Biophysical Characterization of a Novel Phosphopentomutase from the Hyperthermophilic Archaeon Thermococcus kodakarensis
Abstract
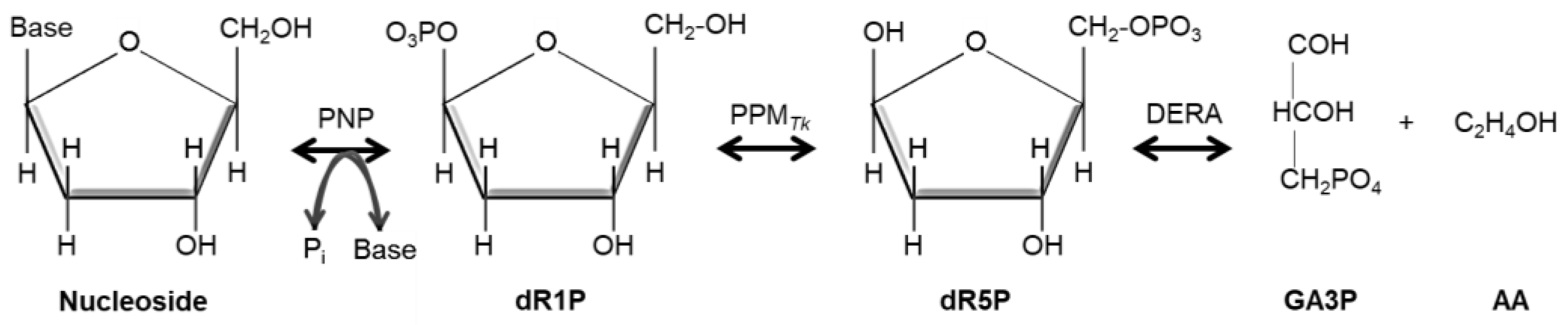
1. Introduction
2. Results
2.1. Domain Structure Analysis of PPMTk
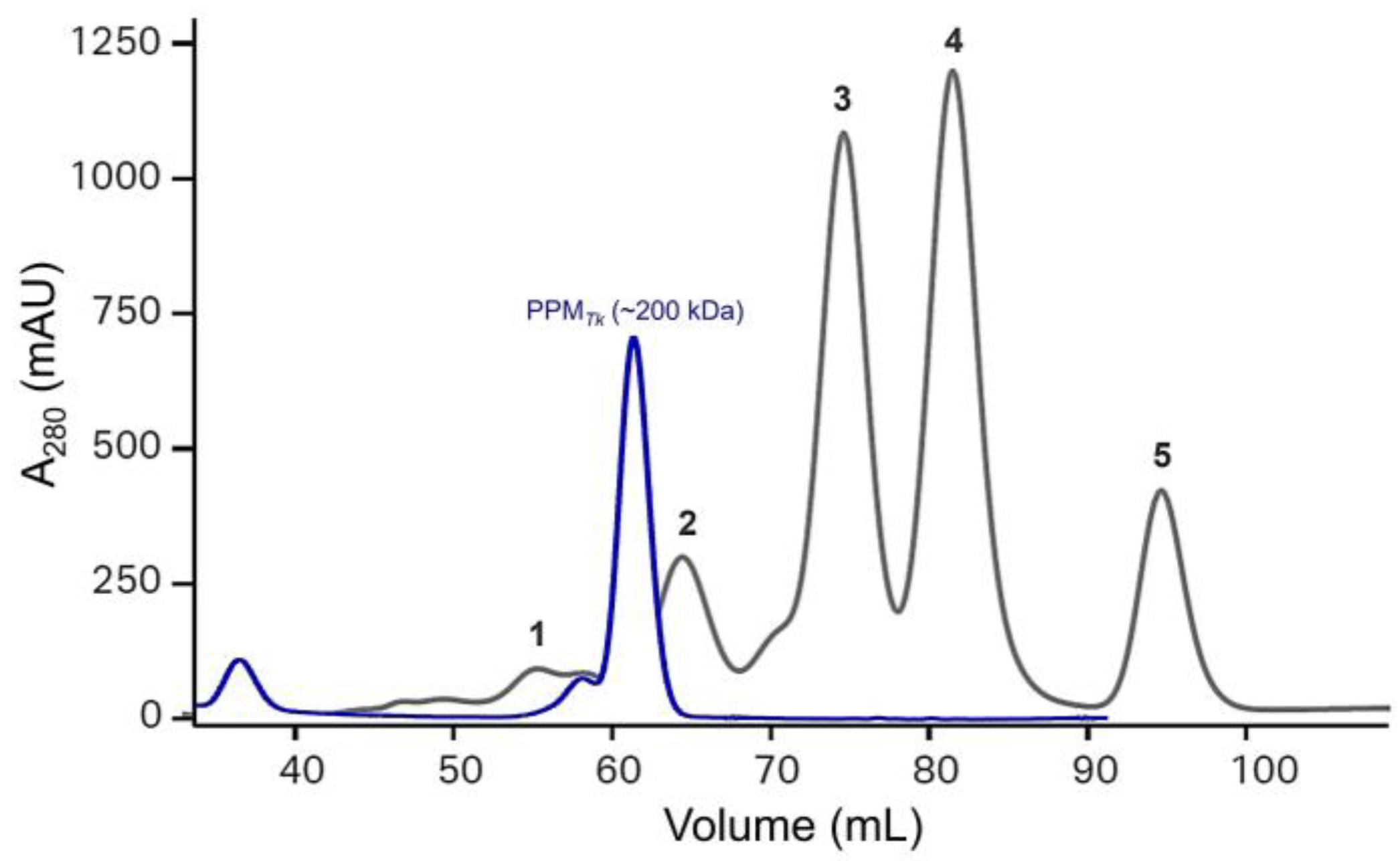
2.2. Vector Construction, Overexpression, and Purification of PPMTk
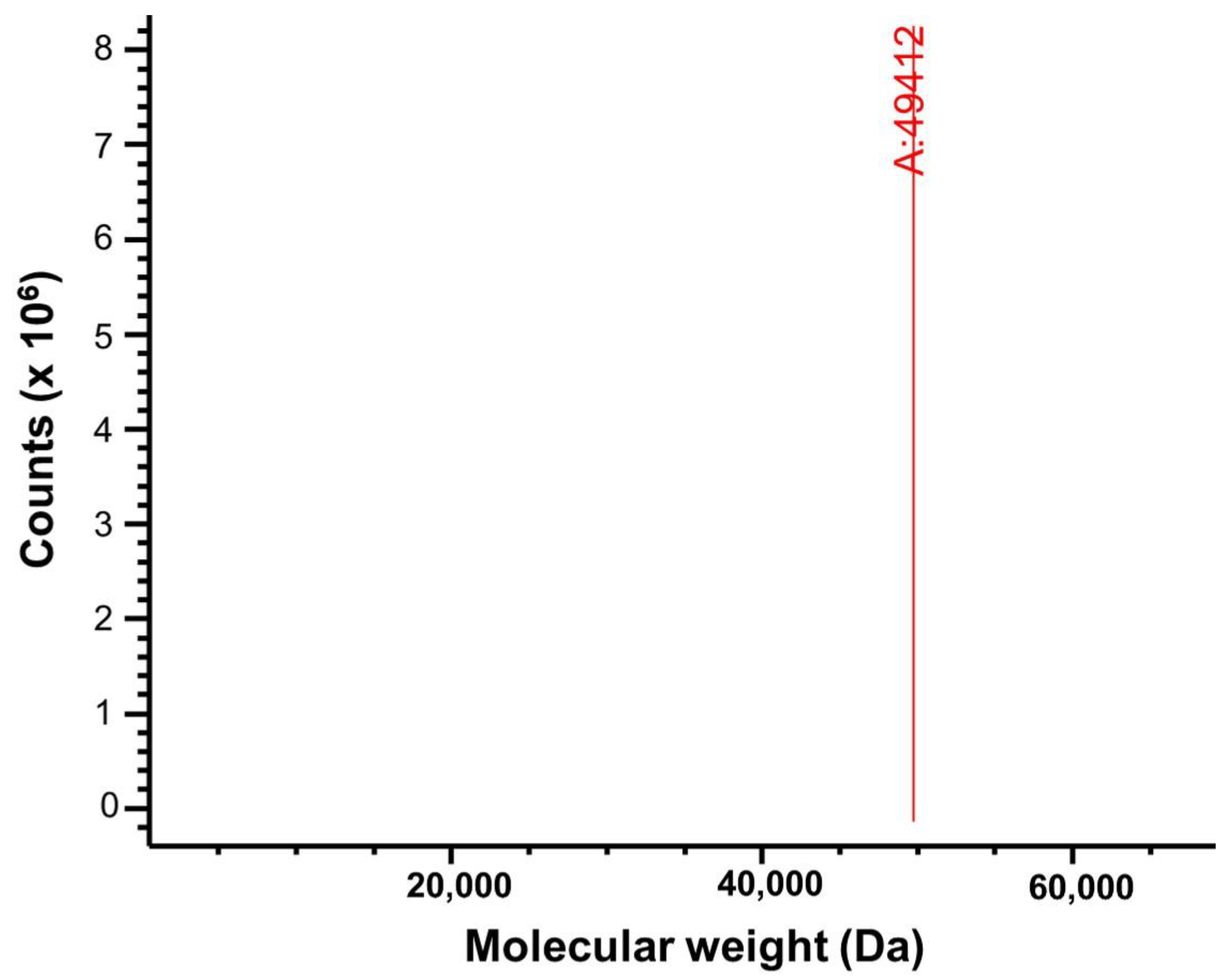
2.3. Electrospray Ionization Mass Spectrometric Analysis of PPMTk
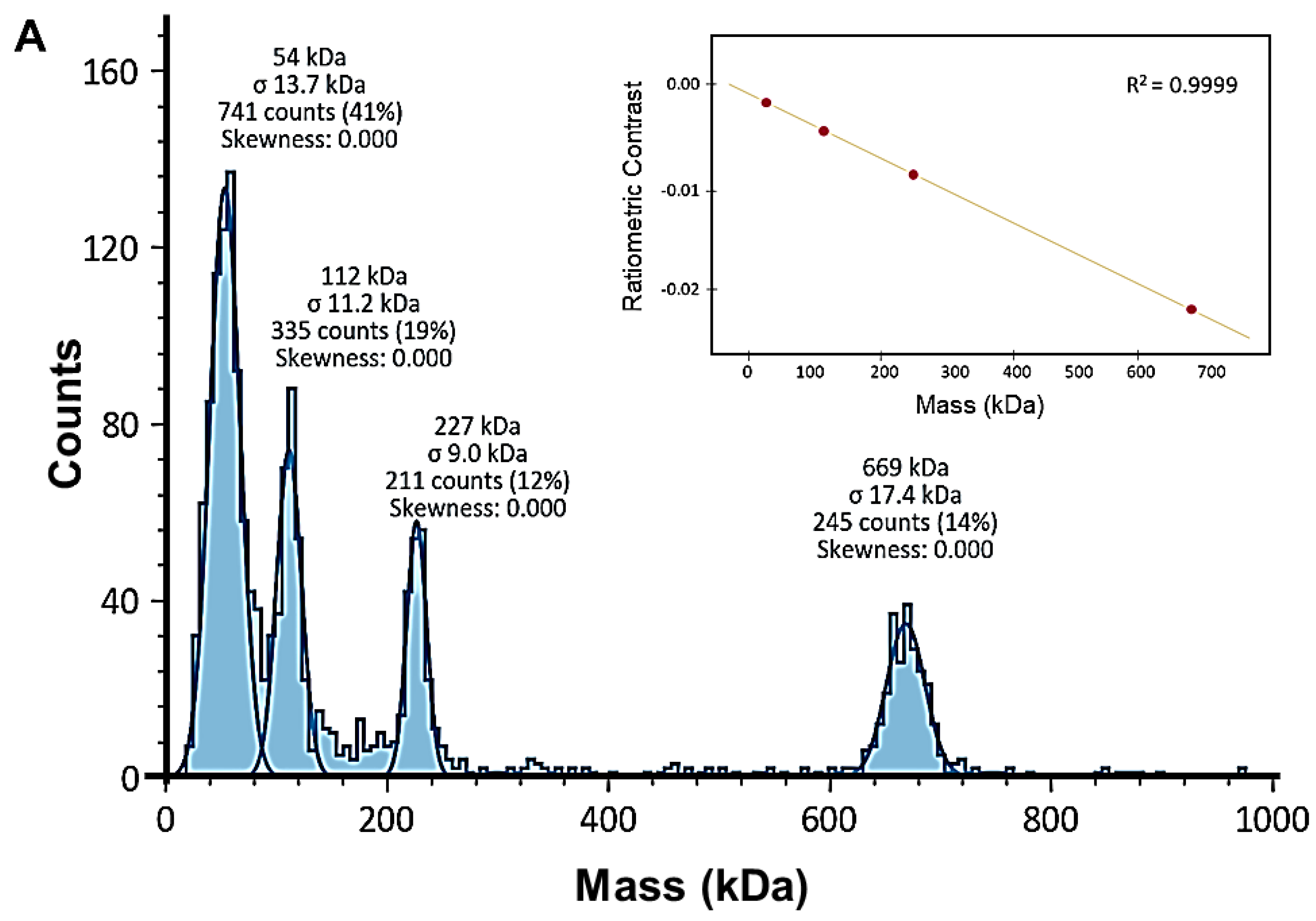
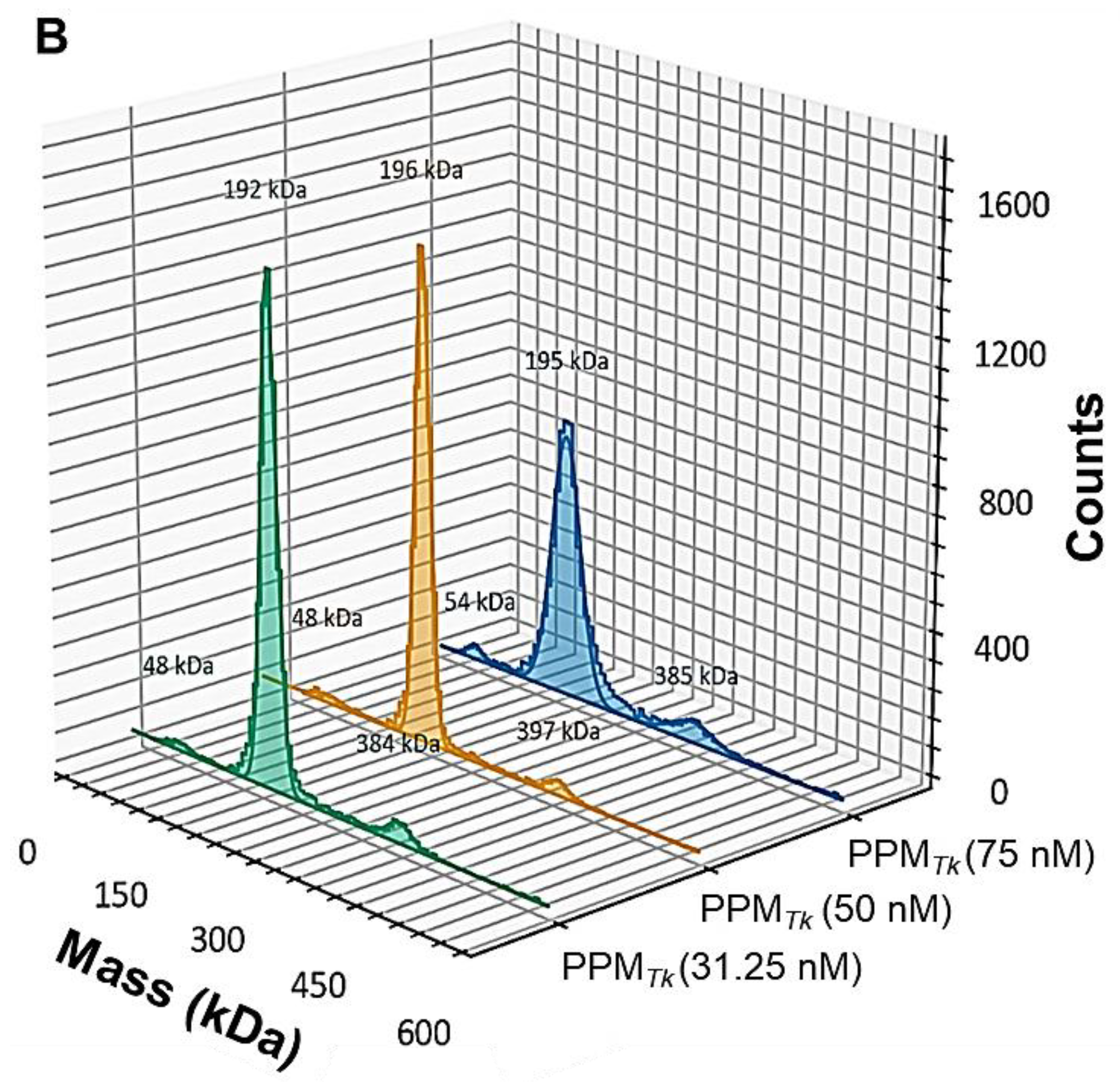
2.4. Mass Photometric Studies of PPMTk
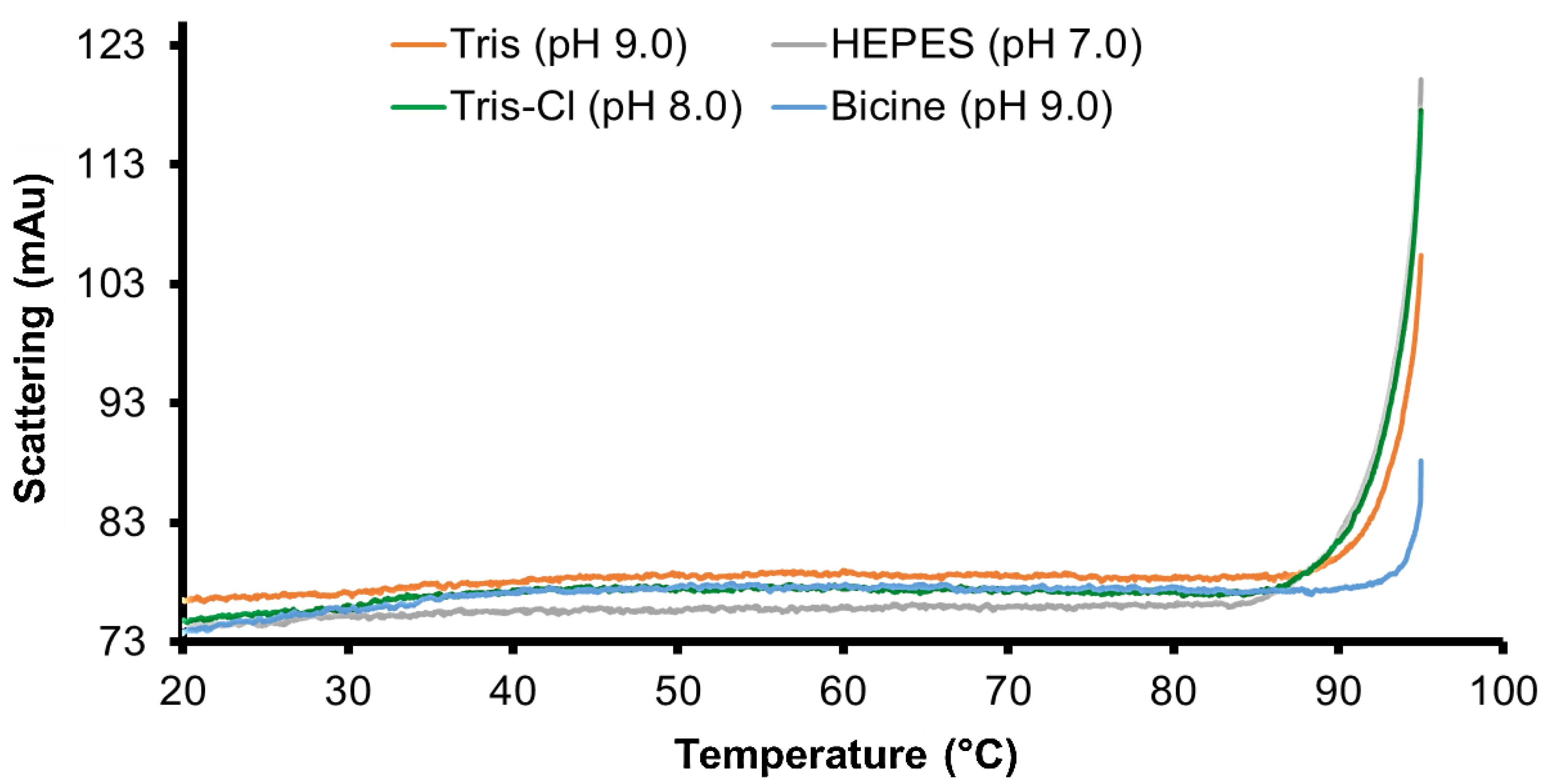
2.5. Differential Scanning Fluorimetry Analysis of PPMTk
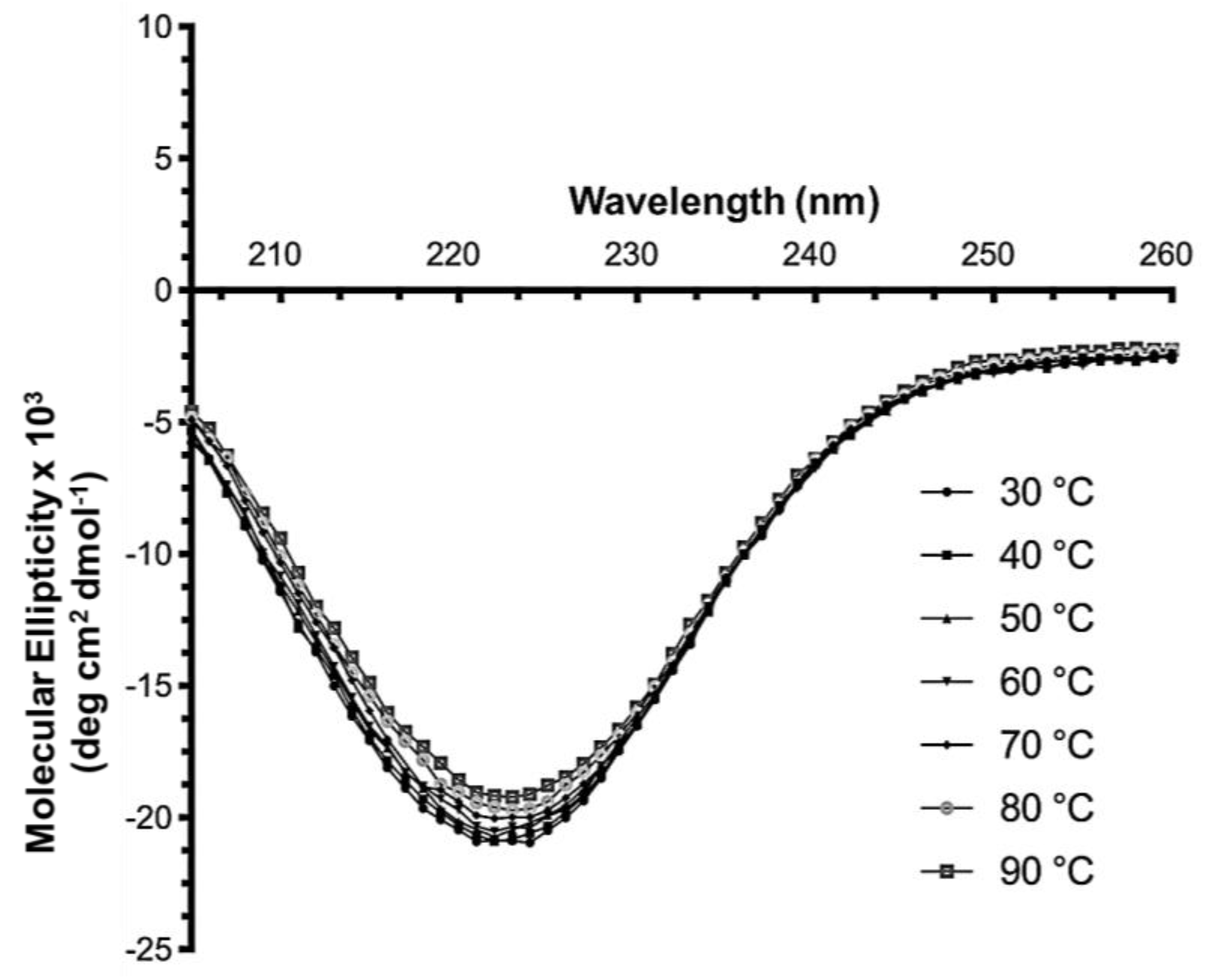
2.6. Secondary Structure Analysis of PPMTk Using CD Spectrometry
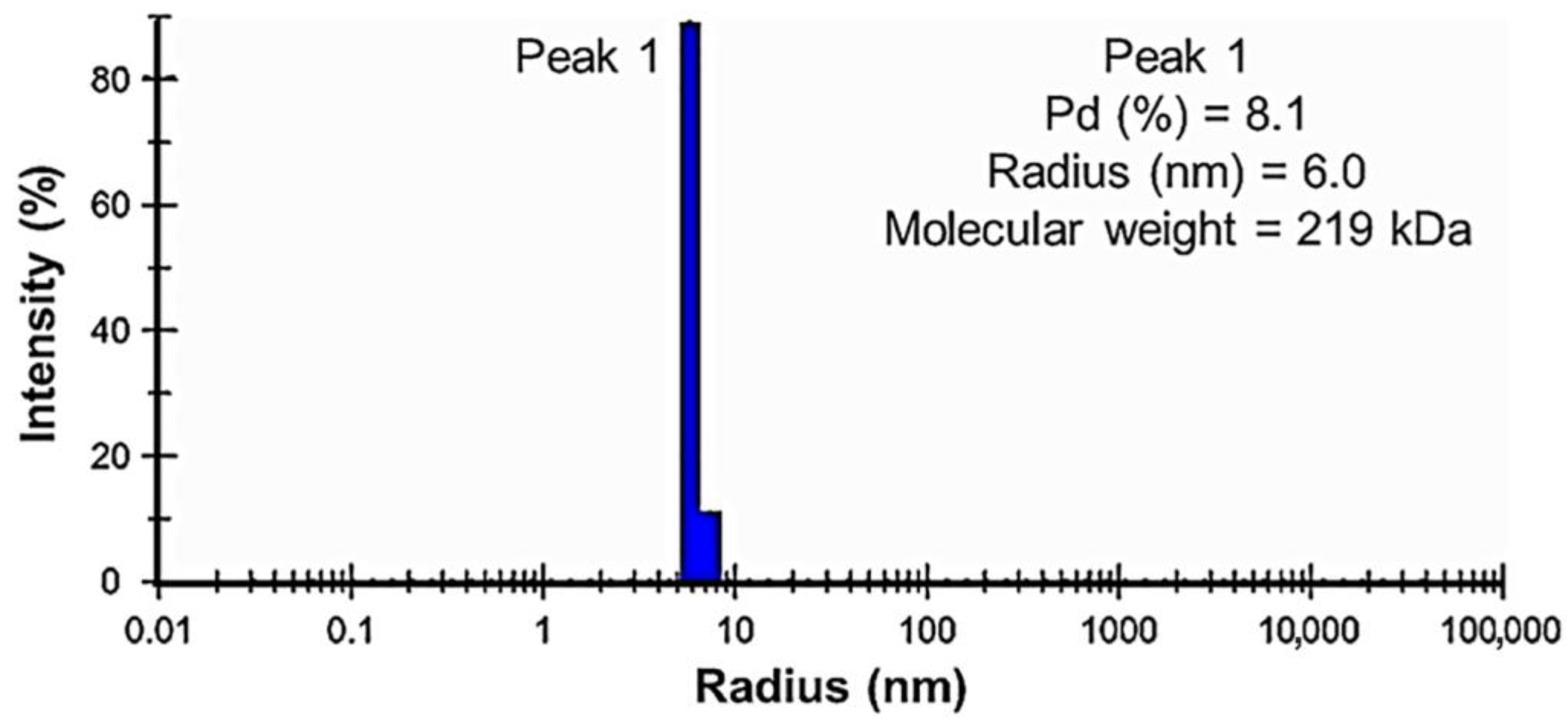
2.7. Dynamic Light Scattering Analysis of PPMTk
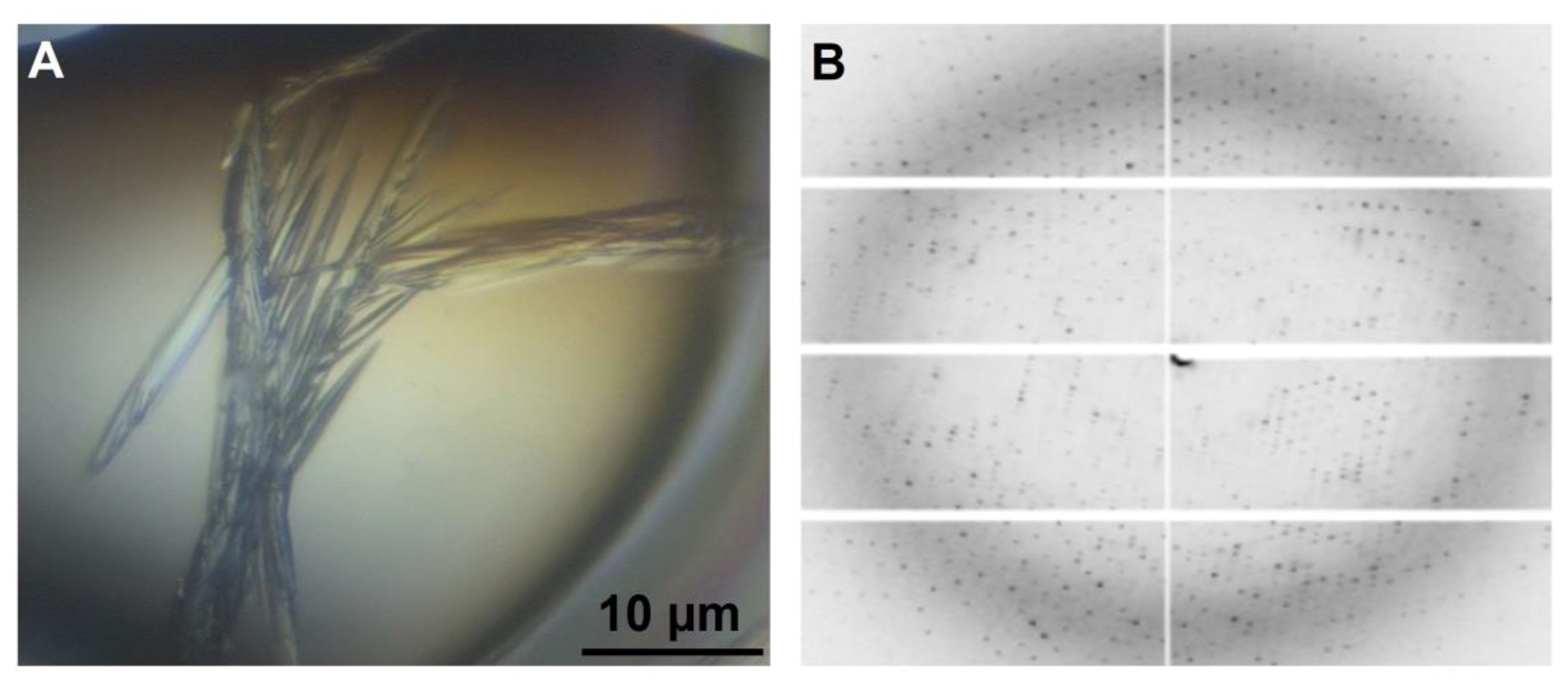
2.8. Crystallization of PPMTk and Diffraction Data Analysis
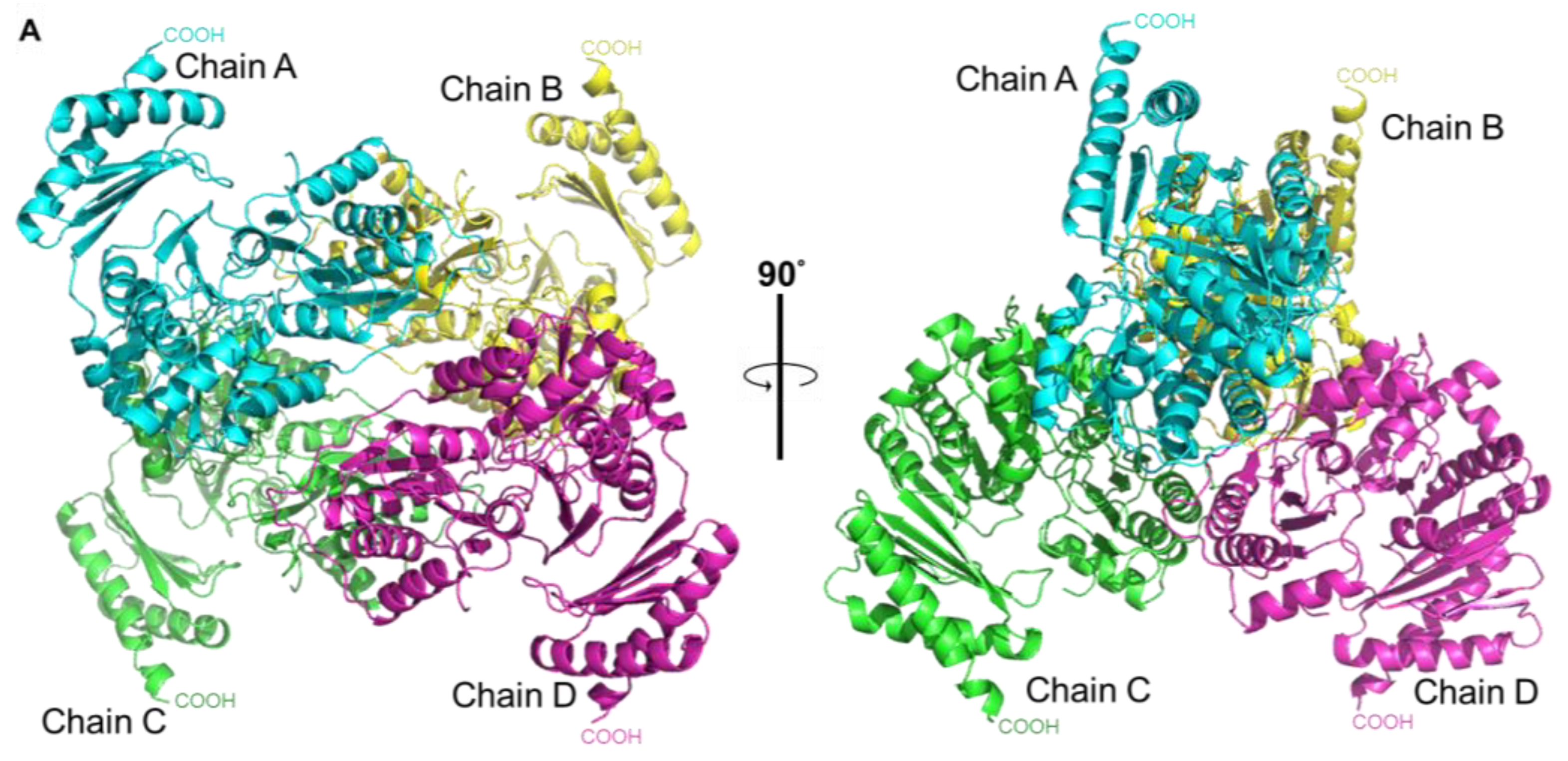
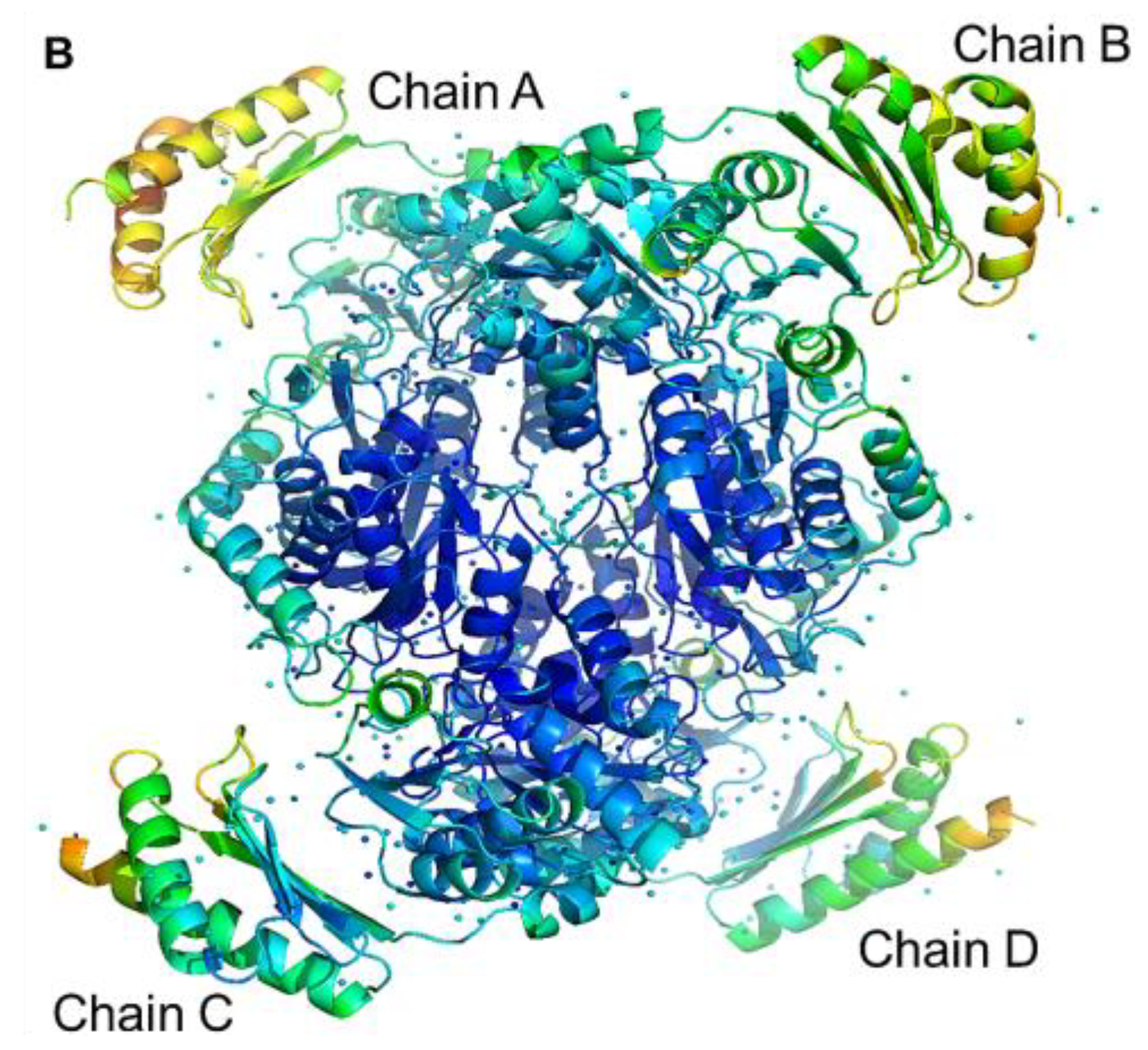
2.9. Architecture of the Tetramer of PPMTk
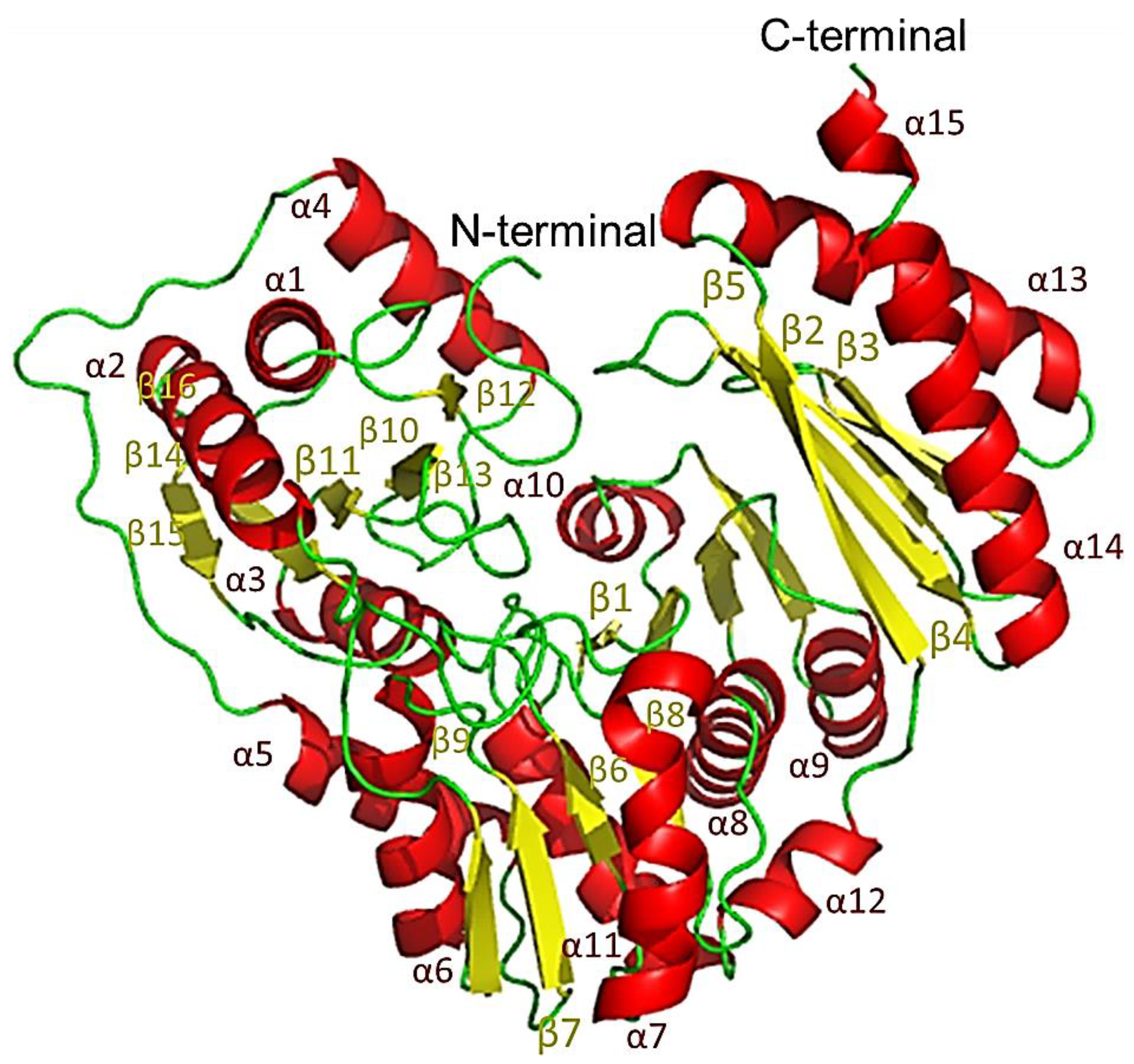
2.10. The Protomer of PPMTk
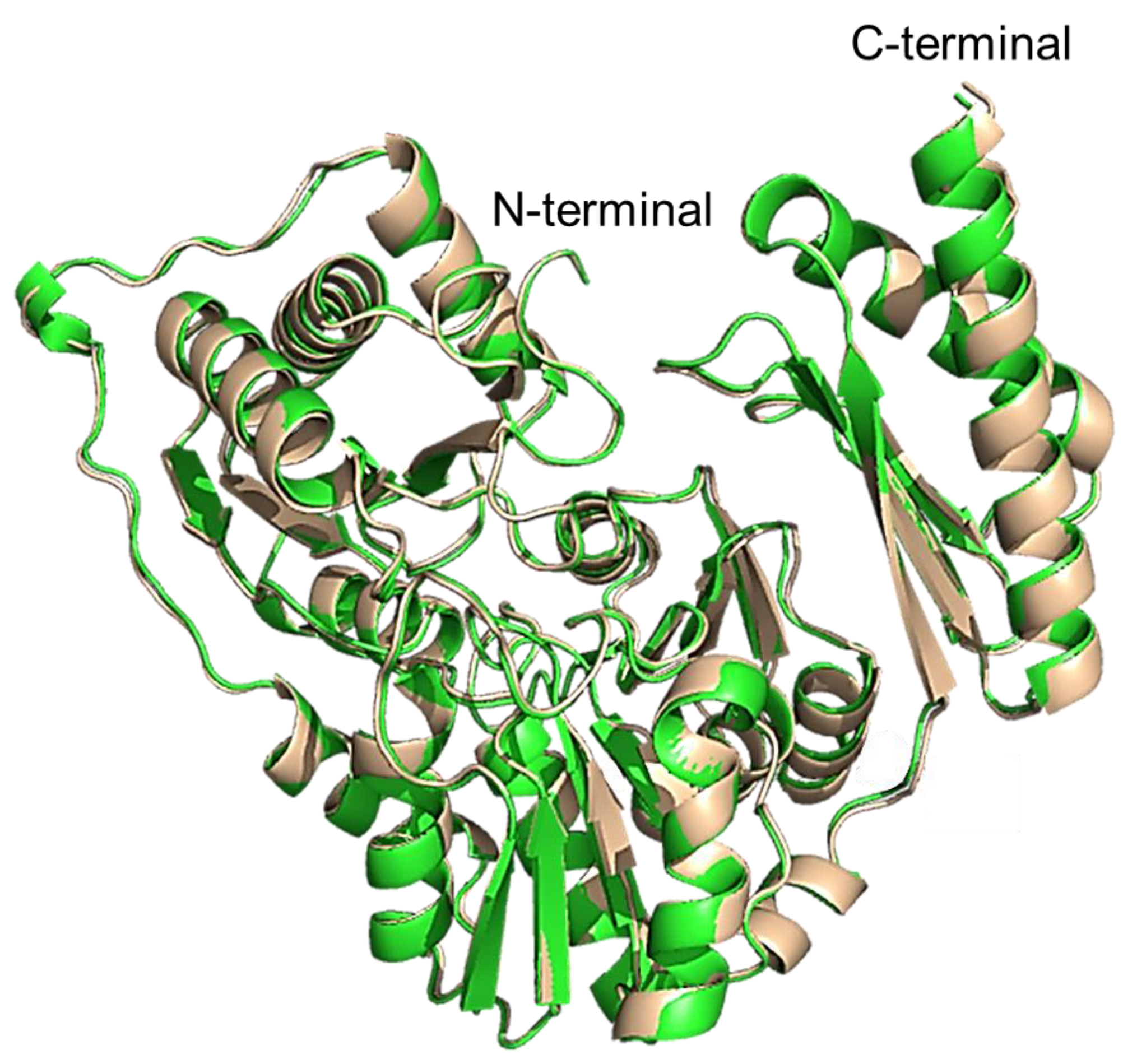
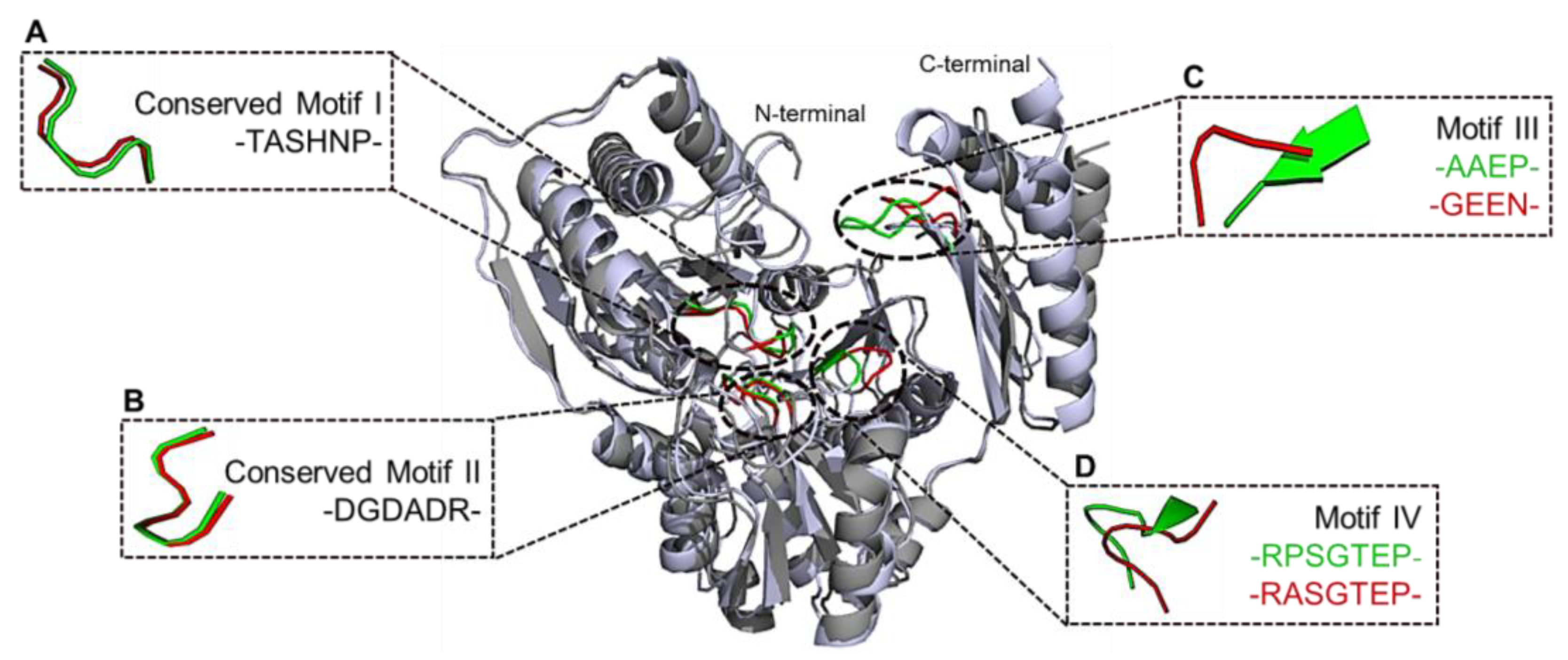
2.11. Structural Comparison
2.12. Comparison of PPMTk with Phosphoglucomutase
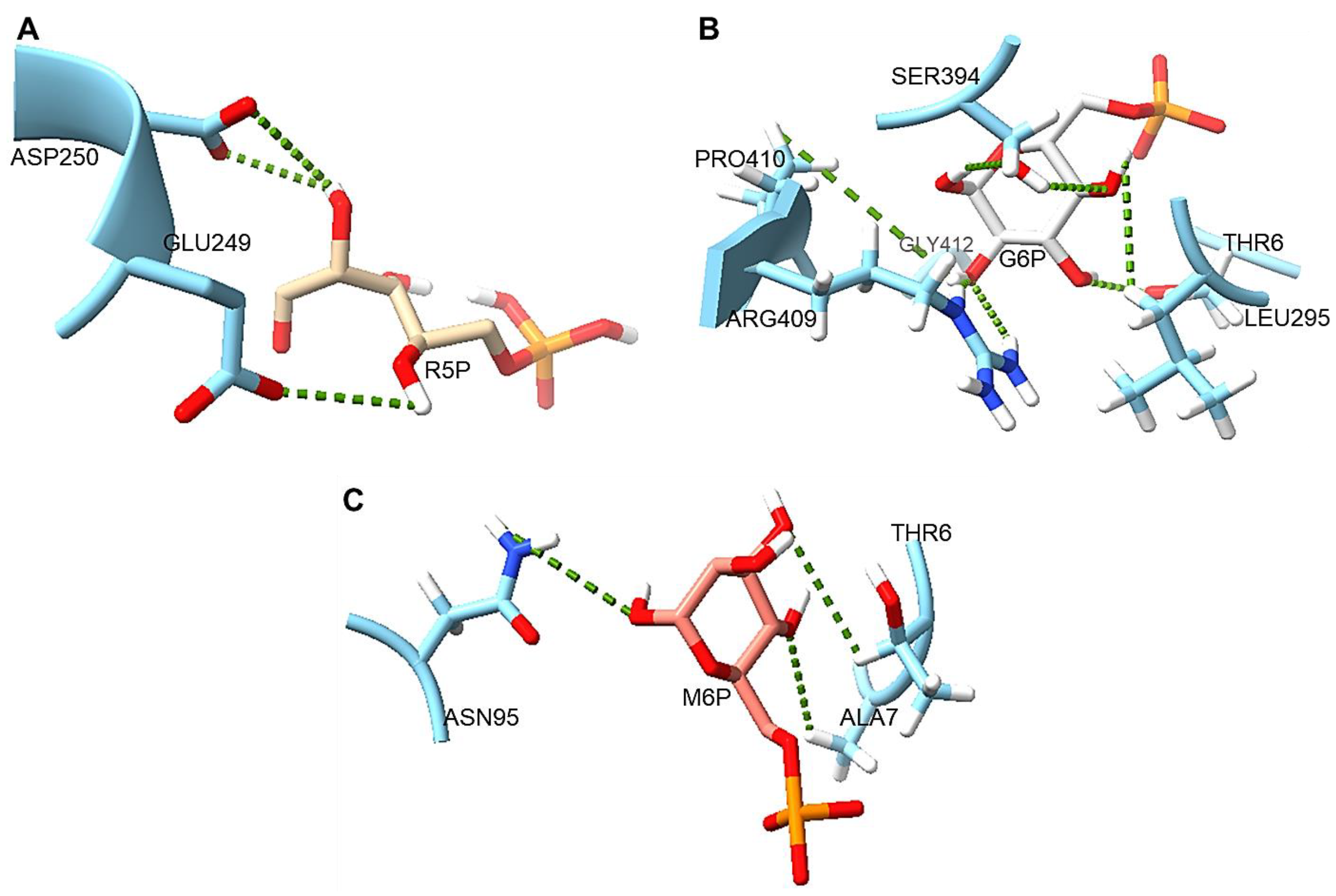
2.13. Protein–Ligand Interaction
3. Discussion
4. Materials and Methods
4.1. Sequence Retrieval, Physicochemical Parameters and Domain Structural Analysis
4.2. Plasmid, Bacterial Strains, and Media
4.3. Construction of the Expression Vector
4.4. Optimization of the Expression of Tk1777 Gene in E. coli
4.5. Cell Lysis and Partial Protein Purification
4.6. Purification of Recombinant PPMTk
4.7. Estimation of Molecular Weight by Gel Filtration Chromatography
4.8. Determination of Molecular Weight Using ESI-MS
4.9. Mass Photometric Analysis of PPMTk
4.10. Differential Scanning Fluorimetry Analysis
4.11. Secondary Structure Analysis by CD Spectrometry
4.12. Dynamic Light Scattering Analysis
4.13. Crystallization, Diffraction Data Collection, and Data Processing
4.14. In Silico Analysis of PPMTk
4.15. Structure Deposition
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rytter, H.; Jamet, A.; Ziveri, J.; Ramond, E.; Coureuil, M.; Lagouge-Roussey, P.; Euphrasie, D.; Tros, F.; Goudin, N.; Chhuon, C. The pentose phosphate pathway constitutes a major metabolic hub in pathogenic Francisella. PLoS Pathog. 2021, 17, 1–32. [Google Scholar] [CrossRef]
- Bräsen, C.; Esser, D.; Rauch, B.; Siebers, B. Carbohydrate metabolism in archaea: Current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 2014, 78, 89–175. [Google Scholar] [CrossRef] [PubMed]
- Orita, I.; Sato, T.; Yurimoto, H.; Kato, N.; Atomi, H.; Imanaka, T.; Sakai, Y. The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakarensis. J. Bacteriol. 2006, 188, 4698–4704. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Utashima, S.; Yoshii, Y.; Hirata, K.; Kanda, S.; Onoda, Y.; Jin, J.-Q.; Xiao, S.; Minami, R.; Fukushima, H.; et al. A non-carboxylating pentose bisphosphate pathway in halophilic archaea. Commun. Biol. 2022, 5, 1290. [Google Scholar] [CrossRef] [PubMed]
- Rashid, N.; Imanaka, H.; Fukui, T.; Atomi, H.; Imanaka, T. Presence of a novel phosphopentomutase and a 2-deoxyribose 5-phosphate aldolase reveals a metabolic link between pentoses and central carbon metabolism in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 2004, 186, 4185–4191. [Google Scholar] [CrossRef] [PubMed]
- Videira, P.A.; Cortes, L.L.; Fialho, A.M.; Sá-Correia, I. Identification of the pgmG gene, encoding a bifunctional protein with phosphoglucomutase and phosphomannomutase activities, in the gellan gum-producing strain Sphingomonas paucimobilis ATCC 31461. Appl. Environ. Microbiol. 2000, 66, 2252–2258. [Google Scholar] [CrossRef]
- Iverson, T.M.; Panosian, T.D.; Birmingham, W.R.; Nannemann, D.P.; Bachmann, B.O. Molecular differences between a mutase and a phosphatase: Investigations of the activation step in Bacillus cereus phosphopentomutase. Biochemistry 2012, 51, 1964–1975. [Google Scholar] [CrossRef]
- Birmingham, W.R.; Starbird, C.A.; Panosian, T.D.; Nannemann, D.P.; Iverson, T.M.; Bachmann, B.O. Bioretrosynthetic construction of a didanosine biosynthetic pathway. Nat. Chem. Biol. 2014, 10, 392–399. [Google Scholar] [CrossRef]
- Paysan, L.T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, 418–427. [Google Scholar] [CrossRef]
- Waman, V.P.; Bordin, N.; Alcraft, R.; Vickerstaff, R.; Rauer, C.; Chan, Q.; Sillitoe, I.; Yamamori, H.; Orengo, C. CATH 2024: CATH-AlphaFlow doubles the number of structures in CATH and reveals nearly 200 new folds. J. Mol. Biol. 2024, 436, 168551–168560. [Google Scholar] [CrossRef]
- Levin, S.; Almo, S.C.; Satir, B.H. Functional diversity of the phosphoglucomutase superfamily: Structural implications. Protein Eng. 1999, 12, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Mio, T.; Yamada-Okabe, T.; Arisawa, M.; Yamada-Okabe, H. Functional cloning and mutational analysis of the human cDNA for phosphoacetylglucosamine mutase: Identification of the amino acid residues essential for the catalysis. Biochim. Biophys. Acta 2000, 1492, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Mazumdar, S. Electrospray ionization mass spectrometry: A technique to access the information beyond the molecular weight of the analyte. Int. J. Anal. Chem. 2012, 2012, 1–40. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, 439–444. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Krissinel, E. Enhanced fold recognition using efficient short fragment clustering. J. Mol. Biochem. 2012, 1, 76–85. [Google Scholar] [PubMed]
- Holm, L.; Kääriäinen, S.; Rosenström, P.; Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 2008, 24, 2780–2781. [Google Scholar] [CrossRef]
- Kawamura, T.; Tsuge, M.; Watanabe, N.; Tanaka, I. Crystal Structure of Pyrococcus horikoshii phosphomannomutase/phosphoglucomutase complexed with Mg2+. Unpublished work, 2005. [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Stierand, K.; Rarey, M. PoseView—molecular interaction patterns at a glance. J. Cheminform. 2010, 2, P50. [Google Scholar] [CrossRef]
- Rashid, N.; Kanai, T.; Atomi, H.; Imanaka, T. Among multiple phosphomannomutase gene orthologues, only one gene encodes a protein with phosphoglucomutase and phosphomannomutase activities in Thermococcus kodakaraensis. J. Bacteriol. 2004, 186, 6070–6076. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Panosian, T.; Nannemann, D.; Watkins, G.; Phelan, V.; McDonald, W.; Wadzinski, B.; Bachmann, B.; Iverson, T. Bacillus cereus phosphopentomutase is an alkaline phosphatase family member that exhibits an altered entry point into the catalytic cycle. J. Biol. Chem. 2011, 286, 8043–8054. [Google Scholar] [CrossRef] [PubMed]
- Tung, M.; Gallagher, D.T. The biomolecular cystallization database version 4: Expanded content and new features. Acta Crystallogr. D Biol. Crystallogr. 2008, D65, 18–23. [Google Scholar]
- Wu, D.; Piszczek, G. Standard protocol for mass photometry experiments. Eur. Biophys. J. 2021, 50, 403–409. [Google Scholar] [CrossRef]
- Wu, T.; Hornsby, M.; Zhu, L.; Yu, J.C.; Shokat, K.M.; Gestwicki, J.E. Protocol for performing and optimizing differential scanning fluorimetry experiments. STAR Protoc. 2023, 4, 102688. [Google Scholar] [CrossRef] [PubMed]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, D66, 125–132. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Agirre, J.; Atanasova, M.; Bagdonas, H.; Ballard, C.B.; Basle, A.; Beilsten-Edmands, J.; Borges, R.J.; Brown, D.G.; Burgos-Marmol, J.J.; Berrisford, J.M.; et al. The CCP4 suite: Integrative software for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2023, D79, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011, D67, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Huey, R.; Morris, G.M.; Forli, S. Using AutoDock 4 and AutoDock Vina with AutoDockTools: A tutorial. Scripps Res. Inst. Mol. Graph. Lab. 2012, 10550, 1000. [Google Scholar]
- Kochnev, Y.; Hellemann, E.; Cassidy, K.C.; Durrant, J.D. Webina: An open-source library and web app that runs AutoDock Vina entirely in the web browser. Bioinformatics 2020, 36, 4513–4515. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 33. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]

| Data Collection | |
|---|---|
| Source | Photon Factory beamline BL-1A |
| Detector | Eiger X 4M |
| Wavelength (Å) | 1.90 |
| Data collection temperature (K) | 100 |
| Resolution range (Å) | 50.0–2.39 (2.53–2.39) * |
| Space group | P21 |
| a, b, c (Å); β (°) | 79.77, 97.58, 128.15; 94.0 |
| Total reflections | 539,005 (86105) |
| Unique reflections | 77,057 (12,137) |
| Multiplicity | 7.0 (7.9) |
| Completeness (%) | 98.9 (97.0) |
| Mean I/σ(I) | 10.5 (1.6) |
| Rmerge | 0.135 (1.12) |
| CC1/2 | 0.998 (0.70) |
| Refinement | |
| Reflections used in refinement | 74,699 (5367) |
| Reflections used for Rfree | 2310 (166) |
| Rwork | 0.201 (0.373) |
| Rfree | 0.274 (0.391) |
| Matthews coefficient (Å) | 2.46 |
| Solvent content (%) | 49.97 |
| No. of atoms | |
| Protein | 13,829 |
| Ligands | 48 |
| Water | 420 |
| No. of protein residues | 1780 |
| R.M.S. deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.72 |
| Ramachandran plot | |
| Favored (%) | 93.4 |
| Outliers (%) | 1.2 |
| Rotamer outliers (%) | 6.1 |
| Clashscore | 6.9 (98th percentile) |
| B factors (Å2) | |
| Average | 50.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naz, Z.; Lubkowski, J.; Saleem, M.; Aslam, M.; Rahman, M.; Wlodawer, A.; Rashid, N. Biophysical Characterization of a Novel Phosphopentomutase from the Hyperthermophilic Archaeon Thermococcus kodakarensis. Int. J. Mol. Sci. 2024, 25, 12893. https://doi.org/10.3390/ijms252312893
Naz Z, Lubkowski J, Saleem M, Aslam M, Rahman M, Wlodawer A, Rashid N. Biophysical Characterization of a Novel Phosphopentomutase from the Hyperthermophilic Archaeon Thermococcus kodakarensis. International Journal of Molecular Sciences. 2024; 25(23):12893. https://doi.org/10.3390/ijms252312893
Chicago/Turabian StyleNaz, Zahra, Jacek Lubkowski, Muhammad Saleem, Mehwish Aslam, Moazur Rahman, Alexander Wlodawer, and Naeem Rashid. 2024. "Biophysical Characterization of a Novel Phosphopentomutase from the Hyperthermophilic Archaeon Thermococcus kodakarensis" International Journal of Molecular Sciences 25, no. 23: 12893. https://doi.org/10.3390/ijms252312893
APA StyleNaz, Z., Lubkowski, J., Saleem, M., Aslam, M., Rahman, M., Wlodawer, A., & Rashid, N. (2024). Biophysical Characterization of a Novel Phosphopentomutase from the Hyperthermophilic Archaeon Thermococcus kodakarensis. International Journal of Molecular Sciences, 25(23), 12893. https://doi.org/10.3390/ijms252312893








