MicroRNA Nobel Prize: Timely Recognition and High Anticipation of Future Products—A Prospective Analysis
Abstract
1. Introduction
2. MiRNA Nobel Prize
3. Nomenclature
4. Biogenesis
- Transcription of miRNA Genes: Most miRNAs are transcribed by RNA polymerase II (Pol II) as primary miRNAs (pri-miRNAs), several kilobases long and capped, polyadenylated, and structured with stem–loop formations. Some miRNAs, however, are transcribed by RNA polymerase III. These pri-miRNAs can originate from independent miRNA genes, protein-coding genes’ introns, or polycistronic clusters containing multiple miRNA sequences [19];
- Nuclear Processing of pri-miRNA: Once transcribed, pri-miRNAs undergo processing within the nucleus. A microprocessor complex, composed of the RNase III enzyme Drosha and its cofactor DiGeorge syndrome critical region gene 8 (DGCR8), cleaves the pri-miRNA at the stem–loop region, releasing a shorter precursor miRNA (pre-miRNA) of approximately 70 nucleotides. This cleavage step is crucial for defining the miRNA’s 5′ and 3′ ends [20];
- Nuclear Export of pre-miRNA: The pre-miRNA is then exported from the nucleus to the cytoplasm. Exportin-5, a Ran-GTP-dependent nuclear transport receptor, recognizes the double-stranded stem structure of the pre-miRNA and facilitates its export across the nuclear membrane. The high-affinity interaction between Exportin-5 and pre-miRNA ensures that only properly processed pre-miRNAs are transported out of the nucleus [21];
- Cytoplasmic Processing of pre-miRNA: Once in the cytoplasm, the pre-miRNA is further processed by the RNase III enzyme Dicer, which cleaves the loop structure of the pre-miRNA, yielding a miRNA duplex of about 22 nucleotides. Dicer partners with the trans-activator RNA-binding protein (TRBP) and Argonaute (AGO) proteins to form the RNA-induced silencing complex (RISC). One strand of the miRNA duplex, the guide strand, is predominantly loaded onto AGO proteins in RISC, while the complementary passenger strand is degraded [22];
- Functional Maturation and Targeting: The mature miRNA-RISC complex functions in gene silencing. The miRNA guides RISC to target mRNAs, where it typically binds to the 3′ UTRs of target transcripts through imperfect base-pairing. If the complementarity is high, this binding leads to either translational repression or mRNA cleavage. The extent of complementarity between the miRNA and its target determines the mode of the gene silencing [6].
4.1. Biogenesis in Plants
4.2. Evolution
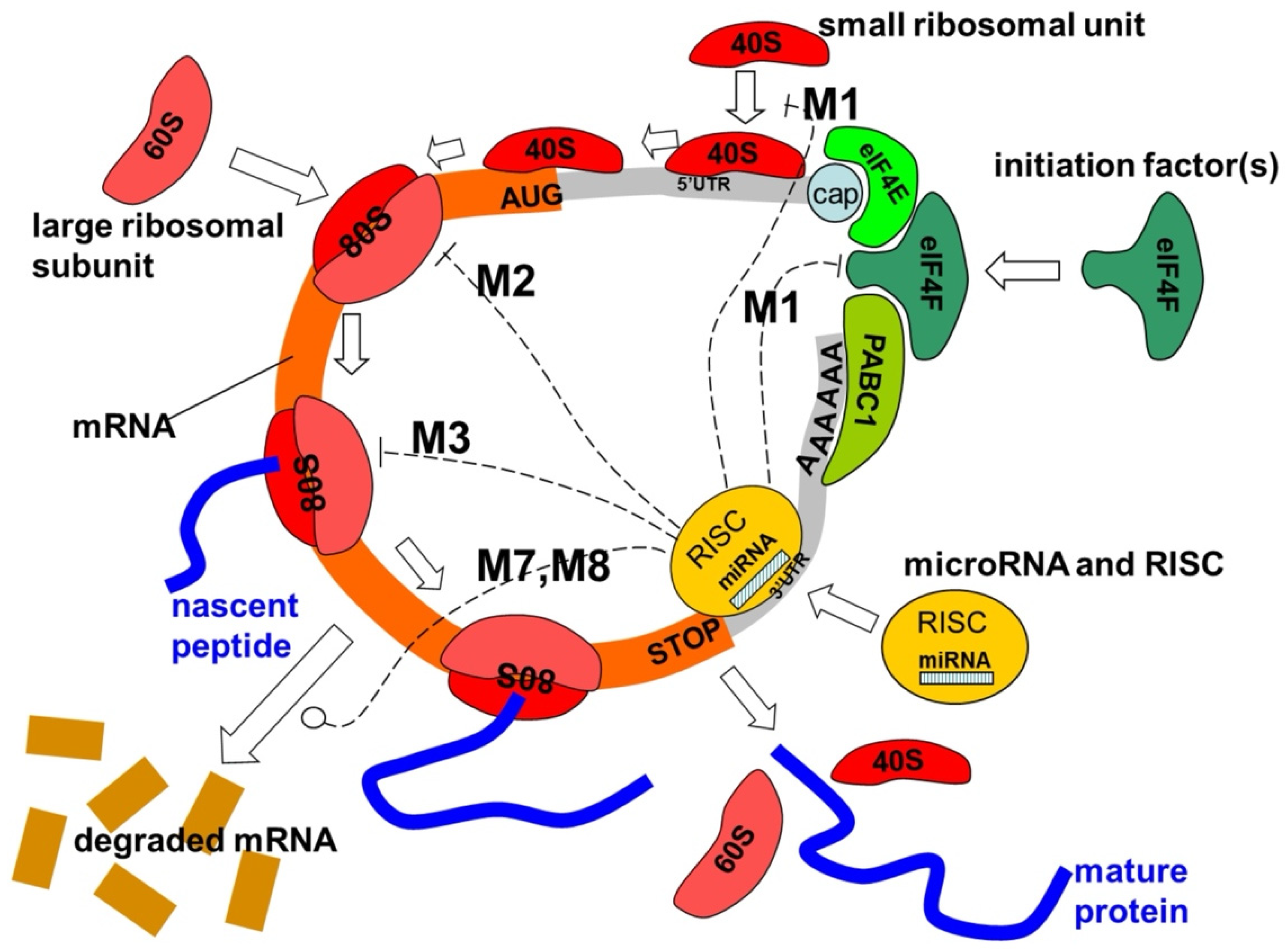
5. Mechanisms of miRNA Action
- Cap-40S initiation inhibition;
- 60S ribosomal unit joining inhibition;
- Elongation inhibition;
- Ribosome drop-off (premature termination);
- Co-translational nascent protein degradation;
- Sequestration in P-bodies [44];
- MRNA decay (destabilization) by shortening its poly(A) tail;
- MRNA cleavage, where the mRNA strand is cleaved into two pieces;
- Transcriptional inhibition through microRNA-mediated chromatin reorganization, followed by gene silencing;
5.1. RNA-Induced Silencing Complex (RISC)
5.2. Mode of Silencing and Regulatory Loops
5.3. Turnover
5.4. Cellular Functions
5.5. Exosomes
6. Experimental Methods
7. Applications
7.1. Biomarkers
7.2. Cancer
7.3. Cardiovascular Diseases
7.4. Neurodegenerative Diseases
7.5. Autoimmune Diseases
7.6. Viral Infections
7.7. Alcoholism
7.8. Aging
- miR-146a: This miRNA can inhibit IL-6 and IL-1β, two key inflammatory cytokines upregulated in aged, senescent cells. Targeting these cytokines with miR-146a mimics could reduce inflammation and slow tissue degeneration;
- miR-29: This miRNA can inhibit collagen-degrading enzymes, such as matrix metalloproteinases (MMPs), that are involved in the breakdown of the extracellular matrix in aging tissues. MiR-29 mimics could maintain tissue structure and reduce fibrosis, which becomes more common with age [157];
- miR-375: This miRNA inhibits IGF-1R, a receptor in the IGF pathway that promotes growth and can contribute to age-related diseases, like cancer. Overexpression of miR-375 could reduce IGF-1R activity and help to mitigate these risks [158];
- miR-34a: This miRNA has been shown to suppress p53 and Bcl-2, proteins involved in apoptosis and stress responses, respectively. Modulating the levels of miR-34a may help to balance apoptosis in aging cells, potentially reducing unwanted cell death in key tissues, like the brain or heart [159];
- miR-103/107: These miRNAs target caveolin-1, a regulator of insulin signaling. Inhibiting these miRNAs could enhance insulin sensitivity, which typically decreases with age, helping to prevent or manage metabolic diseases [153].
7.9. Obesity
7.10. Hemostasis
7.11. Others
7.12. Plants
7.13. MiRNAs in Precision Medicine
7.14. MiRNAs in Regenerative Medicine
7.15. Synthetic Biology and miRNA-Based Therapeutics
7.16. MiRNAs in Gene Editing
7.17. Tissue-Specific miRNA
- MiR-1, miR-133, and miR-206 are known as “myo-miRs” and play crucial roles in regulating muscle development (myogenesis), differentiation, and repair [192]. They suppress genes involved in muscle atrophy and promote the growth of new muscle fibers after injury or exercise;
- MiR-1 and miR-133 are also critical in the heart for controlling cardiac muscle differentiation;
- MiR-208a and miR-499 are mainly involved in cardiac contractility and protecting the heart against hypertrophy;
- MiR-122 is liver-specific and is crucial in cholesterol and fatty acid metabolism. It is also essential for liver development and function. MiR-21 and miR-199a are involved in liver fibrosis and hepatocyte proliferation [193];
- MiR-124 is one of the most abundant miRNAs in the brain and is involved in neuronal differentiation and neuroprotection;
- MiR-9 helps to regulate neurogenesis, while miR-132 and miR-128 are essential for synaptic plasticity and cognitive function;
- MiR-143 and miR-103 regulate adipocyte differentiation and insulin sensitivity, making them necessary in fat metabolism and energy homeostasis. MiR-155 is involved in inflammatory regulation within adipose tissue;
- MiR-375 is crucial for insulin secretion and pancreatic beta cell function. It regulates glucose homeostasis and pancreatic cell differentiation, contributing to overall metabolic health;
- MiR-192 and miR-194 are essential for kidney development and maintaining nephron integrity;
- MiR-29 and miR-21 are involved in kidney fibrosis and injury response, playing critical roles in kidney diseases;
- MiR-21 and miR-126 are involved in lung development, inflammation, and repair processes, particularly during fibrosis;
- MiR-29b regulates collagen deposition and is essential in lung fibrotic disorders. MiR-26a, miR-29b, and miR-214 play critical roles in bone development by regulating osteoblast differentiation and bone formation. These miRNAs are targets for promoting bone repair and regeneration;
- MiR-203 and miR-205 regulate keratinocyte differentiation, maintain skin integrity, and promote wound healing. MiR-31 supports epidermal proliferation and repair after injury;
- MiR-150, miR-155, and miR-223 are critical regulators in hematopoietic cells, controlling the differentiation of immune cells and playing roles in inflammation and immune responses;
- MiR-210 and miR-141 regulate placental development and adaptation to hypoxia;
- MiR-517 is involved in trophoblast function and placental growth;
- MiR-34c and miR-449a are involved in spermatogenesis and regulating Sertoli and Leydig cells;
- MiR-202 plays a role in male fertility and testicular development.
7.18. Targeted Delivery Using Tissue-Specific Promoters
7.18.1. Ligand-Conjugated Nanoparticles for Receptor-Mediated Targeting
7.18.2. Cell-Penetrating Peptides (CPPs)
7.18.3. Aptamers for Cell-Specific Targeting
7.18.4. Virus-Like Particles (VLPs) for Tissue-Specific Targeting
7.18.5. Lentiviruses and Retroviruses
7.18.6. CRISPR/Cas9 and Aptamers for Enhanced Targeting
7.18.7. CRISPR-Based miRNA Activation Systems
8. Delivery
8.1. Ex Vivo Manipulation
8.2. In Vivo Administration
8.3. Course of Action
- Delivery System: If delivered via nanoparticles, liposomes, or viral vectors, the miRNA may be protected from immediate degradation, allowing for it to stay active longer. However, nucleases in the bloodstream or tissues rapidly degrade miRNAs delivered without protection;
- Tissue Type: Different tissues have varying miRNA turnover rates. For example, miRNAs in the liver quickly degrade because of high metabolic activity, while miRNAs in the brain or muscle may last longer because of different enzymatic environments;
- Endosomal Escape: For the miRNA to be active, it must escape the endosome after cellular uptake. If the delivery system is inefficient at facilitating endosomal escape, the miRNA may be degraded within the cell before it can act on its target mRNA;
- Half-life of MiRNA: Endogenous miRNAs typically have a half-life of 12–24 h, varying depending on the cellular environment. Synthetic miRNA mimics may be modified to enhance stability, but they still typically exhibit transient effects unless continuously delivered.
- Stable Expression Vectors: In ex vivo manipulation, miRNAs can be introduced using viral vectors (e.g., lentiviruses or AAVs) that integrate into the host cell’s genome, enabling continuous miRNA expression after the cells are transplanted back into the patient. In this case, the host cells would produce the miRNA indefinitely, offering a permanent or long-term solution;
- Lentiviral vectors, which integrate into the genome, can install miRNAs permanently in dividing cells, such as those in the liver or blood. However, this approach risks insertional mutagenesis (disruption of essential genes), making it less ideal for permanent in vivo miRNA therapy unless particular targeting strategies are employed;
- CRISPR-Based MiRNA Activation: Another ex vivo approach involves using CRISPR-based gene editing to permanently activate or repress specific miRNA genes within a cell’s genome. This would ensure the miRNA remains permanently active in the cells reintroduced to the patient. CRISPR/Cas9-based gene editing offers a theoretical path toward permanent miRNA installation in vivo. Using CRISPR/Cas9 to activate or repress the expressions of endogenous miRNAs directly, it may be possible to modulate miRNA expression within specific tissues permanently. For instance, CRISPRa (CRISPR activation) can upregulate the expressions of miRNAs that promote beneficial effects, such as those involved in muscle regeneration or cardioprotection. This approach would include delivering the CRISPR machinery and the guide RNA to specific tissues;
- Similarly, CRISPRi (CRISPR interference) can permanently repress miRNAs that cause harmful effects, such as miRNAs that promote fibrosis or inflammation. CRISPR can be delivered in vivo using lipid nanoparticles, plasmids, or other delivery vehicles that allow for the Cas9 protein and guide RNA (gRNA) to target specific cells, creating a permanent change in the genome. This approach provides a potential long-term solution for genetic diseases or tissue-specific modifications.
8.4. Synthetic mRNA Therapy with Self-Amplifying Systems
8.5. Non-Viral Gene Therapy Systems for Long-Term Expression
- Immune Response: One major challenge with permanent in vivo miRNA expression is the risk of triggering an immune response. Viral and non-viral delivery systems may activate the immune system, leading to vector clearance or potential damage to the host tissue. This is particularly relevant for viral vectors that persist in the body for long periods;
- Insertional Mutagenesis: If miRNA constructs are delivered using integrating viral vectors (such as lentiviruses), there is a risk of insertional mutagenesis, where the viral genome integrates into a critical region of the host’s genome, potentially disrupting essential genes and causing adverse effects, including cancer;
- Unintended Off-Target Effects: Permanently installing miRNA in a tissue could lead to off-target gene regulation, where the miRNA affects genes beyond the intended target. Although miRNAs are generally specific, they can target multiple genes, which raises the possibility of unintended consequences over the long term.
8.6. Nanoparticle-Based Delivery (Repeated Administration)
8.7. Cell-Based Therapies
9. Discovery
- Isolate the total RNA from the tissue or cells of interest;
- Use specialized protocols to enrich small RNAs, including miRNAs (typically using size-selection methods);
- Prepare a small RNA library from the isolated RNA by ligating specific adapters to the 5′ and 3′ ends of small RNAs;
- Perform high-throughput sequencing on the small-RNA library. This generates millions of short reads corresponding to small RNAs present in the sample;
- Align the sequenced reads to the reference genome to identify known and potentially new miRNAs. Novel miRNAs can be identified by detecting sequences that match the criteria for miRNA precursor structures (such as the formation of a hairpin secondary structure);
- Once candidate miRNAs are identified through sequencing, northern blotting can be used to validate their expressions. This method detects small RNA molecules based on size and allows researchers to confirm the presence and abundance of a novel miRNA in a specific tissue or developmental stage;
- QPCR can be used to confirm the expression levels of newly discovered miRNAs.
- The presence of hairpin structures in precursor miRNAs (pre-miRNAs);
- The conservation of sequences across species;
- The minimum free energy (MFE) of the predicted secondary structure to determine if it will likely form a stable hairpin.
- miRDeep: A widely used algorithm that identifies novel miRNAs by aligning small-RNA reads to the genome and predicting precursor structures;
- miRBase: A comprehensive miRNA database that includes information on known miRNAs and can be used to cross-check potential novel miRNAs;
- RNAfold: A tool used to predict the secondary structure of RNA sequences, helping to determine if a candidate sequence forms a stable hairpin structure typical of miRNA precursors.
9.1. Comparative Genomics
- Align sequences from closely related species to find conserved regions of small RNAs;
- Analyze the conservation of the predicted secondary structures of the miRNA precursors.
9.2. Machine-Learning Models
10. Regulatory Process
11. Challenges and Limitations
11.1. Off-Target Effects and Immune Responses
11.2. MiRNA Stability and Degradation
12. Intellectual Property
Examples of miRNA Patents
13. Commercial Products
14. Conclusions
- miRNA Rejuvenation of Therapeutic Protein Expression: In therapeutic protein production, controlling the consistent and long-term expressions of proteins, especially in cases like gene therapies, is essential for achieving a sustained therapeutic effect. miRNAs could be used to rejuvenate or regulate the expressions of proteins significantly when their production naturally declines because of aging or disease [252];
- Enhancing Protein Expression in Aging Tissues: Aging often leads to a decline in the production of key proteins, which can contribute to diseases, such as sarcopenia, neurodegeneration, and metabolic disorders. MiRNA therapy could be designed to rejuvenate these therapeutic proteins’ expressions by suppressing protein production inhibitors or activating the necessary transcription factors. For example, the downregulation of miR-34a, which is associated with aging, could be employed to increase the expression of sirtuins (proteins involved in longevity and metabolism) or factors that promote muscle regeneration, like IGF-1 (insulin-like growth factor) [253];
- Controlling Protein Production in Gene Therapy: Ensuring that the therapeutic gene is expressed at the right time and in the right amount is crucial. MiRNA-based systems could precisely control protein expression by designing synthetic miRNA circuits that respond to environmental or cellular signals. For instance, miRNA switches could be used to control the expression of therapeutic proteins only when needed, reducing side effects and improving the overall safety profile of gene therapies. This could be particularly beneficial in conditions requiring intermittent treatment, such as intermittent hormone deficiencies or enzyme replacement therapies [254];
- miRNAs in Enhancing Protein Stability and Folding: miRNAs can be used to regulate chaperone proteins involved in folding therapeutic proteins. In cystic fibrosis, the correct folding of proteins is critical, and mutations can lead to misfolding. MiRNA-based therapies could target pathways that enhance the production of molecular chaperones, improving the folding and function of therapeutic proteins [255];
- MiRNA-Based Vaccines: Vaccines are traditionally designed to elicit immune responses against specific pathogens by presenting the body with an antigen (such as a weakened virus or protein subunits) [256]. However, miRNA technology could open new frontiers in vaccine development, offering several advantages, including enhanced specificity, long-lasting immune responses, and the potential to create personalized vaccines. One of the most exciting applications of miRNA technology in vaccines involves creating vaccines that induce host cells to produce antigens from within. By designing a vaccine that introduces miRNAs targeting viral RNA or mRNA encoding viral proteins, host cells can produce specific viral antigens, stimulating the immune system more naturally. This approach could benefit rapidly mutating viruses, such as influenza or SARS-CoV-2. A synthetic miRNA-based vaccine could target conserved regions of the viral genome, minimizing the effects of viral mutations. This would provide longer-lasting immunity and eliminate the need for frequent updates to vaccine formulations. Cancer vaccines stimulate the immune system to recognize and attack tumor-specific antigens. MiRNA technology could enable the creation of personalized cancer vaccines by targeting tumor-specific mutations.
- Tumors often downregulate or mutate proteins that suppress tumor growth, such as p53, or proteins that would usually alert the immune system to cancerous cells. MiRNA-based vaccines could enhance the expression of these proteins in tumor cells, increasing the likelihood that the immune system will detect and destroy them [257]. Another futuristic application is using miRNAs to boost the presentation of antigens in traditional vaccines. Antigen-presenting cells (APCs) play a crucial role in the immune response by displaying antigens to T-cells. MiRNAs could enhance these cells’ function by upregulating key pathways involved in antigen processing and presentation. For instance, miR-155 is involved in immune cell activation, and its upregulation could enhance the effectiveness of vaccine-induced immune responses. By co-delivering miRNAs that boost antigen presentation with traditional vaccines, it may be possible to create vaccines that provide more robust, longer-lasting immunity [258];
- miRNA in Plant-Based Vaccines: MiRNAs could also play an essential role in producing plant-based vaccines, which use plants to produce antigens that can be administered orally or through traditional injection. Researchers could enhance the yield and stability of antigen production in plants by manipulating miRNAs in plant systems. For example, miRNAs that regulate protein synthesis or stress responses in plants could be targeted to increase the efficiency of vaccine antigen production in edible crops, such as lettuce or tomatoes. This approach could pave the way for inexpensive and easily scalable vaccines suitable for global use, particularly in resource-limited settings [259].
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Salloway, S.; Chalkias, S.; Barkhof, F.; Burkett, P.; Barakos, J.; Purcell, D.; Suhy, J.; Forrestal, F.; Tian, Y.; Umans, K.; et al. Amyloid-Related Imaging Abnormalities in 2 Phase 3 Studies Evaluating Aducanumab in Patients with Early Alzheimer Disease. JAMA Neurol. 2022, 79, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.; Thakur, N.; Monga, I.; Thakur, A.; Kumar, M. VIRmiRNA: A comprehensive resource for experimentally validated viral miRNAs and their targets. Database (Oxford) 2014, 2014, bau103. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Burki, T. 2024 Nobel Prize awarded for work on microRNAs. Lancet 2024, 404, 1507–1508. [Google Scholar] [CrossRef]
- Ruvkun, G. Molecular biology. Glimpses of a tiny RNA world. Science 2001, 294, 797–799. [Google Scholar] [CrossRef] [PubMed]
- van Rooij, E.; Olson, E.N. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Investig. 2007, 117, 2369–2376. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Meyer, J.; Borkhardt, A.; Tuschl, T. New microRNAs from mouse and human. RNA 2003, 9, 175–179. [Google Scholar] [CrossRef]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.W.; Bruford, E.A. Naming ‘junk’: Human non-protein coding RNA (ncRNA) gene nomenclature. Hum. Genom. 2011, 5, 90. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Kim, V.N.; Nam, J.W. Genomics of microRNA. Trends Genet. 2006, 22, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- MacRae, I.J.; Zhou, K.; Doudna, J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007, 14, 934–940. [Google Scholar] [CrossRef]
- Miller, T.M.; Pestronk, A.; David, W.; Rothstein, J.; Simpson, E.; Appel, S.H.; Andres, P.L.; Mahoney, K.; Allred, P.; Alexander, K.; et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: A phase 1, randomised, first-in-man study. Lancet Neurol. 2013, 12, 435–442. [Google Scholar] [CrossRef]
- Lelandais-Brière, C.; Sorin, C.; Declerck, M.; Benslimane, A.; Crespi, M.; Hartmann, C. Small RNA diversity in plants and its impact in development. Curr. Genom. 2010, 11, 14–23. [Google Scholar] [CrossRef]
- Axtell, M.J.; Westholm, J.O.; Lai, E.C. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011, 12, 221. [Google Scholar] [CrossRef]
- Wheeler, B.M.; Heimberg, A.M.; Moy, V.N.; Sperling, E.A.; Holstein, T.W.; Heber, S.; Peterson, K.J. The deep evolution of metazoan microRNAs. Evol. Dev. 2009, 11, 50–68. [Google Scholar] [CrossRef]
- Pashkovskiy, P.P.; Ryazansky, S.S. Biogenesis, evolution, and functions of plant microRNAs. Biochemistry 2013, 78, 627–637. [Google Scholar] [CrossRef]
- Heimberg, A.M.; Sempere, L.F.; Moy, V.N.; Donoghue, P.C.J.; Peterson, K.J. MicroRNAs and the advent of vertebrate morphological complexity. Proc. Natl. Acad. Sci. USA 2008, 105, 2946–2950. [Google Scholar] [CrossRef]
- Nozawa, M.; Miura, S.; Nei, M. Origins and Evolution of MicroRNA Genes in Drosophila Species. Genome Biol. Evol. 2010, 2, 180–189. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Sung, G.-H.; Spatafora, J.W.; Carrington, J.C. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 2004, 36, 1282–1290. [Google Scholar] [CrossRef]
- Warthmann, N.; Das, S.; Lanz, C.; Weigel, D. Comparative Analysis of the MIR319a MicroRNA Locus in Arabidopsis and Related Brassicaceae. Mol. Biol. Evol. 2008, 25, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Peterson, K.J.; Dietrich, M.R.; McPeek, M.A. MicroRNAs and metazoan macroevolution: Insights into canalization, complexity, and the Cambrian explosion. Bioessays 2009, 31, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Jogdeo, S.; Kasschau, K.D.; Sullivan, C.M.; Chapman, E.J.; Laubinger, S.; Smith, L.M.; Dasenko, M.; Givan, S.A.; Weigel, D.; et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 2010, 22, 1074–1089. [Google Scholar] [CrossRef] [PubMed]
- Caravas, J.; Friedrich, M. Of mites and millipedes: Recent progress in resolving the base of the arthropod tree. Bioessays 2010, 32, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Kenny, N.J.; Namigai, E.K.O.; Marlétaz, F.; Hui, J.H.L.; Shimeld, S.M. Draft genome assemblies and predicted microRNA complements of the intertidal lophotrochozoans Patella vulgata (Mollusca, Patellogastropoda) and Spirobranchus (Pomatoceros) lamarcki (Annelida, Serpulida). Mar. Genom. 2015, 24, 139–146. [Google Scholar] [CrossRef]
- Cock, J.M.; Sterck, L.; Rouzé, P.; Scornet, D.; Allen, A.E.; Amoutzias, G.; Anthouard, V.; Artiguenave, F.; Aury, J.-M.; Badger, J.H.; et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 2010, 465, 617–621. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and Functional Diversification of MIRNA Genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007, 17, 118–126. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Morozova, N.; Zinovyev, A.; Nonne, N.; Pritchard, L.L.; Gorban, A.N.; Harel-Bellan, A. Kinetic signatures of microRNA modes of action. RNA 2012, 18, 1635–1655. [Google Scholar] [CrossRef]
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, B.; Wu, T.; Skogerbø, G.; Zhu, X.; Guo, X.; He, S.; Chen, R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol. Biol. 2009, 10, 12. [Google Scholar] [CrossRef]
- Hawkins, P.G.; Morris, K.V. RNA and transcriptional modulation of gene expression. Cell Cycle 2008, 7, 602–607. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef]
- Shaw, G.; Kamen, R. A conserved AU sequence from the 3′; untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 1986, 46, 659–667. [Google Scholar] [CrossRef]
- Jing, Q.; Huang, S.; Guth, S.; Zarubin, T.; Motoyama, A.; Chen, J.; Di Padova, F.; Lin, S.-C.; Gram, H.; Han, J. Involvement of MicroRNA in AU-Rich Element-Mediated mRNA Instability. Cell 2005, 120, 623–634. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Kai, Z.S.; Pasquinelli, A.E. MicroRNA assassins: Factors that regulate the disappearance of miRNAs. Nat. Struct. Mol. Biol. 2010, 17, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Großhans, H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 2009, 461, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-J.; Reyes, J.L.; Chua, N.-H.; Gaasterland, T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004, 5, R65. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Taira, K. MicroRNA-196 inhibits HOXB8 expression in myeloid differentiation of HL60 cells. Nucleic Acids Symp. Ser. 2004, 48, 211–212. [Google Scholar] [CrossRef]
- Moxon, S.; Jing, R.; Szittya, G.; Schwach, F.; Rusholme Pilcher, R.L.; Moulton, V.; Dalmay, T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008, 18, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Mazière, P.; Enright, A.J. Prediction of microRNA targets. Drug Discov. Today 2007, 12, 452–458. [Google Scholar] [CrossRef]
- Williams, A.E. Functional aspects of animal microRNAs. Cell. Mol. Life Sci. 2008, 65, 545–562. [Google Scholar] [CrossRef]
- Eulalio, A.; Huntzinger, E.; Nishihara, T.; Rehwinkel, J.; Fauser, M.; Izaurralde, E. Deadenylation is a widespread effect of miRNA regulation. RNA 2009, 15, 21–32. [Google Scholar] [CrossRef]
- Stark, A.; Brennecke, J.; Bushati, N.; Russell, R.B.; Cohen, S.M. Animal MicroRNAs Confer Robustness to Gene Expression and Have a Significant Impact on 3′UTR Evolution. Cell 2005, 123, 1133–1146. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi Pier, P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Lehr, D.; Ohm, P. Playing with the Data: What Legal Scholars Should Learn About Machine Learning. U.C. Davis Law Rev. 2017, 51, 653. [Google Scholar]
- Jansen, F.; Yang, X.; Proebsting, S.; Hoelscher, M.; Przybilla, D.; Baumann, K.; Schmitz, T.; Dolf, A.; Endl, E.; Franklin, B.S.; et al. MicroRNA Expression in Circulating Microvesicles Predicts Cardiovascular Events in Patients with Coronary Artery Disease. J. Am. Heart Assoc. 2014, 3, e001249. [Google Scholar] [CrossRef]
- Briolay, T.; Petithomme, T.; Fouet, M.; Nguyen-Pham, N.; Blanquart, C.; Boisgerault, N. Delivery of cancer therapies by synthetic and bio-inspired nanovectors. Mol. Cancer 2021, 20, 55. [Google Scholar] [CrossRef]
- Cuman, C.; Van Sinderen, M.; Gantier, M.P.; Rainczuk, K.; Sorby, K.; Rombauts, L.; Osianlis, T.; Dimitriadis, E. Human Blastocyst Secreted microRNA Regulate Endometrial Epithelial Cell Adhesion. eBioMedicine 2015, 2, 1528–1535. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, Q.; You, W.; Chen, M.; Xia, J. MiRNAs as Biomarkers of Myocardial Infarction: A Meta-Analysis. PLoS ONE 2014, 9, e88566. [Google Scholar] [CrossRef]
- Miranda, K.C.; Bond, D.T.; McKee, M.; Skog, J.; Păunescu, T.G.; Da Silva, N.; Brown, D.; Russo, L.M. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010, 78, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhu, D.; Huang, L.; Zhang, J.; Bian, Z.; Chen, X.; Liu, Y.; Zhang, C.Y.; Zen, K. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS ONE 2012, 7, e46957. [Google Scholar] [CrossRef]
- Lv, L.-L.; Cao, Y.-H.; Ni, H.-F.; Xu, M.; Liu, D.; Liu, H.; Chen, P.-S.; Liu, B.-C. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. Am. J. Physiol.-Ren. Physiol. 2013, 305, F1220–F1227. [Google Scholar] [CrossRef]
- Iavello, A.; Frech, V.S.L.; Gai, C.; Deregibus, M.C.; Quesenberry, P.J.; Camussi, G. Role of Alix in miRNA packaging during extracellular vesicle biogenesis. Int. J. Mol. Med. 2016, 37, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.D.; Lin, F.M.; Wu, W.Y.; Liang, C.; Huang, W.C.; Chan, W.L.; Tsai, W.T.; Chen, G.Z.; Lee, C.J.; Chiu, C.M.; et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011, 39, D163–D169. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chou, C.H.; Chang, N.W.; Shrestha, S.; Hsu, S.D.; Lin, Y.L.; Lee, W.H.; Yang, C.D.; Hong, H.C.; Wei, T.Y.; Tu, S.J.; et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016, 44, D239–D247. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Fromm, B.; Domanska, D.; Høye, E.; Ovchinnikov, V.; Kang, W.; Aparicio-Puerta, E.; Johansen, M.; Flatmark, K.; Mathelier, A.; Hovig, E.; et al. MirGeneDB 2.0: The metazoan microRNA complement. Nucleic Acids Res. 2019, 48, D132–D141. [Google Scholar] [CrossRef]
- Wang, H. A Review of Nanotechnology in microRNA Detection and Drug Delivery. Cells 2024, 13, 1277. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Lizano, E.; Houben, A.J.S.; Bezdan, D.; Báñez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; González, J.; Chen, K.C.; et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 2014, 15, R57. [Google Scholar] [CrossRef] [PubMed]
- Boeckel, J.-N.; Reis, S.M.; Leistner, D.; Thomé, C.E.; Zeiher, A.M.; Fichtlscherer, S.; Keller, T. From heart to toe: Heart’s contribution on peripheral microRNA levels. Int. J. Cardiol. 2014, 172, 616–617. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-G.; Calin, G.A.; Volinia, S.; Croce, C.M. MicroRNA expression profiling using microarrays. Nat. Protoc. 2008, 3, 563–578. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Shingara, J.; Keiger, K.; Shelton, J.; Laosinchai-Wolf, W.; Powers, P.; Conrad, R.; Brown, D.; Labourier, E. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA 2005, 11, 1461–1470. [Google Scholar] [CrossRef]
- Buermans, H.P.J.; Ariyurek, Y.; van Ommen, G.; den Dunnen, J.T.; t’Hoen, P.A.C. New methods for next generation sequencing based microRNA expression profiling. BMC Genom. 2010, 11, 716. [Google Scholar] [CrossRef] [PubMed]
- Flynt, A.S.; Li, N.; Thatcher, E.J.; Solnica-Krezel, L.; Patton, J.G. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 2007, 39, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Dorsett, Y.; Tuschl, T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 2004, 10, 544–550. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; Rebhan, M.A.E.; Crivelli, S.E.M.; Denzler, R.; Stoffel, M.; Hall, J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2013, 42, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.-Y.; Giraldez, A.J.; Schier, A.F. Target Protectors Reveal Dampening and Balancing of Nodal Agonist and Antagonist by miR-430. Science 2007, 318, 271–274. [Google Scholar] [CrossRef]
- Lagendijk, A.K.; Moulton, J.D.; Bakkers, J. Revealing details: Whole mount microRNA in situ hybridization protocol for zebrafish embryos and adult tissues. Biol. Open 2012, 1, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Arora, A.; Wengel, J.; Maiti, S. Thermodynamic, Counterion, and Hydration Effects for the Incorporation of Locked Nucleic Acid Nucleotides into DNA Duplexes. Biochemistry 2006, 45, 7347–7355. [Google Scholar] [CrossRef]
- Nielsen, J.A.; Lau, P.; Maric, D.; Barker, J.L.; Hudson, L.D. Integrating microRNA and mRNA expression profiles of neuronal progenitors to identify regulatory networks underlying the onset of cortical neurogenesis. BMC Neurosci. 2009, 10, 98. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2007, 36, D154–D158. [Google Scholar] [CrossRef]
- Artmann, S.; Jung, K.; Bleckmann, A.; Beissbarth, T. Detection of simultaneous group effects in microRNA expression and related target gene sets. PLoS ONE 2012, 7, e38365. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2008, 37, D98–D104. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A. Trials and Tribulations of MicroRNA Therapeutics. Int. J. Mol. Sci. 2024, 25, 1469. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Reddy, P.H. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2016, 1862, 1617–1627. [Google Scholar] [CrossRef] [PubMed]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef]
- Yu, B.; Jiang, Y.; Wang, X.; Wang, S. An integrated hypothesis for miR-126 in vascular disease. Med. Res. Arch. 2020, 8, 2133. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- D’Alessandra, Y.; Devanna, P.; Limana, F.; Straino, S.; Di Carlo, A.; Brambilla, P.G.; Rubino, M.; Carena, M.C.; Spazzafumo, L.; De Simone, M.; et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur. Heart J. 2010, 31, 2765–2773. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407. [Google Scholar] [CrossRef]
- Rosenfeld, N.; Aharonov, R.; Meiri, E.; Rosenwald, S.; Spector, Y.; Zepeniuk, M.; Benjamin, H.; Shabes, N.; Tabak, S.; Levy, A.; et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008, 26, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Di, K.; Fan, B.; Wu, J.; Gu, X.; Sun, Y.; Khan, A.; Li, P.; Li, Z. MicroRNAs in extracellular vesicles: Sorting mechanisms, diagnostic value, isolation, and detection technology. Front. Bioeng. Biotechnol. 2022, 10, 948959. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, M.M.J.; Krauskopf, J.; Ramaekers, J.G.; Kleinjans, J.C.S.; Prickaerts, J.; Briedé, J.J. Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog. Neurobiol. 2020, 185, 101732. [Google Scholar] [CrossRef] [PubMed]
- Felekkis, K.; Papaneophytou, C. The Circulating Biomarkers League: Combining miRNAs with Cell-Free DNAs and Proteins. Int. J. Mol. Sci. 2024, 25, 3403. [Google Scholar] [CrossRef] [PubMed]
- FDA. Nucleic Acid Based Tests. Available online: https://www.fda.gov/medical-devices/in-vitro-diagnostics/nucleic-acid-based-tests (accessed on 21 October 2024).
- Malek, S. Chronic Lymphocytic Leukemia: Methods and Protocols; Humana Press: New York, NY, USA, 2018; pp. 12–373. [Google Scholar]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Kyriakidis, K.; Tsezou, A. MicroRNAs and the Diagnosis of Childhood Acute Lymphoblastic Leukemia: Systematic Review, Meta-Analysis and Re-Analysis with Novel Small RNA-Seq Tools. Cancers 2022, 14, 3976. [Google Scholar] [CrossRef]
- Võsa, U.; Vooder, T.; Kolde, R.; Fischer, K.; Välk, K.; Tõnisson, N.; Roosipuu, R.; Vilo, J.; Metspalu, A.; Annilo, T. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer 2011, 50, 812–822. [Google Scholar] [CrossRef]
- Akçakaya, P.; Ekelund, S.; Kolosenko, I.; Caramuta, S.; Ozata, D.M.; Xie, H.; Lindforss, U.; Olivecrona, H.; Lui, W.-O. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int. J. Oncol. 2011, 39, 311–318. [Google Scholar] [CrossRef]
- Eyking, A.; Reis, H.; Frank, M.; Gerken, G.; Schmid, K.W.; Cario, E. MiR-205 and MiR-373 Are Associated with Aggressive Human Mucinous Colorectal Cancer. PLoS ONE 2016, 11, e0156871. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, Y.; Wei, J.; Jia, W.; Ge, Z.; Zhang, Z.; Liu, X. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer 2013, 13, 469. [Google Scholar] [CrossRef]
- Jones, K.; Nourse, J.P.; Keane, C.; Bhatnagar, A.; Gandhi, M.K. Plasma microRNA are disease response biomarkers in classical Hodgkin lymphoma. Clin. Cancer Res. 2014, 20, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Hickey, R.J.; Malkas, L.H. Therapeutic Targeting of DNA Replication Stress in Cancer. Genes 2023, 14, 1346. [Google Scholar] [CrossRef] [PubMed]
- Embleton, M.J. Deutsche Stiftung für Krebsforschung. In Genes and Antigens in Cancer Cells: The Monoclonal Antibody Approach, Proceedings of the 4th International Expert Meeting of the Deutsche Stiftung fuür Krebsforschung, Bonn, 27–29 June 1983, 4th ed.; Riethmuller, G., Ed.; Karger: Basel, Switzerland; New York, NY, USA, 1984; pp. 9–192. [Google Scholar]
- Jasperson, K.W.; Tuohy, T.M.; Neklason, D.W.; Burt, R.W. Hereditary and Familial Colon Cancer. Gastroenterology 2010, 138, 2044–2058. [Google Scholar] [CrossRef] [PubMed]
- Gibert, B.; Delloye-Bourgeois, C.; Gattolliat, C.-H.; Meurette, O.; Le Guernevel, S.; Fombonne, J.; Ducarouge, B.; Lavial, F.; Bouhallier, F.; Creveaux, M.; et al. Regulation by miR181 Family of the Dependence Receptor CDON Tumor Suppressive Activity in Neuroblastoma. JNCI: J. Natl. Cancer Inst. 2014, 106, dju318. [Google Scholar] [CrossRef]
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415. [Google Scholar] [CrossRef]
- Due, H.; Svendsen, P.; Bødker, J.S.; Schmitz, A.; Bøgsted, M.; Johnsen, H.E.; El-Galaly, T.C.; Roug, A.S.; Dybkær, K. miR-155 as a Biomarker in B-Cell Malignancies. Biomed. Res. Int. 2016, 2016, 9513037. [Google Scholar] [CrossRef]
- Thakur, S.; Sinhari, A.; Jain, P.; Jadhav, H.R. A perspective on oligonucleotide therapy: Approaches to patient customization. Front. Pharmacol. 2022, 13, 1006304. [Google Scholar] [CrossRef]
- Xu, C.-X.; Xu, M.; Tan, L.; Yang, H.; Permuth-Wey, J.; Kruk, P.A.; Wenham, R.M.; Nicosia, S.V.; Lancaster, J.M.; Sellers, T.A.; et al. MicroRNA MiR-214 Regulates Ovarian Cancer Cell Stemness by Targeting p53/Nanog. J. Biol. Chem. 2012, 287, 34970–34978. [Google Scholar] [CrossRef]
- Sayed, A.S.M.; Xia, K.; Salma, U.; Yang, T.; Peng, J. Diagnosis, Prognosis and Therapeutic Role of Circulating miRNAs in Cardiovascular Diseases. Heart Lung Circ. 2014, 23, 503–510. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Gao, X.-M.; Winbanks, C.E.; Boey, E.J.H.; Tham, Y.K.; Kiriazis, H.; Gregorevic, P.; Obad, S.; Kauppinen, S.; Du, X.-J.; et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc. Natl. Acad. Sci. USA 2012, 109, 17615–17620. [Google Scholar] [CrossRef]
- Rink, C.; Khanna, S. MicroRNA in ischemic stroke etiology and pathology. Physiol. Genom. 2011, 43, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Neag, M.A.; Mitre, A.O.; Burlacu, C.C.; Inceu, A.I.; Mihu, C.; Melincovici, C.S.; Bichescu, M.; Buzoianu, A.D. miRNA Involvement in Cerebral Ischemia-Reperfusion Injury. Front. Neurosci. 2022, 16, 901360. [Google Scholar] [CrossRef] [PubMed]
- Fathi, M.; Gharakhanlou, R.; Rezaei, R. The Changes of Heart miR-1 and miR-133 Expressions following Physiological Hypertrophy Due to Endurance Training. Cell J. 2020, 22, 133–140. [Google Scholar] [CrossRef]
- Wu, Q.; Qi, B.; Duan, X.; Ming, X.; Yan, F.; He, Y.; Bu, X.; Sun, S.; Zhu, H. MicroRNA-126 enhances the biological function of endothelial progenitor cells under oxidative stress via PI3K/Akt/GSK3β and ERK1/2 signaling pathways. Bosn. J. Basic Med. Sci. 2021, 21, 71–80. [Google Scholar] [CrossRef]
- Han, D.; Dong, X.; Zheng, D.; Nao, J. MiR-124 and the Underlying Therapeutic Promise of Neurodegenerative Disorders. Front. Pharmacol. 2019, 10, 1555. [Google Scholar] [CrossRef]
- Fan, W.; Liang, C.; Ou, M.; Zou, T.; Sun, F.; Zhou, H.; Cui, L. MicroRNA-146a Is a Wide-Reaching Neuroinflammatory Regulator and Potential Treatment Target in Neurological Diseases. Front. Mol. Neurosci. 2020, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D.; Boldin, M.P.; O’Connell, R.M.; Rao, D.S.; Taganov, K.D. MicroRNAs: New regulators of immune cell development and function. Nat. Immunol. 2008, 9, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, M.; Palmer, M.A.; Wilson, J.A. MicroRNA-122 Regulation of HCV Infections: Insights from Studies of miR-122-Independent Replication. Pathogens 2022, 11, 1005. [Google Scholar] [CrossRef] [PubMed]
- Frattari, G.; Aagaard, L.; Denton, P.W. The role of miR-29a in HIV-1 replication and latency. J. Virus Erad. 2017, 3, 185–191. [Google Scholar] [CrossRef]
- Tuddenham, L.; Jung Jette, S.; Chane-Woon-Ming, B.; Dölken, L.; Pfeffer, S. Small RNA Deep Sequencing Identifies MicroRNAs and Other Small Noncoding RNAs from Human Herpesvirus 6B. J. Virol. 2012, 86, 1638–1649. [Google Scholar] [CrossRef]
- Lewohl, J.M.; Nunez, Y.O.; Dodd, P.R.; Tiwari, G.R.; Harris, R.A.; Mayfield, R.D. Up-regulation of microRNAs in brain of human alcoholics. Alcohol Clin. Exp. Res. 2011, 35, 1928–1937. [Google Scholar] [CrossRef] [PubMed]
- Tapocik, J.D.; Solomon, M.; Flanigan, M.; Meinhardt, M.; Barbier, E.; Schank, J.R.; Schwandt, M.; Sommer, W.H.; Heilig, M. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenom. J. 2013, 13, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Gorini, G.; Nunez, Y.O.; Mayfield, R.D. Integration of miRNA and protein profiling reveals coordinated neuroadaptations in the alcohol-dependent mouse brain. PLoS ONE 2013, 8, e82565. [Google Scholar] [CrossRef]
- Tapocik, J.D.; Barbier, E.; Flanigan, M.; Solomon, M.; Pincus, A.; Pilling, A.; Sun, H.; Schank, J.R.; King, C.; Heilig, M. microRNA-206 in Rat Medial Prefrontal Cortex Regulates BDNF Expression and Alcohol Drinking. J. Neurosci. 2014, 34, 4581–4588. [Google Scholar] [CrossRef]
- Lippai, D.; Bala, S.; Csak, T.; Kurt-Jones, E.A.; Szabo, G. Chronic alcohol-induced microRNA-155 contributes to neuroinflammation in a TLR4-dependent manner in mice. PLoS ONE 2013, 8, e70945. [Google Scholar] [CrossRef]
- Li, J.; Li, J.; Liu, X.; Qin, S.; Guan, Y.; Liu, Y.; Cheng, Y.; Chen, X.; Li, W.; Wang, S.; et al. MicroRNA expression profile and functional analysis reveal that miR-382 is a critical novel gene of alcohol addiction. EMBO Mol. Med. 2013, 5, 1402–1414. [Google Scholar] [CrossRef]
- Maldonado, E.; Morales-Pison, S.; Urbina, F.; Solari, A. Aging Hallmarks and the Role of Oxidative Stress. Antioxidants 2023, 12, 651. [Google Scholar] [CrossRef]
- Berk, Ş. Insulin and IGF-1 extend the lifespan of Caenorhabditis elegans by inhibiting insulin/insulin-like signaling and mTOR signaling pathways: C. elegans-Focused cancer research. Biochem. Biophys. Res. Commun. 2024, 729, 150347. [Google Scholar] [CrossRef]
- Nair, D.R.; Satheesh, K.; Raghavan, A.; Nanditha, A.; Vinitha, R.; Susairaj, P.; Snehalatha, C.; Ramachandran, A. Trend in the clinical profile of type 2 diabetes in India-Study from a diabetes care centre in South India. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1851–1857. [Google Scholar] [CrossRef]
- Chen, H.; Lan, H.-Y.; Roukos, D.H.; Cho, W.C. Application of microRNAs in diabetes mellitus. J. Endocrinol. 2014, 222, R1–R10. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008, 132, 875–886. [Google Scholar] [CrossRef]
- Hata, A.; Kashima, R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 121–134. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef]
- Coppé, J.P.; Desprez, P.Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef]
- Guo, J.; Huang, X.; Dou, L.; Yan, M.; Shen, T.; Tang, W.; Li, J. Aging and aging-related diseases: From molecular mechanisms to interventions and treatments. Signal Transduct. Target. Ther. 2022, 7, 391. [Google Scholar] [CrossRef]
- Yang, H.; Chen, Y.; Wang, J.; Wei, H.; Chen, Y.; Jin, J. Activities of daily living measurement after ischemic stroke: Rasch analysis of the modified Barthel Index. Medicine 2021, 100, e24926. [Google Scholar] [CrossRef]
- Shantikumar, S.; Caporali, A.; Emanueli, C. Role of microRNAs in diabetes and its cardiovascular complications. Cardiovasc. Res. 2012, 93, 583–593. [Google Scholar] [CrossRef]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Urbich, C.; Kuehbacher, A.; Dimmeler, S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc. Res. 2008, 79, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Ruan, R.; Liu, Y.; Zhang, X. Circulating mir-199-3p screens the onset of type 2 diabetes mellitus and the complication of coronary heart disease and predicts the occurrence of major adverse cardiovascular events. BMC Cardiovasc. Disord. 2023, 23, 563. [Google Scholar] [CrossRef]
- Han, R.; Gao, J.; Wang, L.; Hao, P.; Chen, X.; Wang, Y.; Jiang, Z.; Jiang, L.; Wang, T.; Zhu, L.; et al. MicroRNA-146a negatively regulates inflammation via the IRAK1/TRAF6/NF-κB signaling pathway in dry eye. Sci. Rep. 2023, 13, 11192. [Google Scholar] [CrossRef]
- Luo, J.; Wu, J.; Li, Z.; Qin, H.; Wang, B.; Wong, T.S.; Yang, W.; Fu, Q.L.; Lei, W. miR-375 suppresses IGF1R expression and contributes to inhibition of cell progression in laryngeal squamous cell carcinoma. Biomed. Res. Int. 2014, 2014, 374598. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, M.T.; Leone, E.; Amodio, N.; Foresta, U.; Lionetti, M.; Pitari, M.R.; Cantafio, M.E.; Gullà, A.; Conforti, F.; Morelli, E.; et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: In vitro and in vivo evidence. Clin. Cancer Res. 2012, 18, 6260–6270. [Google Scholar] [CrossRef]
- Romao, J.M.; Jin, W.; Dodson, M.V.; Hausman, G.J.; Moore, S.S.; Guan, L.L. MicroRNA regulation in mammalian adipogenesis. Exp. Biol. Med. 2011, 236, 997–1004. [Google Scholar] [CrossRef]
- Teruel-Montoya, R.; Rosendaal, F.R.; Martínez, C. MicroRNAs in hemostasis. J. Thromb. Haemost. 2015, 13, 170–181. [Google Scholar] [CrossRef]
- Zuo, Y.; Qiang, L.; Farmer, S.R. Activation of CCAAT/Enhancer-binding Protein (C/EBP)α; Expression by C/EBPβ; during Adipogenesis Requires a Peroxisome Proliferator-activated Receptor-γ-associated Repression of HDAC1 at the C/ebpα; Gene Promoter. J. Biol. Chem. 2006, 281, 7960–7967. [Google Scholar] [CrossRef]
- Jun-Hao, E.T.; Gupta, R.R.; Shyh-Chang, N. Lin28 and let-7 in the Metabolic Physiology of Aging. Trends Endocrinol. Metab. 2016, 27, 132–141. [Google Scholar] [CrossRef]
- Zhu, H.; Shyh-Chang, N.; Segrè Ayellet, V.; Shinoda, G.; Shah Samar, P.; Einhorn William, S.; Takeuchi, A.; Engreitz Jesse, M.; Hagan John, P.; Kharas Michael, G.; et al. The Lin28/let-7 Axis Regulates Glucose Metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Frost, R.J.A.; Olson, E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc. Natl. Acad. Sci. USA 2011, 108, 21075–21080. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Hata, A. MicroRNA regulation of smooth muscle gene expression and phenotype. Curr. Opin. Hematol. 2012, 19, 224–231. [Google Scholar] [CrossRef]
- Nourse, J.; Danckwardt, S. A novel rationale for targeting FXI: Insights from the hemostatic microRNA targetome for emerging anticoagulant strategies. Pharmacol. Ther. 2021, 218, 107676. [Google Scholar] [CrossRef]
- Berardi, E.; Pues, M.; Thorrez, L.; Sampaolesi, M. miRNAs in ESC differentiation. Am. J. Physiol.-Heart Circ. Physiol. 2012, 303, H931–H939. [Google Scholar] [CrossRef]
- Lu, R.-H.; Jia, S.-Z.; Yang, F.; Qin, C.-B.; Zhang, Y.-R.; Meng, X.-L.; Yan, X.; Feng, J.-C.; Nie, G.-X. The function of miR-122 in the lipid metabolism and immunity of grass carp (Ctenopharyngodon idellus). Aquac. Rep. 2020, 17, 100401. [Google Scholar] [CrossRef]
- Tsai, W.C.; Hsu, S.D.; Hsu, C.S.; Lai, T.C.; Chen, S.J.; Shen, R.; Huang, Y.; Chen, H.C.; Lee, C.H.; Tsai, T.F.; et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Investig. 2012, 122, 2884–2897. [Google Scholar] [CrossRef]
- Pagoni, M.; Cava, C.; Sideris, D.C.; Avgeris, M.; Zoumpourlis, V.; Michalopoulos, I.; Drakoulis, N. miRNA-Based Technologies in Cancer Therapy. J. Pers. Med. 2023, 13, 1586. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.J.; Javed, Z.; Sadia, H.; Mehmood, S.; Akbar, A.; Zahid, B.; Nadeem, T.; Roshan, S.; Varoni, E.M.; Iriti, M.; et al. Targeted therapy using nanocomposite delivery systems in cancer treatment: Highlighting miR34a regulation for clinical applications. Cancer Cell Int. 2023, 23, 84. [Google Scholar] [CrossRef]
- Mencía, Á.; Modamio-Høybjør, S.; Redshaw, N.; Morín, M.; Mayo-Merino, F.; Olavarrieta, L.; Aguirre, L.A.; del Castillo, I.; Steel, K.P.; Dalmay, T.; et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009, 41, 609–613. [Google Scholar] [CrossRef]
- Hughes Anne, E.; Bradley Declan, T.; Campbell, M.; Lechner, J.; Dash Durga, P.; Simpson David, A.; Willoughby Colin, E. Mutation Altering the miR-184 Seed Region Causes Familial Keratoconus with Cataract. Am. J. Hum. Genet. 2011, 89, 628–633. [Google Scholar] [CrossRef] [PubMed]
- De Pontual, L.; Yao, E.; Callier, P.; Faivre, L.; Drouin, V.; Cariou, S.; Van Haeringen, A.; Geneviève, D.; Goldenberg, A.; Oufadem, M.; et al. Germline deletion of the miR-17∼92 cluster causes skeletal and growth defects in humans. Nat. Genet. 2011, 43, 1026–1030. [Google Scholar] [CrossRef]
- Zhou, L.; Miller, C.; Miraglia, L.J.; Romero, A.; Mure, L.S.; Panda, S.; Kay, S.A. A genome-wide microRNA screen identifies the microRNA-183/96/182 cluster as a modulator of circadian rhythms. Proc. Natl. Acad. Sci. USA 2021, 118, e2020454118. [Google Scholar] [CrossRef]
- Volná, A.; Bartas, M.; Pečinka, P.; Špunda, V.; Červeň, J. What Do We Know about Barley miRNAs? Int. J. Mol. Sci. 2022, 23, 14755. [Google Scholar] [CrossRef]
- Curaba, J.; Spriggs, A.; Taylor, J.; Li, Z.; Helliwell, C. miRNA regulation in the early development of barley seed. BMC Plant Biol. 2012, 12, 120. [Google Scholar] [CrossRef]
- Wurm, A.A.; Brilloff, S.; Kolovich, S.; Schäfer, S.; Rahimian, E.; Kufrin, V.; Bill, M.; Carrero, Z.I.; Drukewitz, S.; Krüger, A.; et al. Signaling-induced systematic repression of miRNAs uncovers cancer vulnerabilities and targeted therapy sensitivity. Cell Rep. Med. 2023, 4, 101200. [Google Scholar] [CrossRef] [PubMed]
- Otmani, K.; Lewalle, P. Tumor Suppressor miRNA in Cancer Cells and the Tumor Microenvironment: Mechanism of Deregulation and Clinical Implications. Front. Oncol. 2021, 11, 708765. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, C.P.; Kang, M.H.; Dal-Pra, S.; Mirotsou, M.; Dzau, V.J. MicroRNAs and Cardiac Regeneration. Circ. Res. 2015, 116, 1700–1711. [Google Scholar] [CrossRef]
- Castañón-Cortés, L.G.; Bravo-Vázquez, L.A.; Santoyo-Valencia, G.; Medina-Feria, S.; Sahare, P.; Duttaroy, A.K.; Paul, S. Current advances in the development of microRNA-integrated tissue engineering strategies: A cornerstone of regenerative medicine. Front. Bioeng. Biotechnol. 2024, 12, 1484151. [Google Scholar] [CrossRef]
- Quah, S.; Subramanian, G.; Tan, J.S.L.; Utami, K.H.; Sampath, P. MicroRNAs: A symphony orchestrating evolution and disease dynamics. Trends Mol. Med. 2024. [Google Scholar] [CrossRef]
- Curtale, G.; Rubino, M.; Locati, M. MicroRNAs as Molecular Switches in Macrophage Activation. Front. Immunol. 2019, 10, 799. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yao, Y.; Yan, H.; Wang, R.; Zhang, Z.; Sun, X.; Zhao, L.; Ao, X.; Xie, Z.; Wu, Q. A Tumor-specific MicroRNA Recognition System Facilitates the Accurate Targeting to Tumor Cells by Magnetic Nanoparticles. Mol. Ther. Nucleic Acids 2016, 5, e318. [Google Scholar] [CrossRef]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Shaheen, N.; Shaheen, A.; Osama, M.; Nashwan, A.J.; Bharmauria, V.; Flouty, O. MicroRNAs regulation in Parkinson’s disease, and their potential role as diagnostic and therapeutic targets. NPJ Park. Dis. 2024, 10, 186. [Google Scholar] [CrossRef]
- Hussen, B.M.; Rasul, M.F.; Abdullah, S.R.; Hidayat, H.J.; Faraj, G.S.H.; Ali, F.A.; Salihi, A.; Baniahmad, A.; Ghafouri-Fard, S.; Rahman, M.; et al. Targeting miRNA by CRISPR/Cas in cancer: Advantages and challenges. Mil. Med. Res. 2023, 10, 32. [Google Scholar] [CrossRef]
- Elnashar, A.M.; El-Dien Ibrahim, M.; El-Desoky, M.M.; Ali, O.M.; El-Sayd Mohamed Hassan, M. Female sexual dysfunction in Lower Egypt. BJOG Int. J. Obstet. Gynecol. 2007, 114, 201–206. [Google Scholar] [CrossRef]
- O’Neill, C.P.; Dwyer, R.M. Nanoparticle-Based Delivery of Tumor Suppressor microRNA for Cancer Therapy. Cells 2020, 9, 521. [Google Scholar] [CrossRef]
- Duygu, B.; Juni, R.; Ottaviani, L.; Bitsch, N.; Wit, J.B.M.; de Windt, L.J.; da Costa Martins, P.A. Comparison of different chemically modified inhibitors of miR-199b in vivo. Biochem. Pharmacol. 2019, 159, 106–115. [Google Scholar] [CrossRef]
- Przanowska, R.K.; Sobierajska, E.; Su, Z.; Jensen, K.; Przanowski, P.; Nagdas, S.; Kashatus, J.A.; Kashatus, D.F.; Bhatnagar, S.; Lukens, J.R.; et al. miR-206 family is important for mitochondrial and muscle function, but not essential for myogenesis in vitro. FASEB J. 2020, 34, 7687–7702. [Google Scholar] [CrossRef]
- Li, H.; Liu, T.; Yang, Y.; Cho, W.C.; Flynn, R.J.; Harandi, M.F.; Song, H.; Luo, X.; Zheng, Y. Interplays of liver fibrosis-associated microRNAs: Molecular mechanisms and implications in diagnosis and therapy. Genes Dis. 2023, 10, 1457–1469. [Google Scholar] [CrossRef]
- Zheng, H.; Fu, R.; Wang, J.-T.; Liu, Q.; Chen, H.; Jiang, S.-W. Advances in the Techniques for the Prediction of microRNA Targets. Int. J. Mol. Sci. 2013, 14, 8179–8187. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Nourse, J.; Braun, J.; Lackner, K.; Hüttelmaier, S.; Danckwardt, S. Large-scale identification of functional microRNA targeting reveals cooperative regulation of the hemostatic system. J. Thromb. Haemost. 2018, 16, 2233–2245. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-H.; Gessler, D.J.; Zhan, W.; Gallagher, T.L.; Gao, G. Adeno-associated virus as a delivery vector for gene therapy of human diseases. Signal Transduct. Target. Ther. 2024, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Schröder, L.C.; Frank, D.; Müller, O.J. Transcriptional Targeting Approaches in Cardiac Gene Transfer Using AAV Vectors. Pathogens 2023, 12, 1301. [Google Scholar] [CrossRef]
- Finneran, D.J.; Njoku, I.P.; Flores-Pazarin, D.; Ranabothu, M.R.; Nash, K.R.; Morgan, D.; Gordon, M.N. Toward Development of Neuron Specific Transduction After Systemic Delivery of Viral Vectors. Front. Neurol. 2021, 12, 685802. [Google Scholar] [CrossRef]
- Sinclair, F.; Begum, A.A.; Dai, C.C.; Toth, I.; Moyle, P.M. Recent advances in the delivery and applications of nonviral CRISPR/Cas9 gene editing. Drug Deliv. Transl. Res. 2023, 13, 1500–1519. [Google Scholar] [CrossRef]
- Pelaz, B.; Alexiou, C.; Alvarez-Puebla, R.A.; Alves, F.; Andrews, A.M.; Ashraf, S.; Balogh, L.P.; Ballerini, L.; Bestetti, A.; Brendel, C.; et al. Diverse Applications of Nanomedicine. ACS Nano 2017, 11, 2313–2381. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, T.; Adriani, G.; Kamm, R.D.; Mattu, C.; Chiono, V. MicroRNA delivery through nanoparticles. J. Control. Release 2019, 313, 80–95. [Google Scholar] [CrossRef]
- LeRoith, D.; Holly, J.M.P.; Forbes, B.E. Insulin-like growth factors: Ligands, binding proteins, and receptors. Mol. Metab. 2021, 52, 101245. [Google Scholar] [CrossRef]
- Martín-Sabroso, C.; Torres-Suárez, A.I.; Alonso-González, M.; Fernández-Carballido, A.; Fraguas-Sánchez, A.I. Active Targeted Nanoformulations via Folate Receptors: State of the Art and Future Perspectives. Pharmaceutics 2021, 14, 14. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Chen, Q.; Chen, X.; Han, F.; Chen, Z.; Wang, Y. The blood–brain barrier: Structure, regulation and drug delivery. Signal Transduct. Target. Ther. 2023, 8, 217. [Google Scholar] [CrossRef]
- González-Martínez, I.; Cerro-Herreros, E.; Moreno, N.; García-Rey, A.; Espinosa-Espinosa, J.; Carrascosa-Sàez, M.; Piqueras-Losilla, D.; Arzumanov, A.; Seoane-Miraz, D.; Jad, Y.; et al. Peptide-conjugated antimiRs improve myotonic dystrophy type 1 phenotypes by promoting endogenous MBNL1 expression. Mol. Ther. Nucleic Acids 2023, 34, 102024. [Google Scholar] [CrossRef]
- Nakagawa, H.; Saito, Y. Roles of Natriuretic Peptides and the Significance of Neprilysin in Cardiovascular Diseases. Biology 2022, 11, 1017. [Google Scholar] [CrossRef]
- Zou, L.L.; Ma, J.L.; Wang, T.; Yang, T.B.; Liu, C.B. Cell-penetrating Peptide-mediated therapeutic molecule delivery into the central nervous system. Curr. Neuropharmacol. 2013, 11, 197–208. [Google Scholar] [CrossRef]
- Shraim, A.S.; Abdel Majeed, B.A.; Al-Binni, M.A.; Hunaiti, A. Therapeutic Potential of Aptamer-Protein Interactions. ACS Pharmacol. Transl. Sci. 2022, 5, 1211–1227. [Google Scholar] [CrossRef]
- Asimakidou, E.; Tan, J.K.S.; Zeng, J.; Lo, C.H. Blood-Brain Barrier-Targeting Nanoparticles: Biomaterial Properties and Biomedical Applications in Translational Neuroscience. Pharmaceuticals 2024, 17, 612. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yu, L.; Lin, X.; Liu, X.; Zhang, Y.; Yang, F.; Deng, W. Virus-like Particles as Nanocarriers for Intracellular Delivery of Biomolecules and Compounds. Viruses 2022, 14, 1905. [Google Scholar] [CrossRef]
- Aránega, A.E.; Lozano-Velasco, E.; Rodriguez-Outeiriño, L.; Ramírez de Acuña, F.; Franco, D.; Hernández-Torres, F. MiRNAs and Muscle Regeneration: Therapeutic Targets in Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2021, 22, 4236. [Google Scholar] [CrossRef]
- Bader, J.; Brigger, F.; Leroux, J.-C. Extracellular vesicles versus lipid nanoparticles for the delivery of nucleic acids. Adv. Drug Deliv. Rev. 2024, 215, 115461. [Google Scholar] [CrossRef]
- Nowak, I.; Madej, M.; Secemska, J.; Sarna, R.; Strzalka-Mrozik, B. Virus-Based Biological Systems as Next-Generation Carriers for the Therapy of Central Nervous System Diseases. Pharmaceutics 2023, 15, 1931. [Google Scholar] [CrossRef] [PubMed]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Chan, K.Y.; Lou, S.; Keyes, C.; Wu, J.; Botticello-Romero, N.R.; Zheng, Q.; Johnston, J.; Mills, A.; Brauer, P.P.; et al. An AAV capsid reprogrammed to bind human Transferrin Receptor mediates brain-wide gene delivery. bioRxiv 2023. [Google Scholar] [CrossRef]
- Naso, M.F.; Tomkowicz, B.; Perry, W.L., 3rd; Strohl, W.R. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs 2017, 31, 317–334. [Google Scholar] [CrossRef]
- Taschenberger, G.; Tereshchenko, J.; Kügler, S. A MicroRNA124 Target Sequence Restores Astrocyte Specificity of gfaABC(1)D-Driven Transgene Expression in AAV-Mediated Gene Transfer. Mol. Ther. Nucleic Acids 2017, 8, 13–25. [Google Scholar] [CrossRef]
- Issa, S.S.; Shaimardanova, A.A.; Solovyeva, V.V.; Rizvanov, A.A. Various AAV Serotypes and Their Applications in Gene Therapy: An Overview. Cells 2023, 12, 785. [Google Scholar] [CrossRef]
- Dufait, I.; Liechtenstein, T.; Lanna, A.; Bricogne, C.; Laranga, R.; Padella, A.; Breckpot, K.; Escors, D. Retroviral and lentiviral vectors for the induction of immunological tolerance. Scientifica 2012, 2012, 694137. [Google Scholar] [CrossRef]
- Shalaby, K.; Aouida, M.; El-Agnaf, O. Tissue-Specific Delivery of CRISPR Therapeutics: Strategies and Mechanisms of Non-Viral Vectors. Int. J. Mol. Sci. 2020, 21, 7353. [Google Scholar] [CrossRef]
- Chang, H.; Yi, B.; Ma, R.; Zhang, X.; Zhao, H.; Xi, Y. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci. Rep. 2016, 6, 22312. [Google Scholar] [CrossRef]
- Saiyed, A.N.; Vasavada, A.R.; Johar, S.R.K. Recent trends in miRNA therapeutics and the application of plant miRNA for prevention and treatment of human diseases. Future J. Pharm. Sci. 2022, 8, 24. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, L.; Wang, X.; Jin, H. RNA-based therapeutics: An overview and prospectus. Cell Death Dis. 2022, 13, 644. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.U. Micro-RNAs (miRNAs): Genomic organisation, biogenesis and mode of action. Cell Tissue Res. 2012, 349, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Guo, Y.; Luo, Y.; Mak, M.; Zhang, J.; Xu, W.; Qian, H.; Tao, Z. Visualization of microRNA therapy in cancers delivered by small extracellular vesicles. J. Nanobiotechnol. 2023, 21, 457. [Google Scholar] [CrossRef]
- Segal, M.; Slack, F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin. Drug Discov. 2020, 15, 987–991. [Google Scholar] [CrossRef]
- Ghamlouche, F.; Yehya, A.; Zeid, Y.; Fakhereddine, H.; Fawaz, J.; Liu, Y.-N.; Al-Sayegh, M.; Abou-Kheir, W. MicroRNAs as clinical tools for diagnosis, prognosis, and therapy in prostate cancer. Transl. Oncol. 2023, 28, 101613. [Google Scholar] [CrossRef]
- Dasgupta, I.; Chatterjee, A. Recent Advances in miRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef]
- Maruggi, G.; Ulmer, J.B.; Rappuoli, R.; Yu, D. Self-amplifying mRNA-Based Vaccine Technology and Its Mode of Action. Curr. Top. Microbiol. Immunol. 2021, 440, 31–70. [Google Scholar] [CrossRef]
- Jones, C.H.; Androsavich, J.R.; So, N.; Jenkins, M.P.; MacCormack, D.; Prigodich, A.; Welch, V.; True, J.M.; Dolsten, M. Breaking the mold with RNA—A “RNAissance” of life science. NPJ Genom. Med. 2024, 9, 2. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, X.Y.; Lu, A.; Wang, X.Y.; Jiang, L.X.; Wang, J.C. Non-viral vectors for RNA delivery. J. Control. Release 2022, 342, 241–279. [Google Scholar] [CrossRef]
- Guelfi, G.; Capaccia, C.; Anipchenko, P.; Ciancabilla, F.; Oommen, O.P.; Bufalari, A.; Zerani, M.; Maranesi, M. Mimic miRNA and Anti-miRNA Activated Scaffolds as a Therapeutic Strategy to Promote Bone, Cartilage, and Skin Regeneration. Macromol 2024, 4, 165–189. [Google Scholar] [CrossRef]
- Lu, P.; Ruan, D.; Huang, M.; Tian, M.; Zhu, K.; Gan, Z.; Xiao, Z. Harnessing the potential of hydrogels for advanced therapeutic applications: Current achievements and future directions. Signal Transduct. Target. Ther. 2024, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.I.; Tang, Y.L. Genetic modification of stem cells for transplantation. Adv. Drug Deliv. Rev. 2008, 60, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Chehelgerdi, M.; Behdarvand Dehkordi, F.; Chehelgerdi, M.; Kabiri, H.; Salehian-Dehkordi, H.; Abdolvand, M.; Salmanizadeh, S.; Rashidi, M.; Niazmand, A.; Ahmadi, S.; et al. Exploring the promising potential of induced pluripotent stem cells in cancer research and therapy. Mol. Cancer 2023, 22, 189. [Google Scholar] [CrossRef]
- Stupnikov, A.; Bezuglov, V.; Skakov, I.; Shtratnikova, V.; Pilsner, J.R.; Suvorov, A.; Sergeyev, O. ITAS: Integrated Transcript Annotation for Small RNA. Noncoding RNA 2022, 8, 30. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Dash, P.K.; Gupta, P.; Pradhan, S.K.; Shasany, A.K.; Rai, R. Analysis of Homologous Regions of Small RNAs MIR397 and MIR408 Reveals the Conservation of Microsynteny among Rice Crop-Wild Relatives. Cells 2022, 11, 3461. [Google Scholar] [CrossRef]
- Pakdel, M.H.; Asadi, A.A.; Tavakol, E.; Shariati, V.; Hosseini Mazinani, M. Machine learning-aided microRNA discovery for olive oil quality. PLoS ONE 2024, 19, e0311569. [Google Scholar] [CrossRef]
- Riolo, G.; Cantara, S.; Marzocchi, C.; Ricci, C. miRNA Targets: From Prediction Tools to Experimental Validation. Methods Protoc. 2020, 4, 1. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abdelfattah, A.M.; Park, C.; Choi, M.Y. Update on non-canonical microRNAs. Biomol. Concepts 2014, 5, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.; Huttner, M.; Dueck, A.; Meister, G.; Engelmann, J.C. miRA: Adaptable novel miRNA identification in plants using small RNA sequencing data. BMC Bioinform. 2015, 16, 370. [Google Scholar] [CrossRef] [PubMed]
- Szakats, S.; McAtamney, A.; Wilson, M.J. Identification of novel microRNAs in the embryonic mouse brain using deep sequencing. Mol. Cell. Biochem. 2024, 479, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Keller, A.; Meese, E. The miRNA-target interactions: An underestimated intricacy. Nucleic Acids Res. 2024, 52, 1544–1557. [Google Scholar] [CrossRef]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef]
- Kuhn, D.E.; Martin, M.M.; Feldman, D.S.; Terry, A.V., Jr.; Nuovo, G.J.; Elton, T.S. Experimental validation of miRNA targets. Methods 2008, 44, 47–54. [Google Scholar] [CrossRef]
- Lin, A.; Giuliano, C.J.; Palladino, A.; John, K.M.; Abramowicz, C.; Yuan, M.L.; Sausville, E.L.; Lukow, D.A.; Liu, L.; Chait, A.R.; et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 2019, 11, 8412. [Google Scholar] [CrossRef]
- Zare, M.; Pemmada, R.; Madhavan, M.; Shailaja, A.; Ramakrishna, S.; Kandiyil, S.P.; Donahue, J.M.; Thomas, V. Encapsulation of miRNA and siRNA into Nanomaterials for Cancer Therapeutics. Pharmaceutics 2022, 14, 1620. [Google Scholar] [CrossRef]
- Nguyen, T.P.N.; Kumar, M.; Fedele, E.; Bonanno, G.; Bonifacino, T. MicroRNA Alteration, Application as Biomarkers, and Therapeutic Approaches in Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 4718. [Google Scholar] [CrossRef]
- Fedorczak, A.; Lewiński, A.; Stawerska, R. Involvement of Sirtuin 1 in the Growth Hormone/Insulin-like Growth Factor 1 Signal Transduction and Its Impact on Growth Processes in Children. Int. J. Mol. Sci. 2023, 24, 5406. [Google Scholar] [CrossRef]
- Kitada, T.; DiAndreth, B.; Teague, B.; Weiss, R. Programming gene and engineered-cell therapies with synthetic biology. Science 2018, 359, 1067. [Google Scholar] [CrossRef] [PubMed]
- De Palma, F.D.E.; Raia, V.; Kroemer, G.; Maiuri, M.C. The Multifaceted Roles of MicroRNAs in Cystic Fibrosis. Diagnostics 2020, 10, 1102. [Google Scholar] [CrossRef] [PubMed]
- Yee, P.T.; Poh, C.L. Development of Novel miRNA-based Vaccines and Antivirals against Enterovirus 71. Curr. Pharm. Des. 2016, 22, 6694–6700. [Google Scholar] [CrossRef] [PubMed]
- Cañas, J.A.; Rodrigo-Muñoz, J.M.; Sastre, B.; Gil-Martinez, M.; Redondo, N.; del Pozo, V. MicroRNAs as Potential Regulators of Immune Response Networks in Asthma and Chronic Obstructive Pulmonary Disease. Front. Immunol. 2021, 11, 608666. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, S.J.; Kumar, P. Cross-Talk Between Antigen Presenting Cells and T Cells Impacts Intestinal Homeostasis, Bacterial Infections, and Tumorigenesis. Front. Immunol. 2019, 10, 360. [Google Scholar] [CrossRef]
- Monreal-Escalante, E.; Ramos-Vega, A.; Angulo, C.; Bañuelos-Hernández, B. Plant-Based Vaccines: Antigen Design, Diversity, and Strategies for High Level Production. Vaccines 2022, 10, 100. [Google Scholar] [CrossRef]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef]
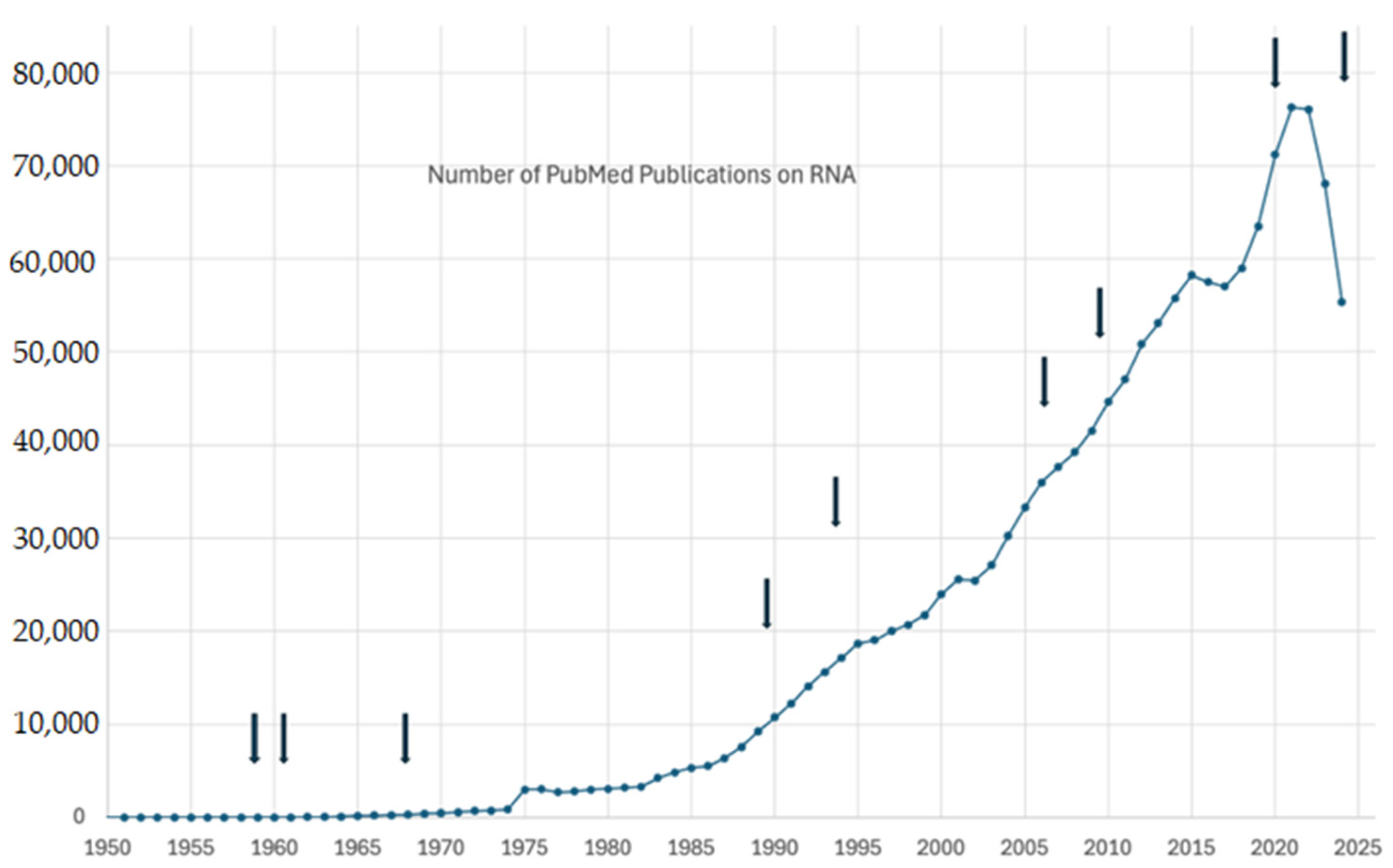


| Type of RNA | Function | Percentage of RNA | Nobel Prize Details |
|---|---|---|---|
| RNA Discovery | Enzyme polynucleotide phosphorylase is responsible for RNA synthesis. | -- | 1959: For discovering the mechanisms of RNA and DNA synthesis |
| Messenger RNA (mRNA) | Serves as the template for protein synthesis during translation | 1–5% | 1961 Nobel Prize in Physiology or Medicine to François Jacob and Jacques Monod for discovering mRNA’s role in protein synthesis |
| Transfer RNA (tRNA) | Carries amino acids to the ribosome during translation | 10–15% | 1968 Nobel Prize in Physiology or Medicine to Robert W. Holley, Har Gobind Khorana, and Marshall W. Nirenberg for their interpretation of the genetic code and its function in protein synthesis, including tRNA discovery |
| RNA as a Catalyst | Catalysis function of RNA | n/a | 1989: Sidney Altman and Thomas R. Cech for discovering that RNA can act as a catalyst, leading to the understanding of ribozymes |
| Small Nuclear RNA (snRNA) | Involved in the splicing of pre-mRNA by forming the spliceosome complex | <0.1% | 1993 Nobel Prize in Physiology or Medicine to Richard J. Roberts and Phillip A. Sharp for discovering split genes and RNA splicing involving snRNA |
| Small Interfering RNA (siRNA) | Involved in RNA interference, degrading complementary mRNA to regulate gene expression; primarily involved in defense against viruses | <0.1% | 2006 Nobel Prize in Physiology or Medicine to Andrew Fire and Craig Mello for discovering RNA interference (RNAi), of which siRNA is a significant component |
| Ribosomal RNA (rRNA) | Forms the structural and catalytic components of ribosomes, essential for protein synthesis | 80–90% | 2009 Nobel Prize in Chemistry to Venkatraman Ramakrishnan, Thomas A. Steitz, and Ada E. Yonath for studies of the structure and function of the ribosome |
| CRISPR-Cas9 using RNA | Gene-editing technology using RNA to guide DNA modification | n/a | 2020: Jennifer A. Doudna and Emmanuelle Charpentier for developing CRISPR-Cas9, a gene-editing technology |
| MicroRNA [1] | It regulates gene expression by binding to target mRNAs and promoting their degradation or inhibiting translation. | <0.1% | 2024 Nobel Prize in Physiology or Medicine to Victor Ambros and Gary Ruvkun for the discovery of microRNA and its role in posttranscriptional gene regulation, revealing a new layer of gene regulation that impacts development and disease |
| Long Noncoding RNA (lncRNA) | Regulates gene expression at various levels, including chromatin remodeling, transcription, and posttranscriptional processing | <1% | No Nobel Prize directly for lncRNA, though it is a rapidly evolving field. |
| Circular RNA (circRNA) | Acts as an miRNA sponge and may regulate gene expression; it is stable because of its circular structure. | <1% | No Nobel Prize is related to circRNA yet, but research is ongoing and rapidly evolving. |
| Piwi-Interacting RNA (piRNA) | Protects the germline by silencing transposable elements; mainly found in reproductive cells | Variable, from <1% to higher than 1% | No Nobel Prize specific to piRNA, but RNA-interference-related discoveries were recognized by the 2006 Nobel Prize. |
| Small Nucleolar RNA (snoRNA) | Guides chemical modifications of other RNAs, particularly rRNA | <0.1% | No Nobel Prize was specific to snoRNA, but it was linked to rRNA function, and it was recognized by the 2009 Nobel Prize in Chemistry for ribosome research. |
| Antisense RNA (asRNA) | Inhibits gene expression by binding to complementary mRNA, preventing translation or inducing degradation | <0.1% | There is no Nobel Prize specifically for antisense RNA, but the field of RNA-based gene silencing has greatly expanded based on earlier work on RNA interference. |
| Database Name | Hyperlink | Description |
|---|---|---|
| miRBase | https://www.mirbase.org/ (accessed on 12 October 2024) | A primary miRNA sequence database that provides annotations, references, and information about miRNA families |
| miRTarBase | https://mirtarbase.cuhk.edu.cn/ (accessed on 12 October 2024) | Contains information on experimentally validated miRNA–target interactions compiled from the literature |
| TargetScan | https://www.targetscan.org/ (accessed on 12 October 2024) | Provides predicted miRNA targets based on conserved binding sites and includes information about miRNA conservation |
| miRDB | http://mirdb.org/ (accessed on 12 October 2024) | A database for predicted miRNA targets using a machine learning approach, with options to query miRNA functions |
| DIANA-miRPath | http://diana.imis.athena-innovation.gr/DianaTools/index.php (accessed on 12 October 2024) | Provides pathway-based miRNA functional analysis by linking miRNA target genes to known pathways |
| HMDD (Human MicroRNA Disease Database) | https://www.cuilab.cn/hmdd (accessed on 12 October 2024) | A manually curated database that collects miRNA–disease associations from the scientific literature |
| miRGator | http://mirgator.kobic.re.kr/ (accessed on 12 October 2024) | An miRNA analysis tool that integrates expression data, functional annotation, and predicted targets |
| miREnvironment | https://www.tools4mirs.org/ (accessed on 12 October 2024) | Focuses on miRNA–environment interactions, allowing users to explore how miRNAs are affected by environmental factors |
| mir2Disease | http://www.mir2disease.org/ (accessed on 12 October 2024) | A curated database that links miRNAs to various human diseases based on experimental evidence |
| OncomiRDB | http://lifeome.net/database/oncomirdb/ (accessed on 12 October 2024) | A specialized database that focuses on miRNAs implicated in cancer biology and therapeutics |
| miRCancer | http://mircancer.ecu.edu/ (accessed on 12 October 2024) | Focuses on miRNAs and their involvement in various types of cancer, with data on expression and regulation |
| miRGeneDB | https://mirgenedb.org/ (accessed on 12 October 2024) | A database providing curated annotations of miRNA genes across different species, focusing on high-quality annotations |
| Criteria | Ex Vivo Manipulation | In Vivo Administration |
|---|---|---|
| Precision and Control | High degree of precision and control because of manipulation in a lab setting | Less control: delivery systems face challenges in targeting specific tissues. |
| Safety and Off-Target Effects | There is a lower risk of off-target effects; cells are screened before reintroduction. | Higher risk of off-target effects and immune responses due to systemic delivery |
| Efficiency and Practicality | It is time consuming, expensive, and more challenging to scale for large populations. | Faster and more accessible to administer; more practical for large-scale applications |
| Applications | Best for cell-based therapies, regenerative medicine, and personalized treatments | Suitable for systemic diseases, cancer therapy, and infectious diseases |
| Regulus Therapeutics |
| RG-012: An miRNA inhibitor targeting miR-21, developed for treating Alport syndrome, a genetic kidney disease. It is currently in Phase II clinical trials. |
| RG-125 (AZD4076): Developed in collaboration with AstraZeneca, targeting miR-103/107 to treat non-alcoholic fatty liver disease (NAFLD) |
| miRagen Therapeutics |
| Cobomarsen (MRG-106): An anti-miR-155 therapy targeting cutaneous T-cell lymphoma (CTCL); it has reached Phase II clinical trials. |
| Remlarsen (MRG-201): An miRNA mimic of miR-29, designed to prevent fibrotic diseases, such as pulmonary and skin fibrosis |
| Alnylam Pharmaceuticals |
| Onpattro (Patisiran): An FDA-approved siRNA-based transthyretin-mediated amyloidosis (ATTR) therapy |
| ALN-PCSsc: An experimental therapy targeting PCSK9 for hypercholesterolemia. |
| Dicerna Pharmaceuticals |
| DCR-PHXC is an RNAi therapeutic targeting HAO1 for primary hyperoxaluria in Phase III trials. |
| DCR-HBVS: An RNAi therapeutic for hepatitis B (HBV) in Phase I/II trials |
| Santaris Pharma (now a part of Roche) |
| Santaris developed miravirsen (SPC3649), an anti-miR-122 therapy for Hepatitis C. It was among the first miRNA-based therapies to enter clinical trials, reaching Phase II. |
| Rosetta Genomics |
| Diagnostic tools using miRNA biomarkers; its miRview® assays are used for the diagnosis of cancers, like lung cancer and mesothelioma. |
| MiRXES |
| GASTROClear, an miRNA biomarker assay for the early detection of gastric cancer. This diagnostic tool has been commercialized in Asia and is considered as a leading non-invasive test for gastric cancer screening. |
| Gene Signal |
| Aganirsen is an anti-miR-21 therapy aimed at treating ocular neovascularization, a process involved in diseases like age-related macular degeneration. |
| Hummingbird Bioscience |
| HMBD-001: This product targets HER3-driven cancers, utilizing RNA interference (RNAi) technology, which modulates gene expression through siRNA or miRNA. This is currently in the preclinical stage. |
| Storm Therapeutics |
| STC-15: An RNA-based therapy targeting specific miRNAs involved in cancer progression. This product is still in the preclinical development stage. |
| Marina Biotech |
| CEQ508: This is an miR-34 mimic designed to treat familial adenomatous polyposis (FAP), a condition that increases the risk for developing colorectal cancer. It is currently in Phase I clinical trials. |
| Silence Therapeutics |
| SLN360: This product is designed to target lipoprotein(a), a known risk factor for cardiovascular diseases, using siRNA-based gene-silencing technology. It is currently in Phase I clinical trials. |
| Viridian Therapeutics |
| VRDN-001: This siRNA-based therapy is in the preclinical phase and aims to treat autoimmune diseases, such as thyroid eye disease, by modulating RNA pathways. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niazi, S.K.; Magoola, M. MicroRNA Nobel Prize: Timely Recognition and High Anticipation of Future Products—A Prospective Analysis. Int. J. Mol. Sci. 2024, 25, 12883. https://doi.org/10.3390/ijms252312883
Niazi SK, Magoola M. MicroRNA Nobel Prize: Timely Recognition and High Anticipation of Future Products—A Prospective Analysis. International Journal of Molecular Sciences. 2024; 25(23):12883. https://doi.org/10.3390/ijms252312883
Chicago/Turabian StyleNiazi, Sarfaraz K., and Matthias Magoola. 2024. "MicroRNA Nobel Prize: Timely Recognition and High Anticipation of Future Products—A Prospective Analysis" International Journal of Molecular Sciences 25, no. 23: 12883. https://doi.org/10.3390/ijms252312883
APA StyleNiazi, S. K., & Magoola, M. (2024). MicroRNA Nobel Prize: Timely Recognition and High Anticipation of Future Products—A Prospective Analysis. International Journal of Molecular Sciences, 25(23), 12883. https://doi.org/10.3390/ijms252312883








