Genome-Wide Identification of the GbUBC Gene Family in Sea-Island Cotton (Gossypium barbadense) and the Active Regulation of Drought Resistance in Cotton by GbUBC23
Abstract
1. Introduction
2. Results
2.1. Identification of the UBC Gene Family in Sea-Island Cotton
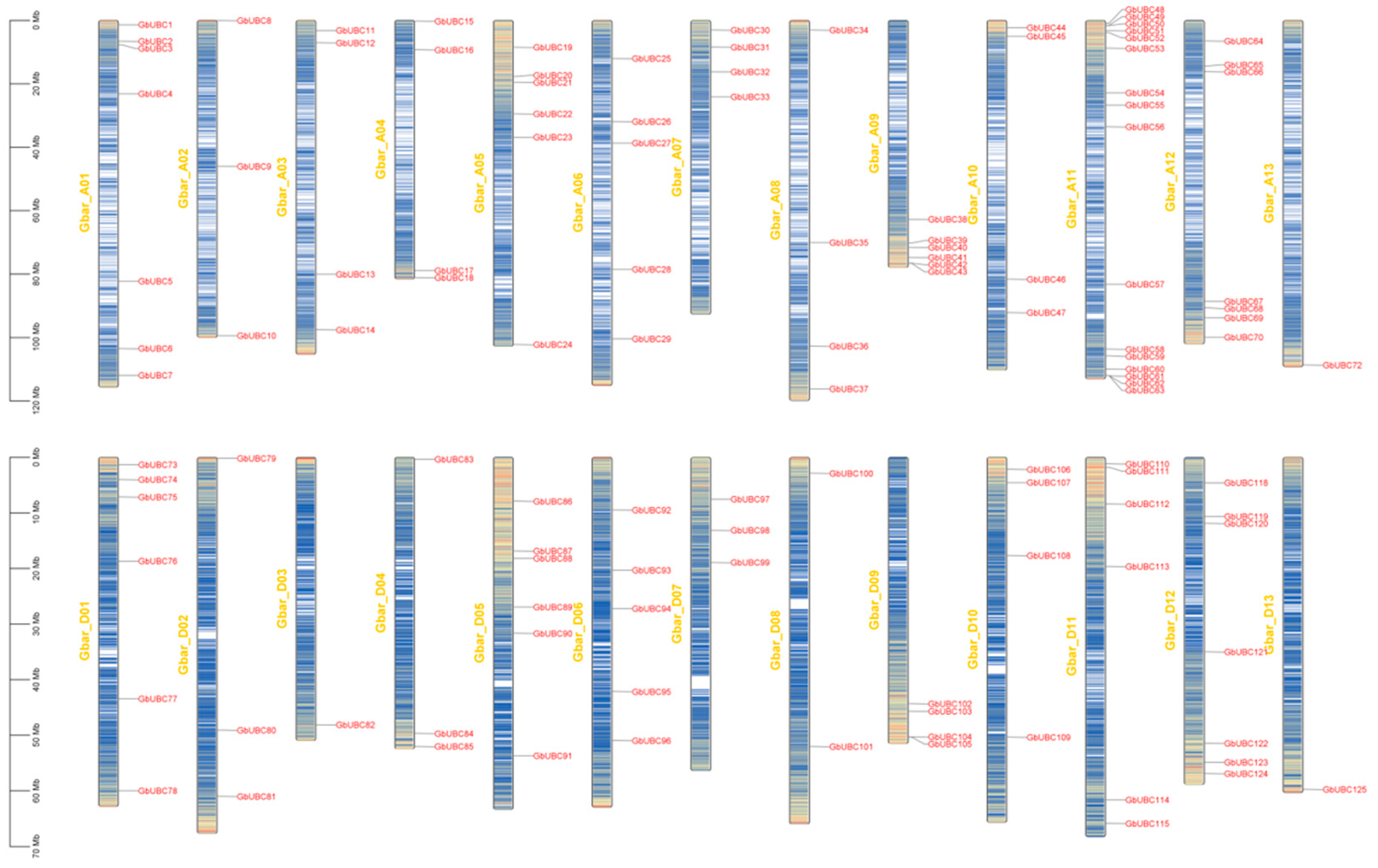
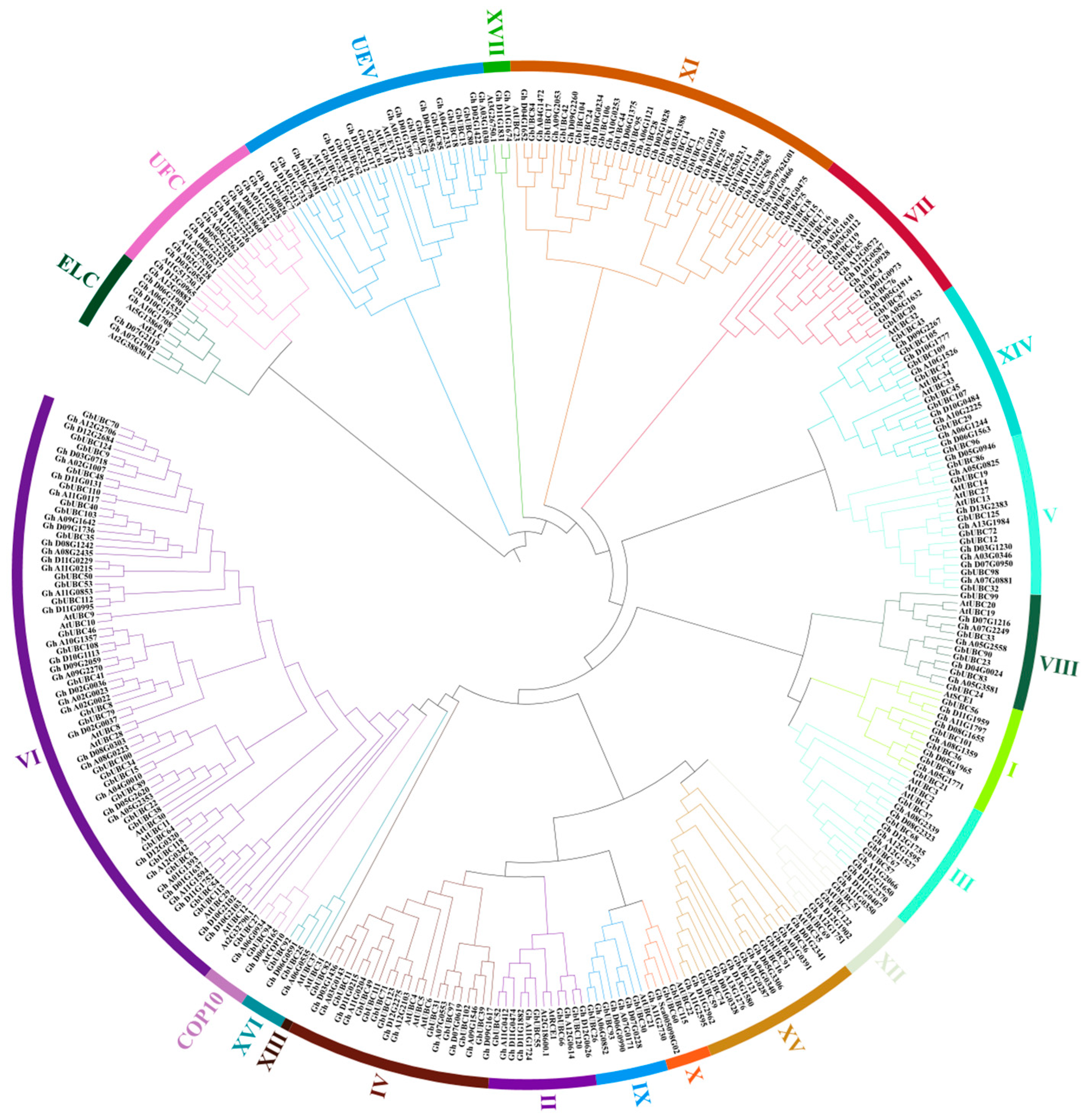
2.2. Chromosome Localization and Phylogenetic Analysis of the GbUBC Genes
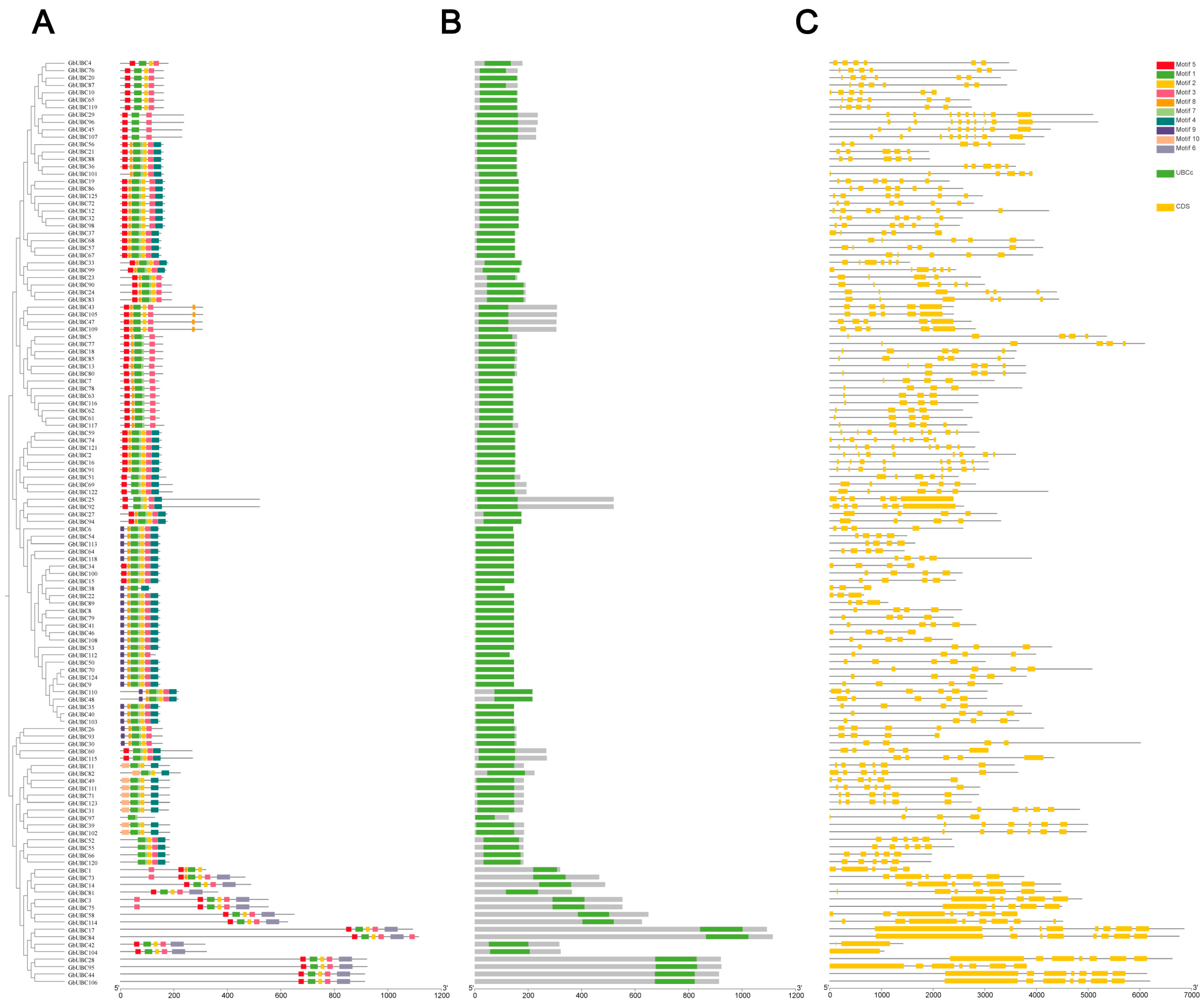
2.3. Structural Analysis of the GbUBC Genes
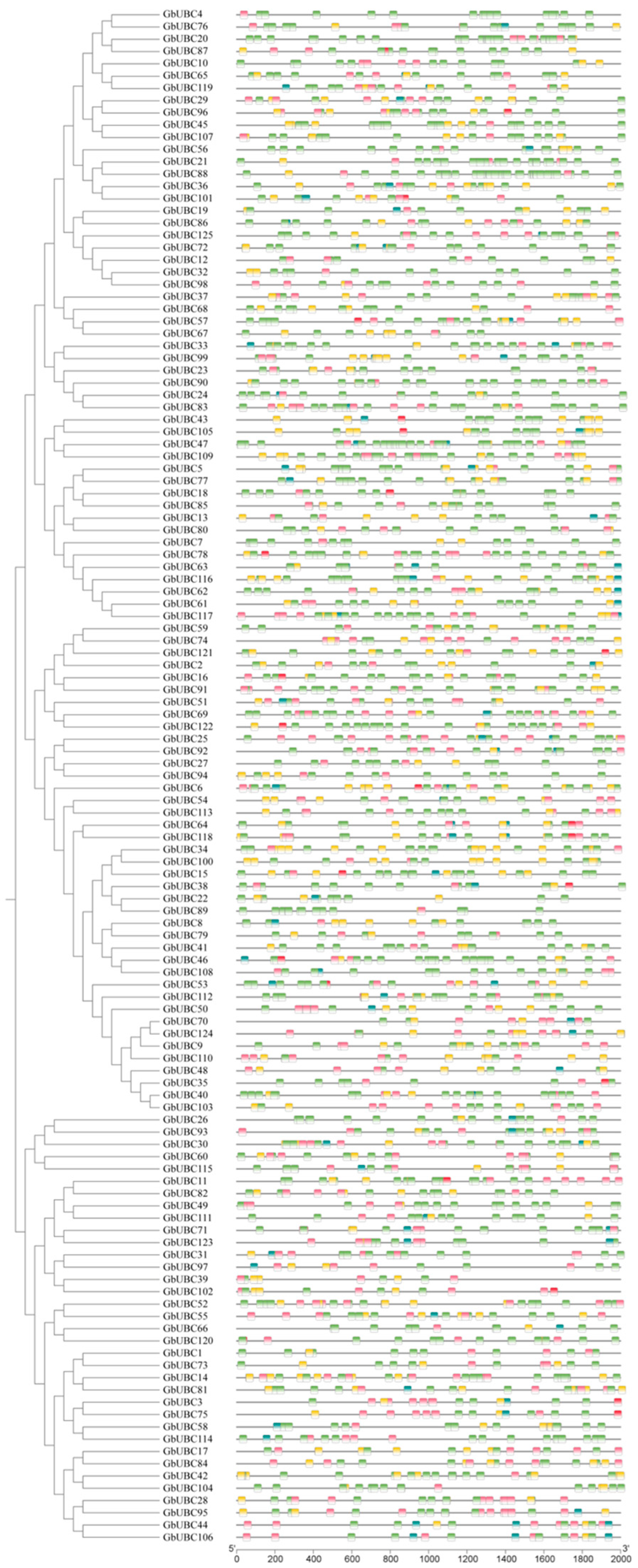
2.4. Analysis of the Cis-Acting Elements of the GbUBC Genes
2.5. Gene Duplication and Collinearity Analysis of the GbUBC Gene Family
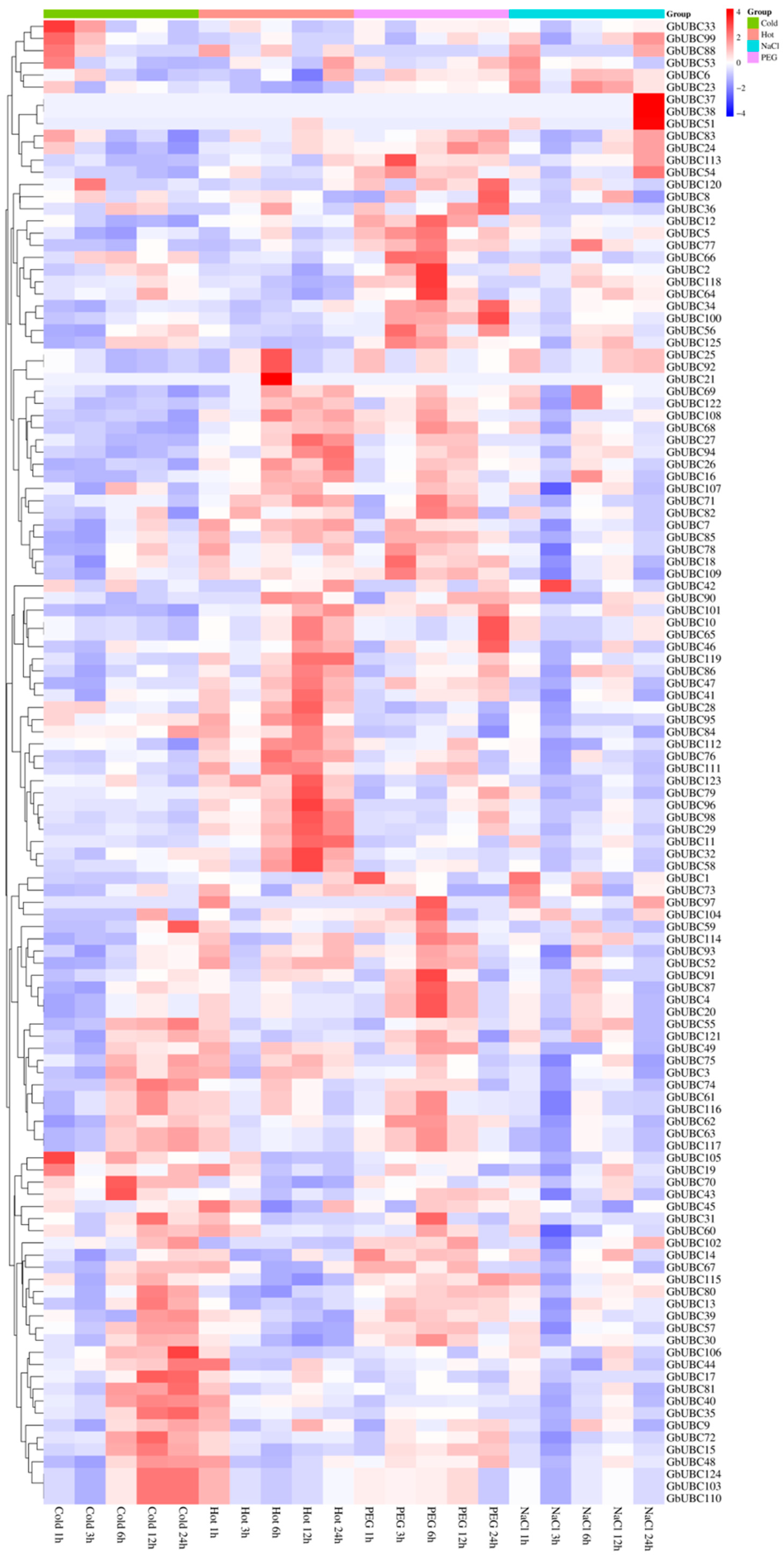
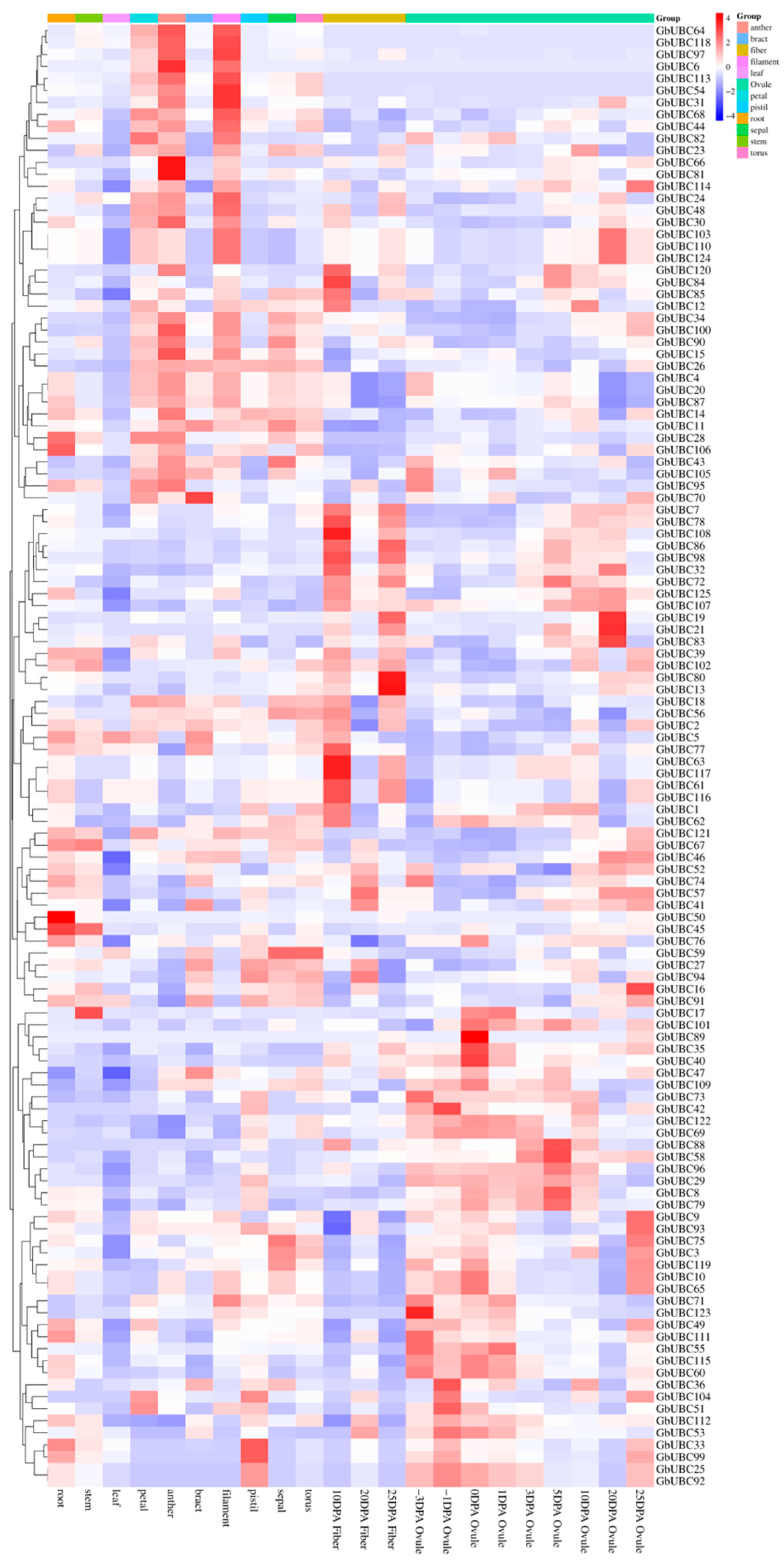
2.6. Expression Analysis of the GbUBC Genes
2.7. GbUBC23 Is Involved in Drought Stress and Is Localized to the Nucleus and Cell Membrane
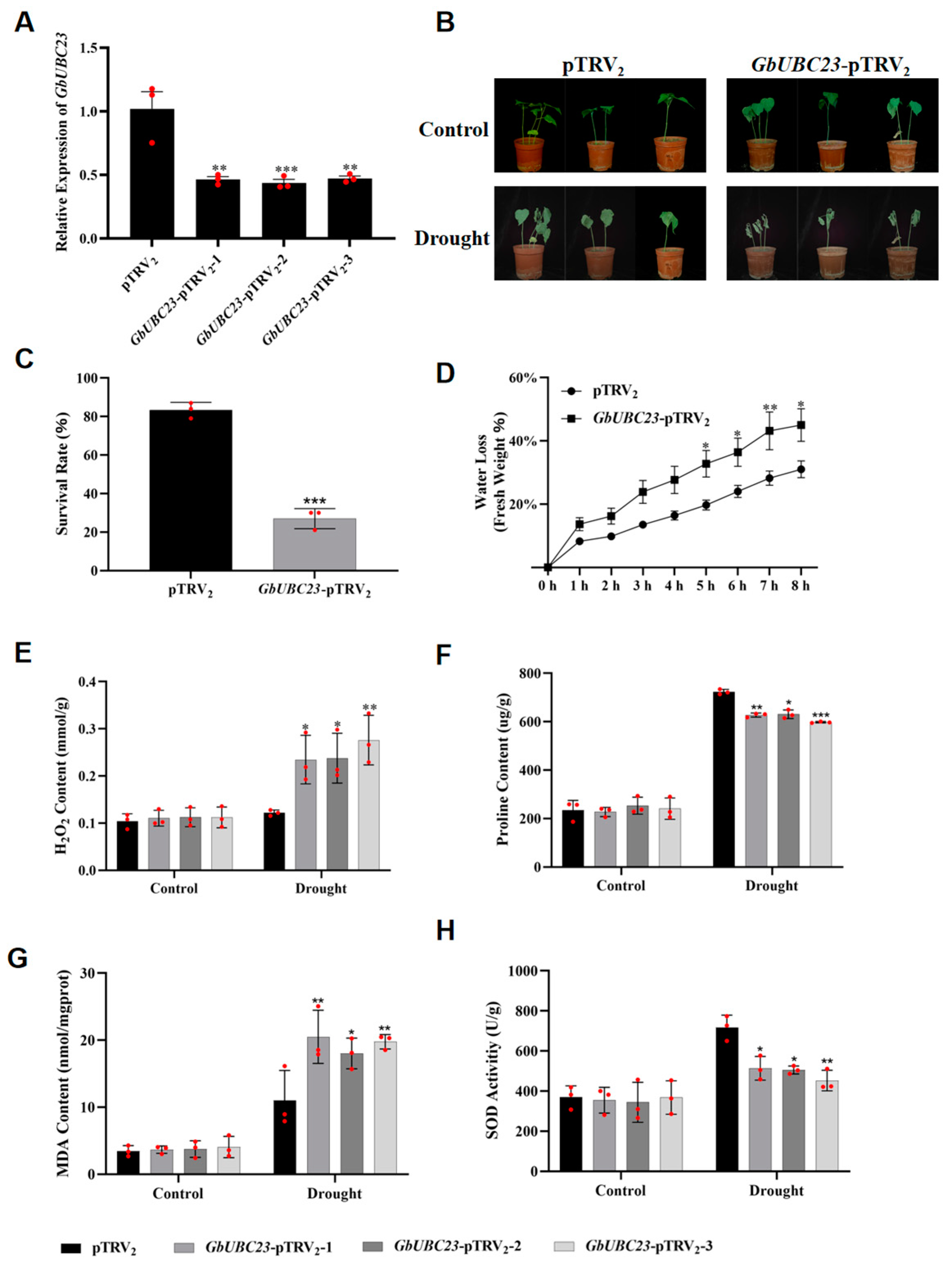
2.8. GbUBC23 Gene Silencing Decreases the Tolerance of Sea-Island Cotton to Drought
2.9. Changes in Drought Stress-Related Gene Expression
3. Discussion
4. Materials and Methods
4.1. Identification of the GbUBC Gene Family in Sea-Island Cotton
4.2. Sequence Alignment and Phylogenetic Analysis
4.3. Gene Structure and Protein-Conserved Motifs
4.4. Analysis of the Cis-Regulatory Elements in the Promoter
4.5. Chromosomal Location, Gene Duplication, and Collinearity Analysis
4.6. GbUBC Gene Expression Profiles
4.7. RNA Extraction and qPCR Analysis
4.8. Subcellular Localization
4.9. Virus-Induced Gene Silencing (VIGS) of GbUBC23
4.10. Plant Materials and Treatments
4.11. Determination of Physiological and Biochemical Indices
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.A.; Goldberg, A.L. The logic of the 26 s proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003, 8, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tang, X.; Qi, X.; Fu, X.; Ghimire, S.; Ma, R.; Li, S.; Zhang, N.; Si, H. The ubiquitin conjugating enzyme: An important ubiquitin transfer plat form in ubiquitin-proteasome system. Int. J. Mol. Sci. 2020, 21, 2894. [Google Scholar] [CrossRef] [PubMed]
- Hicke, L.; Schubert, H.L.; Hill, C.P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005, 6, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Kraft, E.; Stone, S.L.; Ma, L.; Su, N.; Gao, Y.; Lau, O.-S.; Deng, X.-W.; Callis, J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 2005, 139, 1597–1611. [Google Scholar] [CrossRef]
- Wang, P.; Guo, K.; Su, Q.; Deng, J.; Zhang, X.; Tu, L. Histone ubiquitination controls organ size in cotton (Gossypium hirsutum). Plant J. 2022, 110, 1005–1020. [Google Scholar] [CrossRef]
- Gao, W.; Zhang, L.; Zhang, Y.; Zhang, P.; Shahinnia, F.; Chen, T.; Yang, D. Genome-wide identification and expression analysis of the UBC gene family in wheat (Triticum aestivum L.). BMC Plant Biol. 2024, 24, 341. [Google Scholar] [CrossRef]
- Jue, D.; Sang, X.; Shu, B.; Liu, L.; Wang, Y.; Jia, Z.; Zou, Y.; Shi, S. Characterization and expression analysis of genes encoding ubiquitin conjugating domain-containing enzymes in Carica papaya. PLoS ONE 2017, 12, e0171357. [Google Scholar] [CrossRef] [PubMed]
- Jue, D.; Sang, X.; Liu, L.; Shu, B.; Wang, Y.; Xie, J.; Liu, C.; Shi, S. The ubiquitin-conjugating enzyme gene family in longan (Dimocarpus longan Lour.): Genome-wide identification and gene expression during flower induction and abiotic stress responses. Molecules 2018, 23, 662. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, Y.; Xin, H.; Li, S.; Liang, Z. Involvement of ubiquitin-conjugating enzyme (E2 gene family) in ripening process and response to cold and heat stress of Vitis vinifera. Sci. Rep. 2017, 7, 13290. [Google Scholar] [CrossRef] [PubMed]
- E, Z.; Zhang, Y.; Li, T.; Wang, L.; Zhao, H. Characterization of the ubiquitin-conjugating enzyme gene family in rice and evaluation of expression profiles under abiotic stresses and hormone treatments. PLoS ONE 2015, 10, e0122621. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Zhao, Q.; Chen, S. Evolution and expression analysis of the sorghum ubiquitin-conjugating enzyme family. Funct. Plant Biol. 2019, 46, 236–247. [Google Scholar] [CrossRef]
- Liu, W.; Tang, X.; Zhu, X.; Qi, X.; Zhang, N.; Si, H. Genome-wide identification and expression analysis of the E2 gene family in potato. Mol. Biol. Rep. 2019, 46, 777–791. [Google Scholar] [CrossRef]
- Sharma, B.; Bhatt, T.K. Genome-wide identification and expression analysis of E2 ubiquitin-conjugating enzymes in tomato. Sci. Rep. 2017, 7, 8613. [Google Scholar] [CrossRef]
- Dong, C.; Hu, H.; Jue, D.; Zhao, Q.; Chen, H.; Xie, J.; Jia, L. The banana E2 gene family: Genomic identification, characterization, expression profiling analysis. Plant Sci. 2016, 245, 11–24. [Google Scholar] [CrossRef]
- Chen, K.; Tang, W.-S.; Zhou, Y.-B.; Xu, Z.-S.; Chen, J.; Ma, Y.-Z.; Chen, M.; Li, H.-Y. Overexpression of GmUBC9 gene enhances plant drought resistance and affects flowering time via histone H2B monoubiquitination. Front. Plant Sci. 2020, 11, 555794. [Google Scholar] [CrossRef]
- Jue, D.; Sang, X.; Lu, S.; Dong, C.; Zhao, Q.; Chen, H.; Jia, L. Genome-wide identification, phylogenetic and expression analyses of the ubiquitin-conjugating enzyme gene family in Maize. PLoS ONE 2015, 10, e0143488. [Google Scholar] [CrossRef]
- Yao, S.; Xie, M.; Hu, M.; Cui, X.; Wu, H.; Li, X.; Hu, P.; Tong, C.; Yu, X. Genome-wide characterization of ubiquitin-conjugating enzyme gene family explores its genetic effects on the oil content and yield of Brassica napus. Front. Plant Sci. 2023, 14, 1118339. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ménard, R.; Berr, A.; Fuchs, J.; Cognat, V.; Meyer, D.; Shen, W.-H. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009, 57, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Lau, O.S.; Deng, X.W. Effect of Arabidopsis COP10 ubiquitin E2 enhancement activity across E2 families and functional conservation among its canonical homologues. Biochem. J. 2009, 418, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Schmidt, W. A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J. 2010, 62, 330–343. [Google Scholar] [CrossRef]
- Wen, R.; Wang, S.; Xiang, D.; Venglat, P.; Shi, X.; Zang, Y.; Datla, R.; Xiao, W.; Wang, H. UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J. 2014, 80, 424–436. [Google Scholar] [CrossRef]
- Wang, S.; Cao, L.; Wang, H. Arabidopsis ubiquitin-conjugating enzyme UBC22 is required for female gametophyte development and likely involved in Lys11-linked ubiquitination. J. Exp. Bot. 2016, 67, 3277–3288. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, W.; Cai, J.; Zhang, Y.; Qin, G.; Tian, S. Tomato nuclear proteome reveals the involvement of specific E2 ubiquit in-conjugating enzymes in fruit ripening. Genome Biol. 2014, 15, 548. [Google Scholar] [CrossRef]
- Wang, L.; Wen, R.; Wang, J.; Xiang, D.; Wang, Q.; Zang, Y.; Wang, Z.; Huang, S.; Li, X.; Datla, R.; et al. Arabidopsis UBC13 differentially regulates two programmed cell death p athways in responses to pathogen and low-temperature stress. New Phytol. 2019, 221, 919–934. [Google Scholar] [CrossRef]
- Jin, S.; Youn, G.; Kim, S.Y.; Kang, T.; Shin, H.-Y.; Jung, J.-Y.; Seo, P.J.; Ahn, J.H. The CUL3A-LFH1-UBC15 ubiquitin ligase complex mediates short vegetative phase degradation to accelerate flowering at high ambient temperature. Plant Commun. 2024, 5, 100814. [Google Scholar] [CrossRef]
- Cui, F.; Liu, L.; Zhao, Q.; Zhang, Z.; Li, Q.; Lin, B.; Wu, Y.; Tang, S.; Xie, Q. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 2012, 24, 233–244. [Google Scholar] [CrossRef]
- Ahn, M.Y.; Oh, T.R.; Seo, D.H.; Kim, J.H.; Cho, N.H.; Kim, W.T. Arabidopsis group XIV ubiquitin-conjugating enzymes AtUBC32, AtUBC33, and AtUBC34 play negative roles in drought stress response. J. Plant Physiol. 2018, 230, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tian, Z.; Zuo, D.; Wang, Q.; Song, G. GhUBC10-2 mediates GhGSTU17 degradation to regulate salt tolerance in cotton (Gossypium hirsutum). Plant Cell Environ. 2024, 47, 1606–1624. [Google Scholar] [CrossRef] [PubMed]
- Middleton, A.J.; Wright, J.D.; Day, C.L. Regulation of E2s: A role for additional ubiquitin binding sites? J. Mol. Biol. 2017, 429, 3430–3440. [Google Scholar] [CrossRef]
- Bae, H.; Kim, W.T. Classification and interaction modes of 40 rice E2 ubiquitin-conjugating enzymes with 17 rice ARM-U-box E3 ubiquitin ligases. Biochem. Biophys. Res. Commun. 2014, 444, 575–580. [Google Scholar] [CrossRef]
- Yu, F.; Lou, L.; Tian, M.; Li, Q.; Ding, Y.; Cao, X.; Wu, Y.; Belda-Palazon, B.; Rodriguez, P.L.; Yang, S.; et al. ESCRT-I component VPS23A affects ABA signaling by recognizing ABA receptors for endosomal degradation. Mol. Plant 2016, 9, 1570–1582. [Google Scholar] [CrossRef] [PubMed]
- Jeon, E.H.; Pak, J.H.; Kim, M.J.; Kim, H.J.; Shin, S.H.; Lee, J.H.; Kim, D.H.; Oh, J.S.; Oh, B.-J.; Jung, H.W.; et al. Ectopic expression of ubiquitin-conjugating enzyme gene from wild rice, OgUBC1, confers resistance against UV-B radiation and Botrytis infection in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2012, 427, 309–314. [Google Scholar] [CrossRef]
- Zhou, G.-A.; Chang, R.-Z.; Qiu, L.-J. Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis. Plant Mol. Biol. 2010, 72, 357–367. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Wan, H.; Ni, Z. Sea-island cotton (Gossypium barbadense L.) GbTCP5 improves plant adaptation to drought and salt stress by directly activating GbERD7, GbUBC19, and GbGOLS2 expression. Ind. Crops Prod. 2023, 203, 117209. [Google Scholar] [CrossRef]
- Cheng, M.-C.; Kuo, W.-C.; Wang, Y.-M.; Chen, H.-Y.; Lin, T.-P. UBC18 mediates ERF1 degradation under light-dark cycles. New Phytol. 2017, 213, 1156–1167. [Google Scholar] [CrossRef]
- Mao, Z.Z.; Gong, Y.; Shi, G.X.; Li, Y.L.; Yu, D.Y.; Huang, F. Cloning of the soybean E2 ubiquitin-conjugating enzyme GmUBC1 and its expression in Arabidopsis thaliana. Hereditas 2020, 42, 788–798. [Google Scholar]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.K. ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef]
- Mehari, T.G.; Xu, Y.; Magwanga, R.O.; Umer, M.J.; Shiraku, M.L.; Hou, Y.; Wang, Y.; Wang, K.; Cai, X.; Zhou, Z.; et al. Identification and functional characterization of Gh_D01G0514 (GhNAC072) transcription factor in response to drought stress tolerance in cotton. Plant Physiol. Biochys. 2021, 166, 361–375. [Google Scholar] [CrossRef]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef]
- Zhang, B.; Chang, L.; Sun, W.; Ullah, A.; Yang, X. Overexpression of an expansin-like gene, GhEXLB2 enhanced drought tole rance in cotton. Plant Physiol. Bioch. 2021, 162, 468–475. [Google Scholar] [CrossRef]
- Xu, C.; Shan, J.; Liu, T.; Wang, Q.; Ji, Y.; Zhang, Y.; Wang, M.; Xia, N.; Zhao, L. Constans-like 1a positively regulates salt and drought tolerance in soybean. Plant Physiol. 2023, 191, 2427–2446. [Google Scholar] [CrossRef]
- Wang, C.; Ru, J.; Liu, Y.; Li, M.; Zhao, D.; Yang, J.; Fu, J.; Xu, Z. Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef] [PubMed]
- Duan, B.; Xie, X.; Jiang, Y.; Zhu, N.; Zheng, H.; Liu, Y.; Hua, X.; Zhao, Y.; Sun, Y. GhMYB44 enhances stomatal closure to confer drought stress tolerance in cotton and Arabidopsis. Plant Physiol. Bioch. 2023, 198, 107692. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Huang, X.; Ding, L.; Wang, Z.; Tang, D.; Chen, B.; Ao, L.; Liu, Y.; Kang, Z.; Mao, H. TaERF87 and TaAKS1 synergistically regulate TaP5CS1/TaP5CR1-mediated proline biosynthesis to enhance drought tolerance in wheat. New Phytol. 2023, 237, 232–250. [Google Scholar] [PubMed]
- Wan, X.R.; Mo, A.Q.; Liu, S.; Yang, L.X.; Li, L. Constitutive expression of a peanut ubiquitin-conjugating enzyme gene in Arabidopsis confers improved water-stress tolerance through regulation of stress-responsive gene expression. J. Biosci. 2011, 111, 478–484. [Google Scholar]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.-S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-de pendent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K.; et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. USA 2018, 115, 11178–11187. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Mistry, J.; Schuster-Böckler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006, 34, 247–251. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLos Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, 427–432. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, 22–226. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, 458–460. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, 597–603. [Google Scholar] [CrossRef]
- Song, H.; Duan, Z.; Wang, Z.; Li, Y.; Wang, Y.; Li, C.; Mao, W.; Que, Q.; Chen, X.; Li, P. Genome-wide identification, expression pattern and subcellular localization analysis of the JAZ gene family in Toona ciliata. Ind. Crops Prod. 2022, 17, 114582. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for b iological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Tuskan, G.A.; Cheng, M.Z. Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 2006, 142, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, J.; Deng, F.; Wang, W.; Cheng, Y.; Song, L.; Hu, M.; Shen, J.; Xu, Q.; Shen, F. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 2019, 19, 459. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zong, Z.; Chen, J.; Sun, X.; Wang, J.; Yu, Y.; Ni, Z. Genome-Wide Identification of the GbUBC Gene Family in Sea-Island Cotton (Gossypium barbadense) and the Active Regulation of Drought Resistance in Cotton by GbUBC23. Int. J. Mol. Sci. 2024, 25, 12948. https://doi.org/10.3390/ijms252312948
Wang Y, Zong Z, Chen J, Sun X, Wang J, Yu Y, Ni Z. Genome-Wide Identification of the GbUBC Gene Family in Sea-Island Cotton (Gossypium barbadense) and the Active Regulation of Drought Resistance in Cotton by GbUBC23. International Journal of Molecular Sciences. 2024; 25(23):12948. https://doi.org/10.3390/ijms252312948
Chicago/Turabian StyleWang, Yi, Zheng Zong, Junchen Chen, Xue Sun, Jiahui Wang, Yuehua Yu, and Zhiyong Ni. 2024. "Genome-Wide Identification of the GbUBC Gene Family in Sea-Island Cotton (Gossypium barbadense) and the Active Regulation of Drought Resistance in Cotton by GbUBC23" International Journal of Molecular Sciences 25, no. 23: 12948. https://doi.org/10.3390/ijms252312948
APA StyleWang, Y., Zong, Z., Chen, J., Sun, X., Wang, J., Yu, Y., & Ni, Z. (2024). Genome-Wide Identification of the GbUBC Gene Family in Sea-Island Cotton (Gossypium barbadense) and the Active Regulation of Drought Resistance in Cotton by GbUBC23. International Journal of Molecular Sciences, 25(23), 12948. https://doi.org/10.3390/ijms252312948





