Y.X.; Artus-Revel, C.G.; Eglinger, J.; Stadler, M.B.; Giorgetti, L.; Chao, J.A. The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol. Cell 2017, 68, 615–625.e9. [Google Scholar] [CrossRef] [PubMed]
- Mateju, D.; Eichenberger, B.; Voigt, F.; Eglinger, J.; Roth, G.; Chao, J.A. Single-Molecule Imaging Reveals Translation of mRNAs Localized to Stress Granules. Cell 2020, 183, 1801–1812.e13. [Google Scholar] [CrossRef]
- Reineke, L.C.; Neilson, J.R. Differences between acute and chronic stress granules, and how these differences may impact function in human disease. Biochem. Pharmacol. 2019, 162, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.Y.; Zhang, Y.F.; Yi, Q.Q.; Yang, C.W.; Liu, Y.F.; Bai, Y. Time-resolved proteomic profiling reveals compositional and functional transitions across the stress granule life cycle. Nat. Commun. 2023, 14, 7782. [Google Scholar] [CrossRef]
- Curdy, N.; Lanvin, O.; Cadot, S.; Laurent, C.; Fournié, J.J.; Franchini, D.M. Stress Granules in the Post-transcriptional Regulation of Immune Cells. Front. Cell Dev. Biol. 2021, 8, 611185. [Google Scholar] [CrossRef]
- Lamichhane, P.P.; Aditi; Xie, X.; Samir, P. Cell-Type-Specific Effect of Innate Immune Signaling on Stress Granules. Stresses 2024, 4, 411–420. [Google Scholar] [CrossRef]
- Nikolic, J.; Civas, A.; Lama, Z.; Lagaudriere-Gesbert, C.; Blondel, D. Rabies Virus Infection Induces the Formation of Stress Granules Closely Connected to the Viral Factories. PLoS Pathog. 2016, 12, e1005942. [Google Scholar] [CrossRef]
- Ruggieri, A.; Dazert, E.; Metz, P.; Hofmann, S.; Bergeest, J.P.; Mazur, J.; Bankhead, P.; Hiet, M.S.; Kallis, S.; Alvisi, G.; et al. Dynamic Oscillation of Translation and Stress Granule Formation Mark the Cellular Response to Virus Infection. Cell Host Microbe 2012, 12, 71–85. [Google Scholar] [CrossRef]
- Hou, S.M.; Kumar, A.; Xu, Z.K.; Airo, A.M.; Stryapunina, I.; Wong, C.P.; Branton, W.; Tchesnokov, E.; Götte, M.; Power, C.; et al. Zika Virus Hijacks Stress Granule Proteins and Modulates the Host Stress Response. J. Virol. 2017, 91, e00474-17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.X.; Xian, W.; Li, Z.L.; Lu, Q.; Chen, X.D.; Ge, J.L.; Tang, Z.H.; Liu, B.H.; Chen, Z.; Gao, X.; et al. Shigella induces stress granule formation by ADP-riboxanation of the eIF3 complex. Cell Rep. 2024, 43, 113789. [Google Scholar] [CrossRef] [PubMed]
- Vonaesch, P.; Campbell-Valois, F.X.; Dufour, A.; Sansonetti, P.J.; Schnupf, P. Shigella flexneri modulates stress granule composition and inhibits stress granule aggregation. Cell Microbiol. 2016, 18, 982–997. [Google Scholar] [CrossRef]
- Malinowska, M.; Niedzwiedzka-Rystwej, P.; Tokarz-Deptula, B.; Deptula, W. Stress granules (SG) and processing bodies (PB) in viral infections. Acta Biochim. Pol. 2016, 63, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, D.; Nakamura, T.; Kubota, Y.; Takekawa, M. Formation of the NLRP3 inflammasome inhibits stress granule assembly by multiple mechanisms. J. Biochem. 2024, 175, 629–641. [Google Scholar] [CrossRef]
- Place, D.E.; Samir, P.; Malireddi, R.K.S.; Kanneganti, T.D. Integrated stress response restricts macrophage necroptosis. Life Sci. Alliance 2022, 5, e202101260. [Google Scholar] [CrossRef]
- Zheng, Z.Q.; Wang, S.Y.; Xu, Z.S.; Fu, Y.Z.; Wang, Y.Y. SARS-CoV-2 nucleocapsid protein impairs stress granule formation to promote viral replication. Cell Discov. 2021, 7, 38. [Google Scholar] [CrossRef]
- Lindquist, M.E.; Lifland, A.W.; Utley, T.J.; Santangelo, P.J.; Crowe, J.E. Respiratory Syncytial Virus Induces Host RNA Stress Granules To Facilitate Viral Replication. J. Virol. 2010, 84, 12274–12284. [Google Scholar] [CrossRef]
- Montero, H.; Trujillo-Alonso, V. Stress Granules in the Viral Replication Cycle. Viruses 2011, 3, 2328. [Google Scholar] [CrossRef]
- Das, S.; Santos, L.; Failla, A.V.; Ignatova, Z. mRNAs sequestered in stress granules recover nearly completely for translation. RNA Biol. 2022, 19, 877–884. [Google Scholar] [CrossRef]
- Zhai, X.; Wu, S.; Lin, L.X.; Wang, T.Y.; Zhong, X.Y.; Chen, Y.; Xu, W.Z.; Tong, L.; Wang, Y.; Zhao, W.R.; et al. Stress Granule Formation is One of the Early Antiviral Mechanisms for Host Cells Against Coxsackievirus B Infection. Virol. Sin. 2018, 33, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.Y.; Wang, F.L.; Bhujabal, Z.; Peters, R.; Mudd, M.; Duque, T.; Allers, L.; Javed, R.; Salemi, M.; Behrends, C.; et al. Stress granules and mTOR are regulated by membrane atg8ylation during lysosomal damage. J. Cell Biol. 2022, 221, e202207091. [Google Scholar] [CrossRef] [PubMed]
- Buchan, J.R.; Parker, R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Anderson, P. Regulation of Translation by Stress Granules and Processing Bodies. Prog. Mol. Biol. Transl. 2009, 90, 155–185. [Google Scholar] [CrossRef]
- Park, C.; Choi, S.; Kim, Y.E.; Lee, S.; Park, S.H.; Adelstein, R.S.; Kawamoto, S.; Kim, K.K. Stress Granules Contain Rbfox2 with Cell Cycle-related mRNAs. Sci. Rep. 2017, 7, 11211. [Google Scholar] [CrossRef]
- Campos-Melo, D.; Hawley, Z.C.E.; Droppelmann, C.A.; Strong, M.J. The Integral Role of RNA in Stress Granule Formation and Function. Front. Cell Dev. Biol. 2021, 9, 621779. [Google Scholar] [CrossRef]
- Hofmann, S.; Kedersha, N.; Anderson, P.; Ivanov, P. Molecular mechanisms of stress granule assembly and disassembly. BBA-Mol. Cell Res. 2021, 1868, 118876. [Google Scholar] [CrossRef]
- He, X.M.; Yuan, J.; Wang, Y.S. G3BP1 binds to guanine quadruplexes in mRNAs to modulate their stabilities. Nucleic Acids Res. 2021, 49, 11323–11336. [Google Scholar] [CrossRef]
- Waris, S.; Wilce, M.C.J.; Wilce, J.A. RNA Recognition and Stress Granule Formation by TIA Proteins. Int. J. Mol. Sci. 2014, 15, 23377–23388. [Google Scholar] [CrossRef]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.H.; Fang, M.Y.; Luo, E.C.; Krach, F.; Yang, D.J.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604.e13. [Google Scholar] [CrossRef]
- Youn, J.Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532.e11. [Google Scholar] [CrossRef]
- Yang, P.G.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.P.; Messing, J.; Yurtsever, U.; Yang, Z.M.; Wu, J.J.; Li, Y.X.; Pan, Q.F.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e28. [Google Scholar] [CrossRef]
- Gwon, Y.; Maxwell, B.A.; Kolaitis, R.M.; Zhang, P.P.; Kim, H.J.; Taylor, J.P. Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science 2021, 372, eabf6548. [Google Scholar] [CrossRef]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef]
- Kershaw, C.J.; Nelson, M.G.; Lui, J.N.; Bates, C.P.; Jennings, M.D.; Hubbard, S.J.; Ashe, M.P.; Grant, C.M. Integrated multi-omics reveals common properties underlying stress granule and P-body formation. RNA Biol. 2021, 18, 655–673. [Google Scholar] [CrossRef]
- Alluri, R.K.; Li, Z.W.; McCrae, K.R. Stress Granule-Mediated Oxidized RNA Decay in P-Body: Hypothetical Role of ADAR1, Tudor-SN, and STAU1. Front. Mol. Biosci. 2021, 8, 672988. [Google Scholar] [CrossRef]
- Sun, C.L.; Van Gilst, M.; Crowder, C.M. Hypoxia-induced mitochondrial stress granules. Cell Death Dis. 2023, 14, 448. [Google Scholar] [CrossRef]
- Amen, T.; Kaganovich, D. Stress granules inhibit fatty acid oxidation by modulating mitochondrial permeability. Cell Rep. 2021, 35, 109237. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Sun, L.K.; Chai, J.N.; Qi, H.; Zhao, Y.X.; Ma, J.Y.; Xia, M.H.; Hu, X.Q. Stress granules affect the dual PI3K/mTOR inhibitor response by regulating the mitochondrial unfolded protein response. Cancer Cell Int. 2024, 24, 38. [Google Scholar] [CrossRef]
- Arimoto, K.; Fukuda, H.; Imajoh-Ohmi, S.; Saito, H.; Takekawa, M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 2008, 10, 1324–1332. [Google Scholar] [CrossRef]
- Park, Y.J.; Choi, D.W.; Cho, S.W.; Han, J.; Yang, S.; Choi, C.Y. Stress Granule Formation Attenuates RACK1-Mediated Apoptotic Cell Death Induced by Morusin. Int. J. Mol. Sci. 2020, 21, 5360. [Google Scholar] [CrossRef]
- Tsai, N.P.; Ho, P.C.; Wei, L.N. Regulation of stress granule dynamics by Grb7 and FAK signalling pathway. Faseb J. 2009, 23, 500.2. [Google Scholar] [CrossRef]
- Yang, S.; Aulas, A.; Anderson, P.J.; Ivanov, P. Stress granule formation enables anchorage-independence survival in cancer cells. bioRxiv 2024. [Google Scholar] [CrossRef]
- Decker, C.J.; Parker, R. P-Bodies and Stress Granules: Possible Roles in the Control of Translation and mRNA Degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef]
- Fujikawa, D.; Nakamura, T.; Yoshioka, D.; Li, Z.Z.; Moriizumi, H.; Taguchi, M.; Tokai-Nishizumi, N.; Kozuka-Hata, H.; Oyama, M.; Takekawa, M. Stress granule formation inhibits stress-induced apoptosis by selectively sequestering executioner caspases. Curr. Biol. 2023, 33, 1967–1981.e8. [Google Scholar] [CrossRef]
- Zhang, Q.; Sharma, N.R.; Zheng, Z.M.; Chen, M.Z. Viral Regulation of RNA Granules in Infected Cells. Virol. Sin. 2019, 34, 175–191. [Google Scholar] [CrossRef]
- White, J.P.; Lloyd, R.E. Regulation of stress granules in virus systems. Trends Microbiol. 2012, 20, 175–183. [Google Scholar] [CrossRef]
- Cheng, J.Y.; Gao, S.; Zhu, C.; Liu, S.H.; Li, J.Y.; Kang, J.; Wang, Z.Y.; Wang, T. Typical Stress Granule Proteins Interact with the 3′ Untranslated Region of Enterovirus D68 To Inhibit Viral Replication. J. Virol. 2020, 94, 10-1128. [Google Scholar] [CrossRef]
- Albornoz, A.; Carletti, T.; Corazza, G.; Marcello, A. The Stress Granule Component TIA-1 Binds Tick-Borne Encephalitis Virus RNA and Is Recruited to Perinuclear Sites of Viral Replication To Inhibit Viral Translation. J. Virol. 2014, 88, 6611–6622. [Google Scholar] [CrossRef]
- Yoneyama, M.; Jogi, M.; Onomoto, K. Regulation of antiviral innate immune signaling by stress-induced RNA granules. J. Biochem. 2016, 159, 279–286. [Google Scholar] [CrossRef]
- Kim, S.S.; Sze, L.; Liu, C.; Lam, K.P. The stress granule protein G3BP1 binds viral dsRNA and RIG-I to enhance interferon-beta response. J. Biol. Chem. 2019, 294, 6430–6438. [Google Scholar] [CrossRef] [PubMed]
- Reineke, L.C.; Lloyd, R.E. The Stress Granule Protein G3BP1 Recruits Protein Kinase R To Promote Multiple Innate Immune Antiviral Responses. J. Virol. 2015, 89, 2575–2589. [Google Scholar] [CrossRef]
- Paget, M.; Cadena, C.; Ahmad, S.; Wang, H.T.; Jordan, T.X.; Kim, E.; Koo, B.; Lyons, S.M.; Ivanov, P.; tenOever, B.; et al. Stress granules are shock absorbers that prevent excessive innate immune responses to dsRNA. Mol. Cell 2023, 83, 1180–1196.e8. [Google Scholar] [CrossRef]
- Pincus, D.; Oakes, S.A. Unfolding emergency calls stress granules to the ER. Nat. Cell Biol. 2024, 26, 845–846. [Google Scholar] [CrossRef]
- Lee, J.E.; Cathey, P.I.; Wu, H.X.; Parker, R.; Voeltz, G.K. Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science 2020, 367, eaay7108. [Google Scholar] [CrossRef]
- Lopez-Nieto, M.; Sun, Z.; Relton, E.; Safakli, R.; Freibaum, B.D.; Taylor, J.P.; Ruggieri, A.; Smyrnias, I.; Locker, N. Activation of the mitochondrial unfolded protein response regulates the dynamic formation of stress granules. J. Cell Sci. 2024. [Google Scholar] [CrossRef]
- Bussi, C.; Mangiarotti, A.; Vanhille-Campos, C.; Aylan, B.; Pellegrino, E.; Athanasiadi, N.; Fearns, A.; Rodgers, A.; Franzmann, T.M.; Saric, A.; et al. Stress granules plug and stabilize damaged endolysosomal membranes. Nature 2023, 624, E3. [Google Scholar] [CrossRef]
- Liao, Y.; Fernandopulle, M.; Wang, G.; Choi, H.; Hao, L.; Drerup, C.; Patel, R.; Qamar, S.; Nixon-Abell, J.; Shen, Y.; et al. RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Mol. Biol. Cell 2023, 34, 106. [Google Scholar]
- Tiku, V.; Tan, M.W.; Dikic, I. Mitochondrial Functions in Infection and Immunity. Trends Cell Biol. 2020, 30, 263–275. [Google Scholar] [CrossRef]
- Lee, Y.T.; Senturk, M.; Guan, Y.C.; Wang, M.C. Bacteria-organelle communication in physiology and disease. J. Cell Biol. 2024, 223, e202310134. [Google Scholar] [CrossRef]
- Child, J.R.; Chen, Q.; Reid, D.W.; Jagannathan, S.; Nicchitta, C. Recruitment of endoplasmic reticulum-targeted and cytosolic mRNAs into membrane-associated stress granules. Rna 2021, 27, 1241–1256. [Google Scholar] [CrossRef]
- Nicchitta, C.V. An emerging role for the endoplasmic reticulum in stress granule biogenesis. Semin. Cell Dev. Biol. 2024, 156, 160–166. [Google Scholar] [CrossRef]
- Liu, S.Z.; Zhang, X.G.; Yao, X.; Wang, G.; Huang, S.J.; Chen, P.; Tang, M.L.; Cai, J.; Wu, Z.Y.; Zhang, Y.L.; et al. Mammalian IRE1α dynamically and functionally coalesces with stress granules. Nat. Cell Biol. 2024, 26, 917–931. [Google Scholar] [CrossRef]
- Macauslane, K.L.; Pegg, C.L.; Short, K.R.; Schulz, B.L. Modulation of endoplasmic reticulum stress response pathways by respiratory viruses. Crit. Rev. Microbiol. 2024, 50, 750–768. [Google Scholar] [CrossRef]
- Pillich, H.; Loose, M.; Zimmer, K.P.; Chakraborty, T. Diverse roles of endoplasmic reticulum stress sensors in bacterial infection. Mol. Cell Pediatr. 2016, 3, 9. [Google Scholar] [CrossRef]
- Choi, J.A.; Song, C.H. Insights Into the Role of Endoplasmic Reticulum Stress in Infectious Diseases. Front. Immunol. 2020, 10, 3147. [Google Scholar] [CrossRef]
- Laudenbach, B.T.; Krey, K.; Emslander, Q.; Andersen, L.L.; Reim, A.; Scaturro, P.; Mundigl, S.; Dächert, C.; Manske, K.; Moser, M.; et al. NUDT2 initiates viral RNA degradation by removal of 5′-phosphates. Nat. Commun. 2021, 12, 6918. [Google Scholar] [CrossRef]
- Rowell, C.E.R.; Dobrovolny, H.M. Energy Requirements for Loss of Viral Infectivity. Food Environ. Virol. 2020, 12, 281–294. [Google Scholar] [CrossRef]
- Goyal, P.; Rajala, M.S. Reprogramming of glucose metabolism in virus infected cells. Mol. Cell Biochem. 2023, 478, 2409–2418. [Google Scholar] [CrossRef]
- Park, D.W.; Zmijewski, J.W. Mitochondrial Dysfunction and Immune Cell Metabolism in Sepsis. Infect. Chemother. 2017, 49, 10–21. [Google Scholar] [CrossRef]
- Marques, E.; Kramer, R.; Ryan, D.G. Multifaceted mitochondria in innate immunity. NPJ Metab. Health Dis. 2024, 2, 6. [Google Scholar] [CrossRef]
- Andrieux, P.; Chevillard, C.; Cunha-Neto, E.; Nunes, J.P.S. Mitochondria as a Cellular Hub in Infection and Inflammation. Int. J. Mol. Sci. 2021, 22, 11338. [Google Scholar] [CrossRef]
- Marchi, S.; Morroni, G.; Pinton, P.; Galluzzi, L. Control of host mitochondria by bacterial pathogens. Trends Microbiol. 2022, 30, 452–465. [Google Scholar] [CrossRef]
- Maurice, N.M.; Sadikot, R.T. Mitochondrial Dysfunction in Bacterial Infections. Pathogens 2023, 12, 1005. [Google Scholar] [CrossRef]
- Pagán, A.J.; Lee, L.J.; Edwards-Hicks, J.; Moens, C.B.; Tobin, D.M.; Busch-Nentwich, E.M.; Pearce, E.L.; Ramakrishnan, L. mTOR-regulated mitochondrial metabolism limits mycobacterium-induced cytotoxicity. Cell 2022, 185, 3720–3738.e13. [Google Scholar] [CrossRef]
- Thedieck, K.; Holzwarth, B.; Prentzell, M.T.; Boehlke, C.; Kläsener, K.; Ruf, S.; Sonntag, A.G.; Maerz, L.; Grellscheid, S.N.; Kremmer, E.; et al. Inhibition of mTORC1 by Astrin and Stress Granules Prevents Apoptosis in Cancer Cells (vol 154, pg 859, 2013). Cell 2013, 155, 964–966. [Google Scholar] [CrossRef]
- Sachdeva, K.; Sundaramurthy, V. The Interplay of Host Lysosomes and Intracellular Pathogens. Front. Cell Infect. Mi 2020, 10, 595502. [Google Scholar] [CrossRef]
- Bussi, C.; Heunis, T.; Pellegrino, E.; Bernard, E.M.; Bah, N.; Dos Santos, M.S.; Santucci, P.; Aylan, B.; Rodgers, A.; Fearns, A.; et al. Lysosomal damage drives mitochondrial proteome remodelling and reprograms macrophage immunometabolism. Nat. Commun. 2022, 13, 7338. [Google Scholar] [CrossRef]
- Seguin, S.J.; Morelli, F.F.; Vinet, J.; Amore, D.; De Biasi, S.; Poletti, A.; Rubinsztein, D.C.; Carra, S. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ. 2014, 21, 1838–1851. [Google Scholar] [CrossRef] [PubMed]
- Duran, J.; Poolsup, S.; Allers, L.; Lemus, M.R.; Cheng, Q.; Pu, J.; Salemi, M.; Phinney, B.; Jia, J. A mechanism that transduces lysosomal damage signals to stress granule formation for cell survival. bioRxiv 2024. [Google Scholar] [CrossRef]
- Sampaio, N.G.; Chauveau, L.; Hertzog, J.; Bridgeman, A.; Fowler, G.; Moonen, J.P.; Dupont, M.; Russell, R.A.; Noerenberg, M.; Rehwinkel, J. The RNA sensor MDA5 detects SARS-CoV-2 infection. Sci. Rep. 2021, 11, 13638. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Aparicio, M.T.; Ayllón, J.; Leo-Macias, A.; Wolff, T.; García-Sastre, A. Subcellular Localizations of RIG-I, TRIM25, and MAVS Complexes. J. Virol. 2017, 91, e01155. [Google Scholar] [CrossRef] [PubMed]
- Onomoto, K.; Onoguchi, K.; Yoneyama, M. Regulation of RIG-I-like receptor-mediated signaling: Interaction between host and viral factors. Cell Mol. Immunol. 2021, 18, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Toth, A.M.; Li, Z.Q.; Cattaneo, R.; Samuel, C.E. RNA-specific Adenosine Deaminase ADAR1 Suppresses Measles Virus-induced Apoptosis and Activation of Protein Kinase PKR. J. Biol. Chem. 2009, 284, 29350–29356. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.V.; George, C.X.; Welch, M.J.; Liou, L.Y.; Hahm, B.; Lewicki, H.; de la Torre, J.C.; Samuel, C.E.; Oldstone, M.B. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 331–336. [Google Scholar] [CrossRef]
- John, L.; Samuel, C.E. Induction of stress granules by interferon and down-regulation by the cellular RNA adenosine deaminase ADAR1. Virology 2014, 454, 299–310. [Google Scholar] [CrossRef]
- Chathuranga, W.A.G.; Nikapitiya, C.; Kim, J.H.; Chathuranga, K.; Weerawardhana, A.; Dodantenna, N.; Kim, D.J.; Poo, H.; Jung, J.U.; Lee, C.H.; et al. Gadd45 is critical for regulation of type I interferon signaling by facilitating G3BP-mediated stress granule formation. Cell Rep. 2023, 42, 113358. [Google Scholar] [CrossRef]
- Dauber, B.; Poon, D.; dos Santos, T.; Duguay, B.A.; Mehta, N.; Saffran, H.A.; Smiley, J.R. The Herpes Simplex Virus Virion Host Shutoff Protein Enhances Translation of Viral True Late mRNAs Independently of Suppressing Protein Kinase R and Stress Granule Formation. J. Virol. 2016, 90, 6049–6057. [Google Scholar] [CrossRef]
- Reineke, L.C.; Lloyd, R.E. Diversion of stress granules and P-bodies during viral infection. Virology 2013, 436, 255–267. [Google Scholar] [CrossRef]
- Ariumi, Y.; Kuroki, M.; Kushima, Y.; Osugi, K.; Hijikata, M.; Maki, M.; Ikeda, M.; Kato, N. Hepatitis C Virus Hijacks P-Body and Stress Granule Components around Lipid Droplets. J. Virol. 2011, 85, 6882–6892. [Google Scholar] [CrossRef]
- Hargrave, K.E.; MacLeod, M.K.L.; Worrell, J.C. Antigen presenting cells: Professionals, amateurs, and spectators in the long game of lung immunity. Int. J. Biochem. Cell B 2022, 153, 106331. [Google Scholar] [CrossRef] [PubMed]
- Kothandan, V.K.; Kothandan, S.; Kim, D.H.; Byun, Y.; Lee, Y.K.; Park, I.K.; Hwang, S.R. Crosstalk between Stress Granules, Exosomes, Tumour Antigens, and Immune Cells: Significance for Cancer Immunity. Vaccines 2020, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- McCormick, C.; Khaperskyy, D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017, 17, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Curdy, N.; Lanvin, O.; Cerapio, J.P.; Pont, F.; Tosolini, M.; Sarot, E.; Valle, C.; Saint-Laurent, N.; Lhuillier, E.; Laurent, C.; et al. The proteome and transcriptome of stress granules and P bodies during human T lymphocyte activation. Cell Rep. 2023, 42, 112211. [Google Scholar] [CrossRef] [PubMed]
- Yue, T.; Zhan, X.M.; Zhang, D.W.; Jain, R.C.; Wang, K.W.; Choi, J.H.; Misawa, T.; Su, L.J.; Quan, J.X.; Hildebrand, S.; et al. SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity. Science 2021, 372, eaba4220. [Google Scholar] [CrossRef] [PubMed]
- Khedri, M.; Samei, A.; Fasihi-Ramandi, M.; Taheri, R.A. The immunopathobiology of T cells in stress condition: A review. Cell Stress Chaperones 2020, 25, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Voyer, T.L.; Neehus, A.L.; Yang, R.; Ogishi, M.; Rosain, J.; Alroqi, F.; Alshalan, M.; Blumental, S.; Al Ali, F.; Khan, T.; et al. Inherited deficiency of stress granule ZNFX1 in patients with monocytosis and mycobacterial disease. Proc. Natl. Acad. Sci. USA 2021, 118, e2102804118. [Google Scholar] [CrossRef]
- den Boon, J.A.; Diaz, A.; Ahlquist, P. Cytoplasmic Viral Replication Complexes. Cell Host Microbe 2010, 8, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.; Thomsen, A.R. Sensing of RNA Viruses: A Review of Innate Immune Receptors Involved in Recognizing RNA Virus Invasion. J. Virol. 2012, 86, 2900–2910. [Google Scholar] [CrossRef]
- Chau, B.A.; Chen, V.E.S.; Cochrane, A.W.; Parent, L.J.; Mouland, A.J. Liquid-liquid phase separation of nucleocapsid proteins during SARS-CoV-2 and HIV-1 replication. Cell Rep. 2023, 42, 111968. [Google Scholar] [CrossRef]
- Dauber, B.; Wolff, T. Activation of the Antiviral Kinase PKR and Viral Countermeasures. Viruses 2009, 1, 523–544. [Google Scholar] [CrossRef]
- Miller, C.L. Stress granules and virus replication. Future Virol. 2011, 6, 1329–1338. [Google Scholar] [CrossRef]
- Nakagawa, K.; Narayanan, K.; Wada, M.; Makino, S. Inhibition of Stress Granule Formation by Middle East Respiratory Syndrome Coronavirus 4a Accessory Protein Facilitates Viral Translation, Leading to Efficient Virus Replication. J. Virol. 2018, 92, e00902-18. [Google Scholar] [CrossRef]
- Emara, M.M.; Brinton, M.A. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. USA 2007, 104, 9041–9046. [Google Scholar] [CrossRef]
- Ye, X.; Pan, T.; Wang, D.; Fang, L.R.; Ma, J.; Zhu, X.Y.; Shi, Y.L.; Zhang, K.S.; Zheng, H.X.; Chen, H.C.; et al. Foot-and-Mouth Disease Virus Counteracts on Internal Ribosome Entry Site Suppression by G3BP1 and Inhibits G3BP1-Mediated Stress Granule Assembly via Post-Translational Mechanisms. Front. Immunol. 2021, 12, 702530. [Google Scholar] [CrossRef] [PubMed]
- Visser, L.J.; Medina, G.N.; Rabouw, H.H.; de Groot, R.J.; Langereis, M.A.; de los Santos, T.; van Kuppeveld, F.J.M. Foot-and-Mouth Disease Virus Leader Protease Cleaves G3BP1 and G3BP2 and Inhibits Stress Granule Formation. J. Virol. 2019, 93, e00922-18. [Google Scholar] [CrossRef]
- Amorim, R.; Temzi, A.; Griffin, B.D.; Mouland, A.J. Zika virus inhibits eIF2α-dependent stress granule assembly. PLoS Neglected Trop. Dis. 2017, 11, e0005775. [Google Scholar] [CrossRef]
- Bonenfant, G.; Williams, N.; Netzband, R.; Schwarz, M.C.; Evans, M.J.; Pager, C.T. Zika Virus Subverts Stress Granules To Promote and Restrict Viral Gene Expression. J. Virol. 2019, 93, e00520-19. [Google Scholar] [CrossRef]
- Le Sage, V.; Cinti, A.; McCarthy, S.; Amorim, R.; Rao, S.; Daino, G.L.; Tramontano, E.; Branch, D.R.; Mouland, A.J. Ebola virus VP35 blocks stress granule assembly. Virology 2017, 502, 73–83. [Google Scholar] [CrossRef]
- Forrester, J.V. Ebola virus and persistent chronic infection: When does replication cease? Ann. Transl. Med. 2018, 6 (Suppl. 1), S39. [Google Scholar] [CrossRef]
- Nelson, E.V.; Schmidt, K.M.; Deflubé, L.R.; Doğanay, S.; Banadyga, L.; Olejnik, J.; Hume, A.J.; Ryabchikova, E.; Ebihara, H.; Kedersha, N.; et al. Ebola Virus Does Not Induce Stress Granule Formation during Infection and Sequesters Stress Granule Proteins within Viral Inclusions. J. Virol. 2016, 90, 7268–7284. [Google Scholar] [CrossRef]
- Raman, S.N.T.; Liu, G.Q.; Pyo, H.M.; Cui, Y.C.; Xu, F.; Ayalew, L.E.; Tikoo, S.K.; Zhou, Y. DDX3 Interacts with Influenza A Virus NS1 and NP Proteins and Exerts Antiviral Function through Regulation of Stress Granule Formation. J. Virol. 2016, 90, 3661–3675. [Google Scholar] [CrossRef] [PubMed]
- Khaperskyy, D.A.; Hatchette, T.F.; McCormick, C. Influenza A virus inhibits cytoplasmic stress granule formation. Faseb J. 2012, 26, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Khaperskyy, D.A.; Emara, M.M.; Johnston, B.P.; Anderson, P.; Hatchette, T.F.; McCormick, C. Influenza A Virus Host Shutoff Disables Antiviral Stress-Induced Translation Arrest. PLoS Pathog. 2014, 10, e1004217. [Google Scholar] [CrossRef] [PubMed]
- Jayabalan, A.K.; Griffin, D.E.; Leung, A.K.L. Pro-Viral and Anti-Viral Roles of the RNA-Binding Protein G3BP1. Viruses 2023, 15, 449. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Gou, H.W.; Zhou, Y.L.; Wu, C.X.; Ren, X.X.; Wu, X.J.P.; Guan, G.W.; Jin, B.X.; Huang, J.H.; Jin, Z.G.; et al. The SARS-CoV-2 nucleocapsid protein suppresses innate immunity by remodeling stress granules to atypical foci. Faseb J. 2023, 37, e23269. [Google Scholar] [CrossRef]
- Nabeel-Shah, S.; Lee, H.; Ahmed, N.; Burke, G.L.; Farhangmehr, S.; Ashraf, K.; Pu, S.Y.; Braunschweig, U.; Zhong, G.Q.; Wei, H.; et al. SARS-CoV-2 nucleocapsid protein binds host mRNAs and attenuates stress granules to impair host stress response. iScience 2022, 25, 103562. [Google Scholar] [CrossRef]
- Tweedie, A.; Nissan, T. Hiding in Plain Sight: Formation and Function of Stress Granules During Microbial Infection of Mammalian Cells. Front. Mol. Biosci. 2021, 8, 647884. [Google Scholar] [CrossRef]
- Rodrigues, L.O.C.P.; Graça, R.S.F.; Carneiro, L.A.M. Integrated Stress Responses to Bacterial Pathogenesis Patterns. Front. Immunol. 2018, 9, 1306. [Google Scholar] [CrossRef]
- Li, W.Y.; Wang, Y. Stress granules: Potential therapeutic targets for infectious and inflammatory diseases. Front. Immunol. 2023, 14, 1145346. [Google Scholar] [CrossRef]
- Tattoli, I.; Sorbara, M.T.; Vuckovic, D.; Ling, A.; Soares, F.; Carneiro, L.A.M.; Yang, C.; Emili, A.; Philpott, D.J.; Girardin, S.E. Amino Acid Starvation Induced by Invasive Bacterial Pathogens Triggers an Innate Host Defense Program. Cell Host Microbe 2012, 11, 563–575. [Google Scholar] [CrossRef] [PubMed]
- López-Montero, N.; Ramos-Marquès, E.; Risco, C.; García-del Portillo, F. Intracellular Salmonella induces aggrephagy of host endomembranes in persistent infections. Autophagy 2016, 12, 1886–1901. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Fröhlich, K.S.; Mano, M.; Giacca, M.; Vogel, J. A Candidate Approach Implicates the Secreted Salmonella Effector Protein SpvB in P-Body Disassembly. PLoS ONE 2011, 6, e17296. [Google Scholar] [CrossRef] [PubMed]
- Tsutsuki, H.; Yahiro, K.; Ogura, K.; Ichimura, K.; Iyoda, S.; Ohnishi, M.; Nagasawa, S.; Seto, K.; Moss, J.; Noda, M. Subtilase cytotoxin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli induces stress granule formation. Cell Microbiol. 2016, 18, 1024–1040. [Google Scholar] [CrossRef]
- Velásquez, F.; Marín-Rojas, J.; Soto-Rifo, R.; Torres, A.; Del Canto, F.; Valiente-Echeverría, F. Escherichia coli HS and Enterotoxigenic Escherichia coli Hinder Stress Granule Assembly. Microorganisms 2021, 9, 17. [Google Scholar] [CrossRef]
- Abdel-Nour, M.; Carneiro, L.A.M.; Downey, J.; Tsalikis, J.; Outlioua, A.; Prescott, D.; Da Costa, L.S.; Hovingh, E.S.; Farahvash, A.; Gaudet, R.G.; et al. The heme-regulated inhibitor is a cytosolic sensor of protein misfolding that controls innate immune signaling. Science 2019, 365, eaaw4144. [Google Scholar] [CrossRef]
- Nambi, S.; Long, J.E.; Mishra, B.B.; Baker, R.; Murphy, K.C.; Olive, A.J.; Nguyen, H.P.; Shaffer, S.A.; Sassetti, C.M. The oxidative stress network of Mycobacterium tuberculosis reveals coordination between radical detoxification systems. Cell Host Microbe 2015, 17, 829–837. [Google Scholar] [CrossRef]
- Lim, Y.J.; Yi, M.H.; Choi, J.A.; Lee, J.; Han, J.Y.; Jo, S.H.; Oh, S.M.; Cho, H.J.; Kim, D.W.; Kang, M.W.; et al. Roles of endoplasmic reticulum stress-mediated apoptosis in M1-polarized macrophages during mycobacterial infections. Sci. Rep. 2016, 6, 37211. [Google Scholar] [CrossRef]
- Ding, S.Z.; Minohara, Y.; Fan, X.J.; Wang, J.; Reyes, V.E.; Patel, J.; Dirden-Kramer, B.; Boldogh, I.; Ernst, P.B.; Crowe, S.E. Helicobacter pylori Infection Induces Oxidative Stress and Programmed Cell Death in Human Gastric Epithelial Cells. Infect. Immun. 2007, 75, 4030–4039. [Google Scholar] [CrossRef]
- Wang, G.; Olczak, A.; Forsberg, L.S.; Maier, R.J. Oxidative Stress-induced Peptidoglycan Deacetylase in Helicobacter pylori. J. Biol. Chem. 2009, 284, 6790–6800. [Google Scholar] [CrossRef]
- Yong, X.; Tang, B.; Li, B.S.; Xie, R.; Hu, C.J.; Luo, G.; Qin, Y.; Dong, H.; Yang, S.M. Helicobacter pylori virulence factor CagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun. Signal 2015, 13, 30. [Google Scholar] [CrossRef]
- Onomoto, K.; Yoneyama, M.; Fung, G.; Kato, H.; Fujita, T. Antiviral innate immunity and stress granule responses. Trends Immunol. 2014, 35, 420–428. [Google Scholar] [CrossRef]
- Soni, J.; Sinha, S.; Pandey, R. Understanding bacterial pathogenicity: A closer look at the journey of harmful microbes. Front. Microbiol. 2024, 15, 1370818. [Google Scholar] [CrossRef] [PubMed]
- Sjöström, A.E.; Sandblad, L.; Uhlin, B.E.; Wai, S.N. Membrane vesicle-mediated release of bacterial RNA. Sci. Rep. 2015, 5, 15329. [Google Scholar] [CrossRef] [PubMed]
- Simonov, D.; Swift, S.; Blenkiron, C.; Phillips, A.R. Bacterial RNA as a signal to eukaryotic cells as part of the infection process. Discoveries 2016, 4, e70. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, J.D.; Tsai, W.C.; Lloyd, R.E. Multiple Poliovirus Proteins Repress Cytoplasmic RNA Granules. Viruses 2015, 7, 6127–6140. [Google Scholar] [CrossRef]
- Burgess, H.M.; Mohr, I. Defining the Role of Stress Granules in Innate Immune Suppression by the Herpes Simplex Virus 1 Endoribonuclease VHS. J. Virol. 2018, 92, e00829-18. [Google Scholar] [CrossRef]
- Kaufmann, S.H.E.; Dorhoi, A.; Hotchkiss, R.S.; Bartenschlager, R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov. 2018, 17, 35–56. [Google Scholar] [CrossRef]
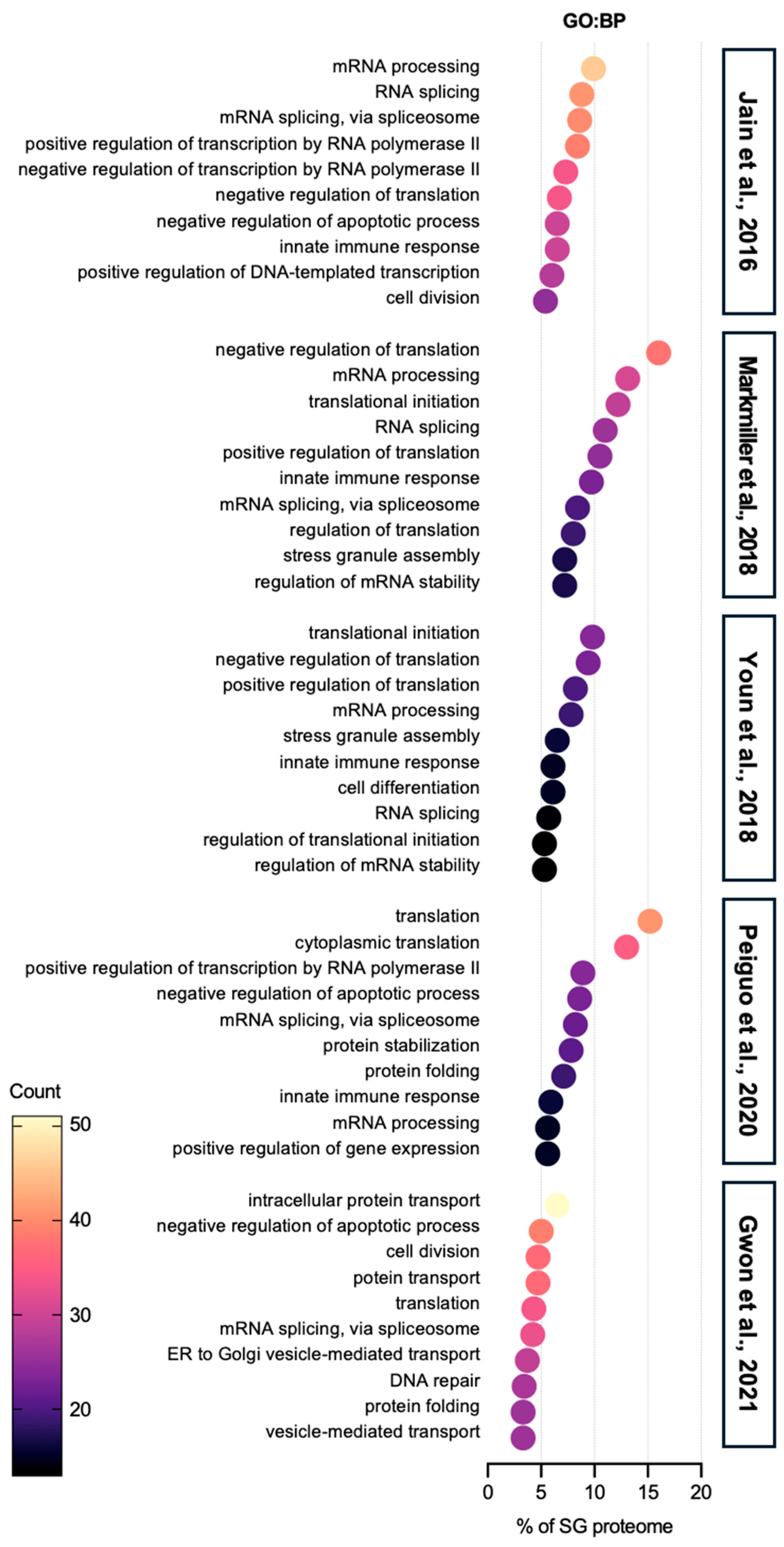

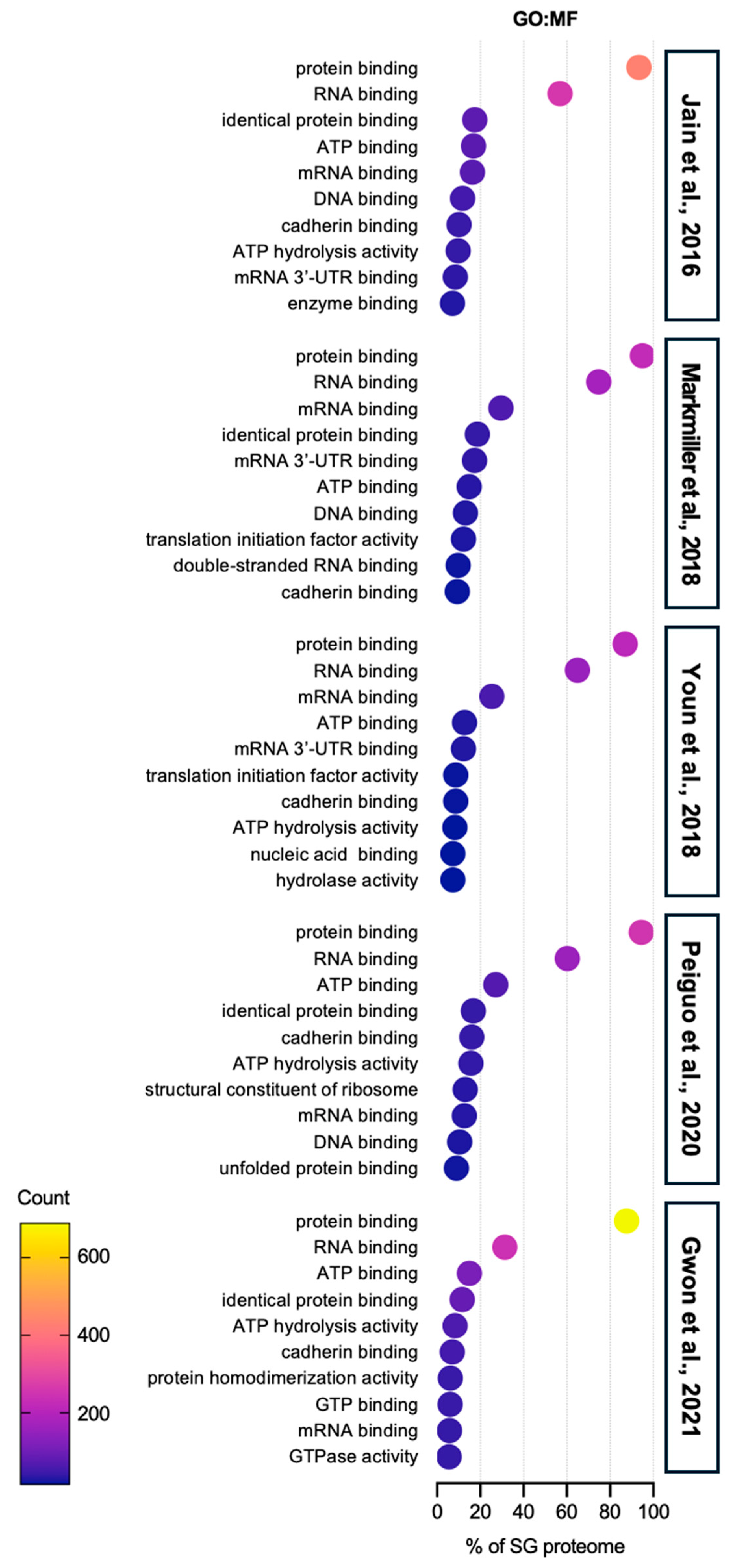
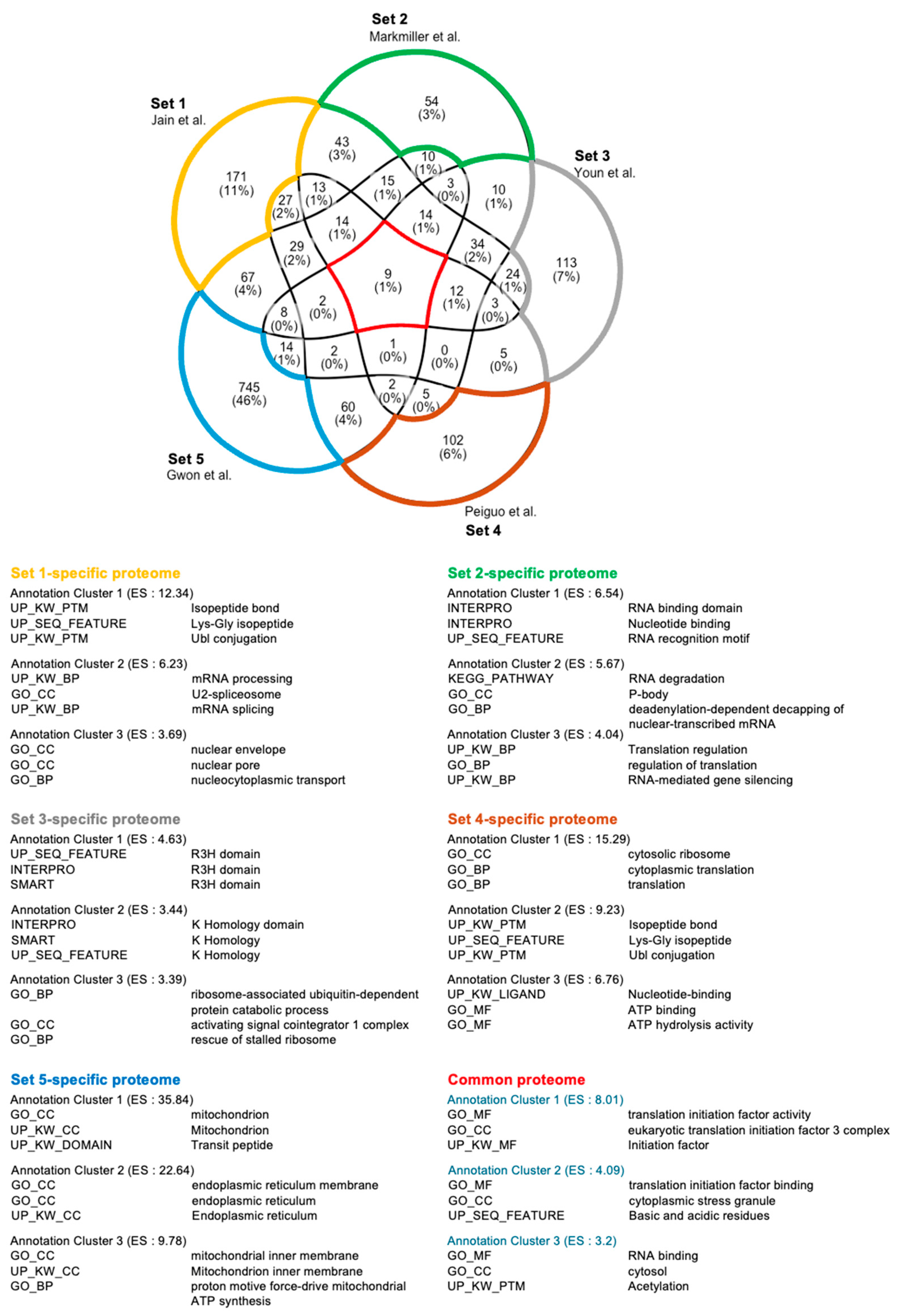
| Enrichment Score (ES) | Term | Counts | Reference SG Proteome | |
|---|---|---|---|---|
| Annotation Cluster 21 (ES = 3.57) | GO:BP | Innate immune response | 30 | [44] |
| GO:BP | Defense response to virus | 18 | ||
| KW:BP | Innate immunity | 28 | ||
| KW:BP | Antiviral defense | 14 | ||
| KW:BP | Immunity | 30 | ||
| Annotation Cluster 12 (ES = 5.59) | GO:BP | Defense response to virus | 17 | [40] |
| KW:BP | Antiviral defense | 14 | ||
| GO:BP | Innate immune response | 23 | ||
| KW:BP | Innate immunity | 23 | ||
| KW:BP | immunity | 24 | ||
| Annotation Cluster 28 (ES = 2.00) | GO:BP | Defense response to virus | 10 | [41] |
| KW:BP | Antiviral defense | 8 | ||
| GO:BP | Innate immune response | 15 | ||
| KW:BP | Innate immunity | 14 | ||
| KW:BP | Immunity | 15 | ||
| Annotation Cluster 22 (ES = 3.08) | GO:BP | DNA duplex unwinding | 9 | [42] |
| KW:BP | Innate immunity | 18 | ||
| GO:BP | Innate immune response | 16 | ||
| KW:BP | Immunity | 19 | ||
| Annotation Cluster 133 (ES = 0.13) | KW:BP | Innate immunity | 20 | [43] |
| GO:BP | Innate immune response | 20 | ||
| KW:BP | Immunity | 21 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Song, C.-H. Stress Granules in Infectious Disease: Cellular Principles and Dynamic Roles in Immunity and Organelles. Int. J. Mol. Sci. 2024, 25, 12950. https://doi.org/10.3390/ijms252312950
Kim J, Song C-H. Stress Granules in Infectious Disease: Cellular Principles and Dynamic Roles in Immunity and Organelles. International Journal of Molecular Sciences. 2024; 25(23):12950. https://doi.org/10.3390/ijms252312950
Chicago/Turabian StyleKim, Jaewhan, and Chang-Hwa Song. 2024. "Stress Granules in Infectious Disease: Cellular Principles and Dynamic Roles in Immunity and Organelles" International Journal of Molecular Sciences 25, no. 23: 12950. https://doi.org/10.3390/ijms252312950
APA StyleKim, J., & Song, C.-H. (2024). Stress Granules in Infectious Disease: Cellular Principles and Dynamic Roles in Immunity and Organelles. International Journal of Molecular Sciences, 25(23), 12950. https://doi.org/10.3390/ijms252312950







