Impact of Short-Term Exposure to Non-Functionalized Polystyrene Nanoparticles on DNA Methylation and Gene Expression in Human Peripheral Blood Mononuclear Cells
Abstract
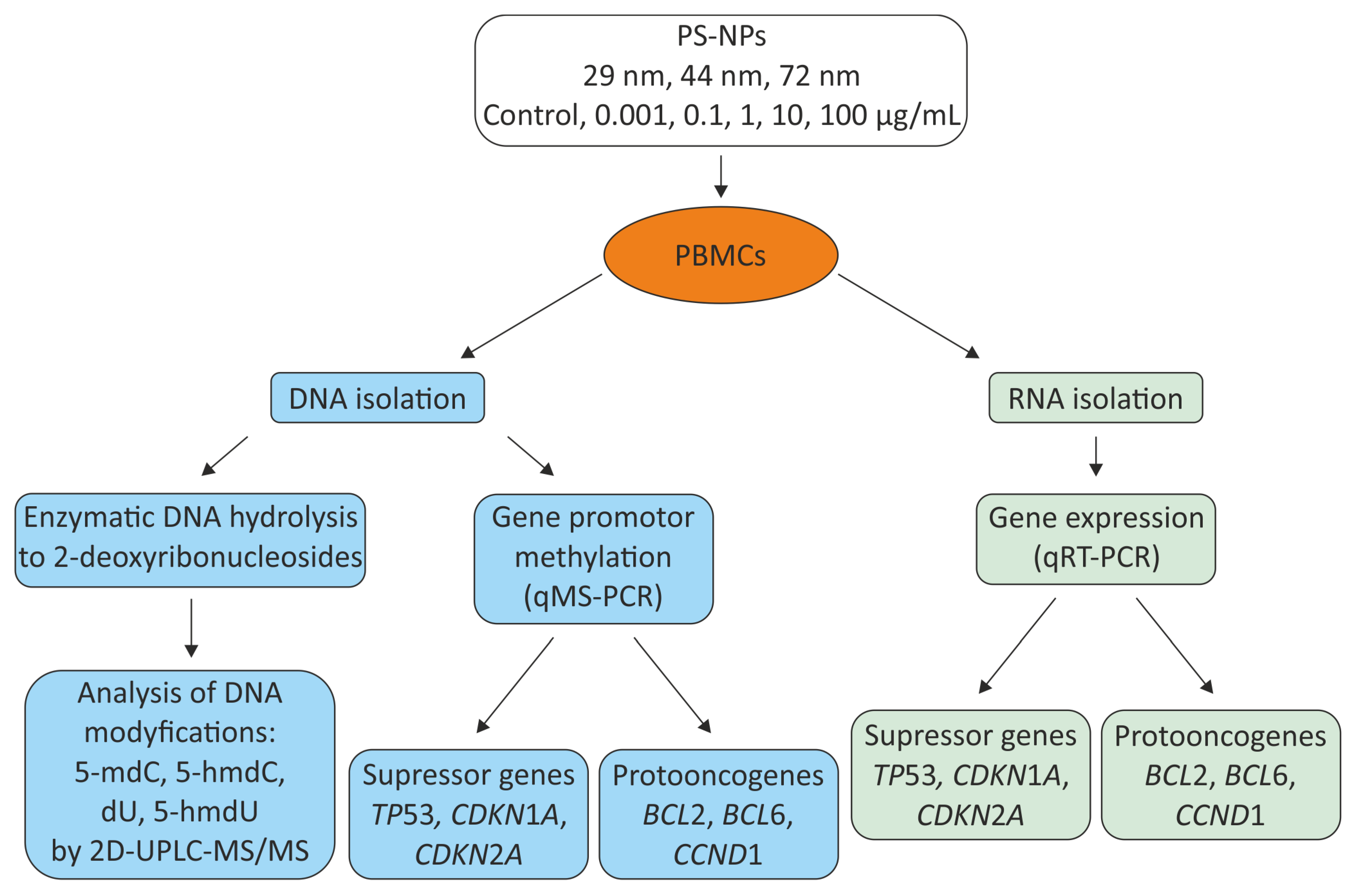
1. Introduction
2. Results
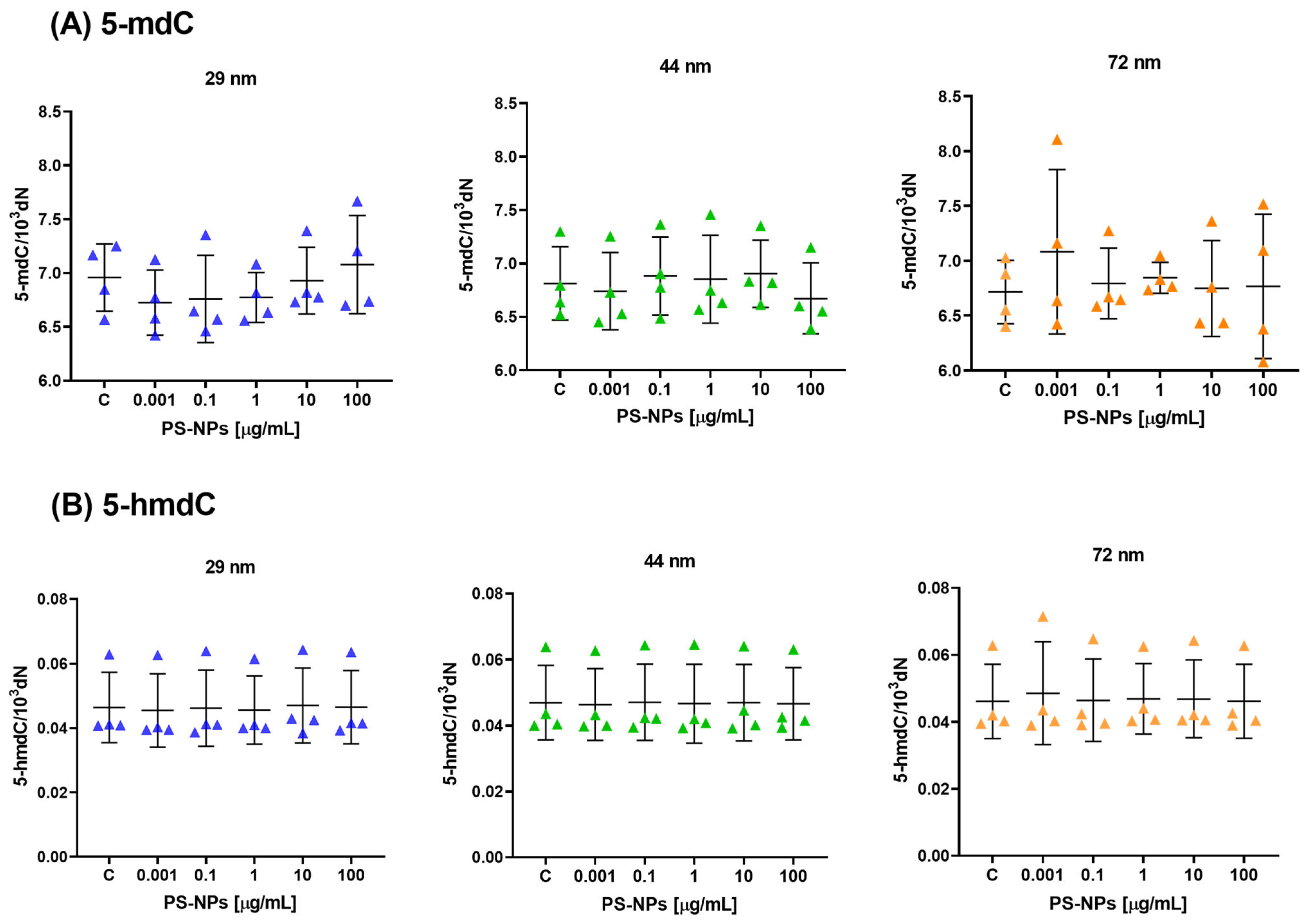
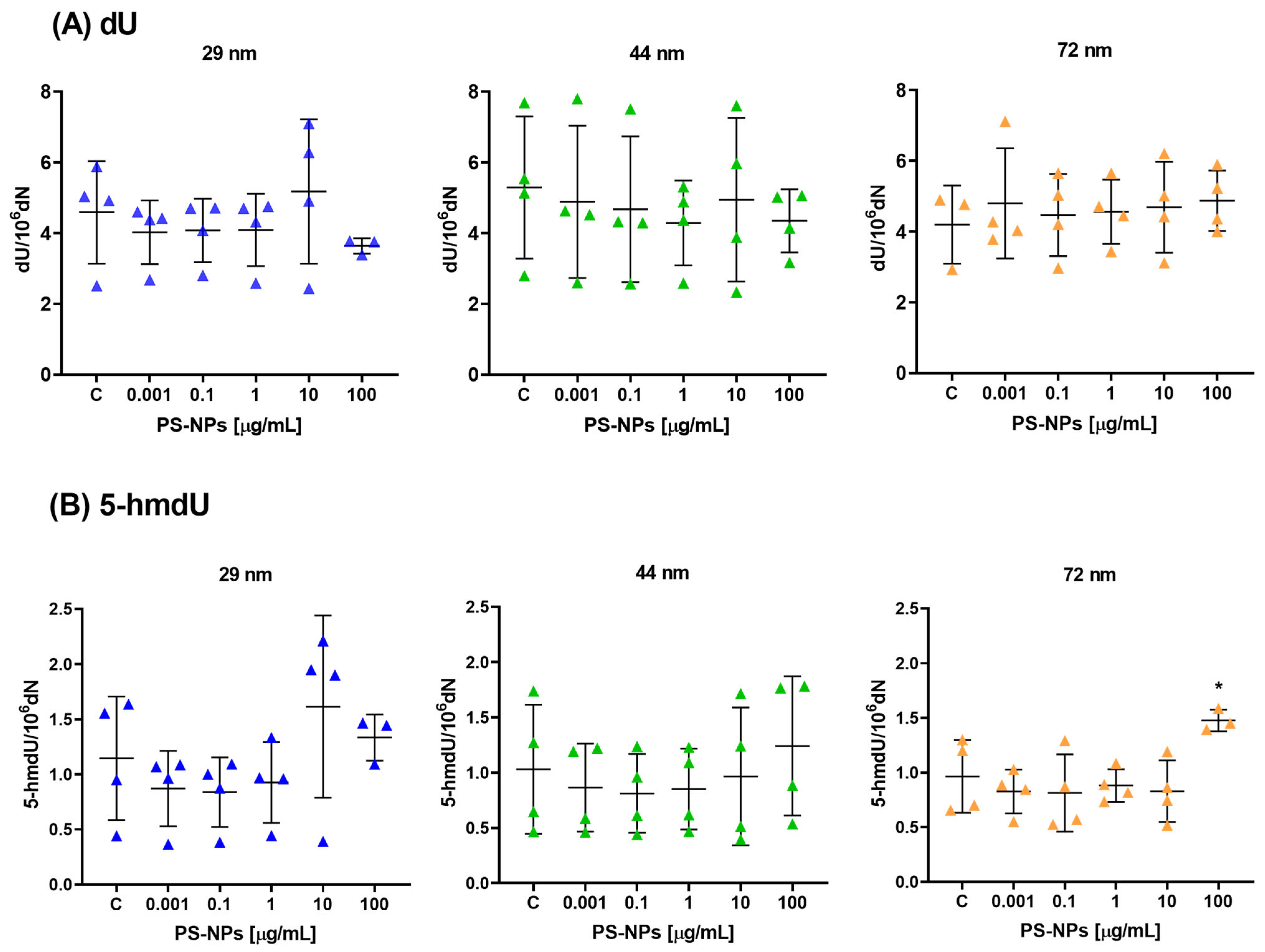
2.1. Effect of PS-NPs on DNA Methylation Determinants (5-mdC, 5-hmdC, dU and 5-hmdU) in Peripheral Blood Mononuclear Cells
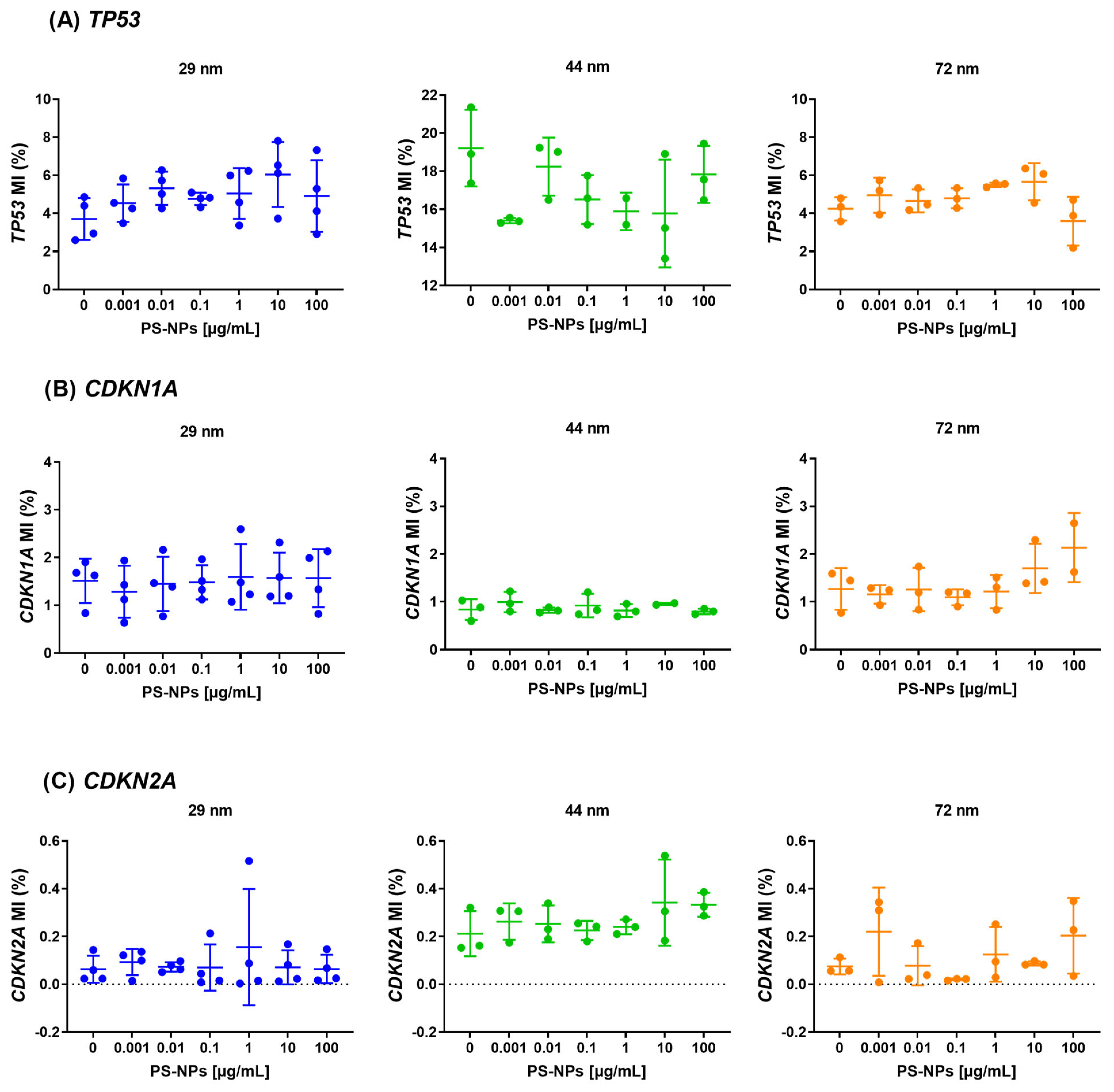
2.2. Analysis of Methylation at Promoter Regions of the Selected Tumor Suppressor Genes
2.3. Analysis of Methylation at Promoter Regions of the Selected Proto-Oncogenes
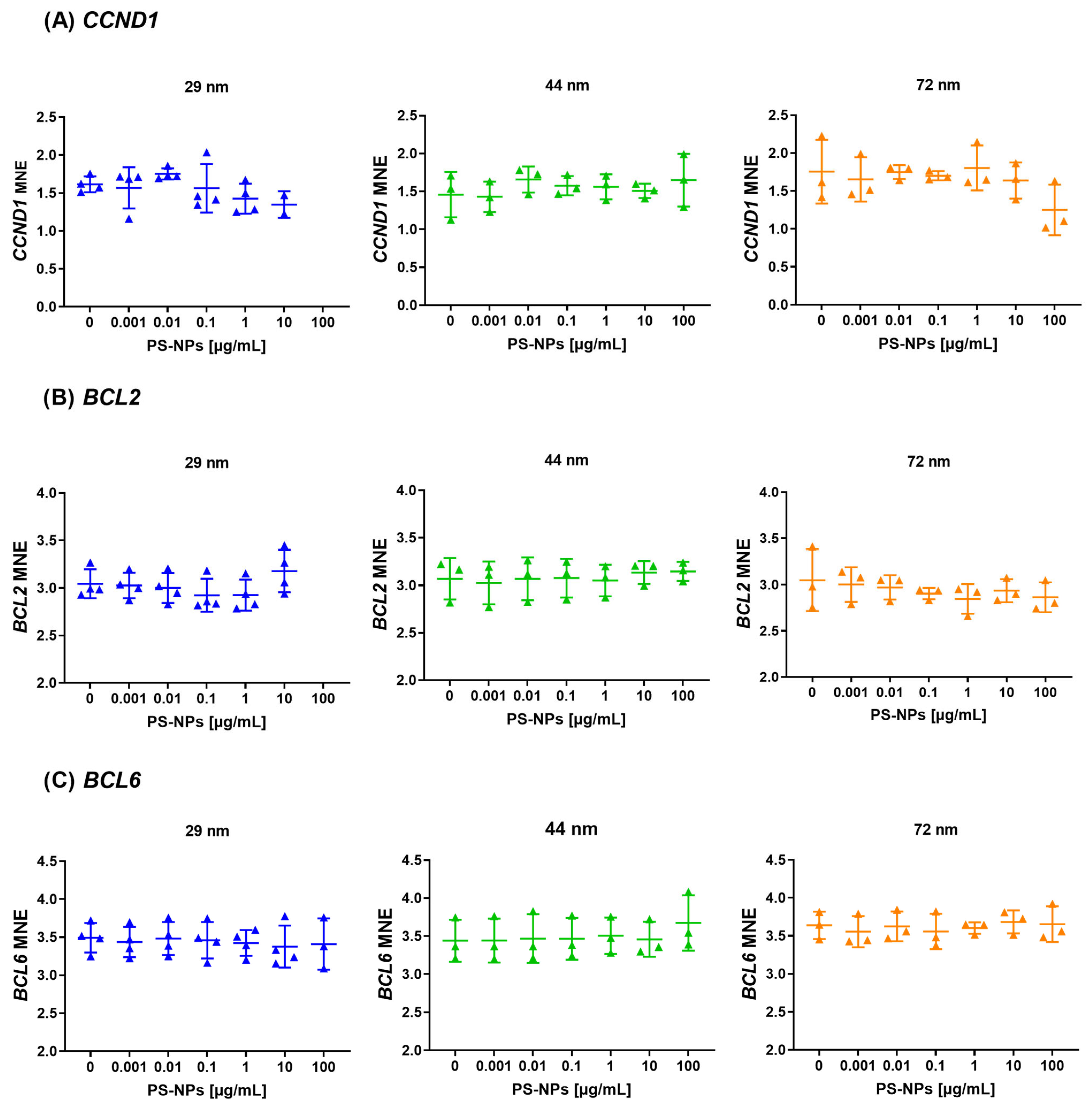
2.4. Analysis of Gene Expression of the Selected Tumor Suppressor Genes
2.5. Analysis of Gene Expression of the Selected Proto-Oncogenes
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Biological Material
4.3. Cells Treatment
4.4. Physico-Chemical Characterization of Non-Functionalized PS-NPs
4.5. DNA Isolation and Enzymatic Hydrolysis to Deoxyribonucleosides
4.6. Determination of Epigenetic Modifications in DNA Isolated from Cells Exposed to PS-NPs
4.7. Methylation at Promoter Regions of Tumor Suppressor Genes (TP53, CDKN2A and CDKN1A) and Proto-Oncogenes (CCND1, BCL2, BCL6)
4.8. Gene Expression
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- OECD OECD Work on Plastics. 2023. Available online: https://www.oecd.org/environment/plastics/ (accessed on 1 October 2024).
- Leslie, H.A.; Brandsma, S.H.; Van Velzen, M.J.M.; Vethaak, A.D. Microplastics En Route: Field Measurements in the Dutch River Delta and Amsterdam Canals, Wastewater Treatment Plants, North Sea Sediments and Biota. Environ. Int. 2017, 101, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Mihai, F.-C.; Gündoğdu, S.; Markley, L.A.; Olivelli, A.; Khan, F.R.; Gwinnett, C.; Gutberlet, J.; Reyna-Bensusan, N.; Llanquileo-Melgarejo, P.; Meidiana, C.; et al. Plastic Pollution, Waste Management Issues, and Circular Economy Opportunities in Rural Communities. Sustainability 2021, 14, 20. [Google Scholar] [CrossRef]
- Albazoni, H.J.; Al-Haidarey, M.J.S.; Nasir, A.S. A Review of Microplastic Pollution: Harmful Effect on Environment and Animals, Remediation Strategies. J. Ecol. Eng. 2024, 25, 140–157. [Google Scholar] [CrossRef] [PubMed]
- Kik, K.; Bukowska, B.; Sicińska, P. Polystyrene Nanoparticles: Sources, Occurrence in the Environment, Distribution in Tissues, Accumulation and Toxicity to Various Organisms. Environ. Pollut. 2020, 262, 114297. [Google Scholar] [CrossRef]
- International Organization for Standardization Plastics. Environmental Aspects. State of Knowledge and Methodologies. 2023. Available online: https://www.iso.org/obp/ui/en/#iso:std:iso:tr:21960:ed-1:v1:en (accessed on 10 June 2022).
- UE Definition of a Nanomaterial. 2017. Available online: https://environment.ec.europa.eu/news/chemicals-commission-revises-definition-nanomaterials-2022-06-10_en (accessed on 10 June 2022).
- Barboza, L.G.A.; Dick Vethaak, A.; Lavorante, B.R.B.O.; Lundebye, A.-K.; Guilhermino, L. Marine Microplastic Debris: An Emerging Issue for Food Security, Food Safety and Human Health. Mar. Pollut. Bull. 2018, 133, 336–348. [Google Scholar] [CrossRef]
- Chae, Y.; Kim, D.; Kim, S.W.; An, Y.-J. Trophic Transfer and Individual Impact of Nano-Sized Polystyrene in a Four-Species Freshwater Food Chain. Sci. Rep. 2018, 8, 284. [Google Scholar] [CrossRef]
- Cocci, P.; Gabrielli, S.; Pastore, G.; Minicucci, M.; Mosconi, G.; Palermo, F.A. Microplastics Accumulation in Gastrointestinal Tracts of Mullus Barbatus and Merluccius Merluccius Is Associated with Increased Cytokine Production and Signaling. Chemosphere 2022, 307, 135813. [Google Scholar] [CrossRef]
- Jenner, L.C.; Rotchell, J.M.; Bennett, R.T.; Cowen, M.; Tentzeris, V.; Sadofsky, L.R. Detection of Microplastics in Human Lung Tissue Using μFTIR Spectroscopy. Sci. Total Environ. 2022, 831, 154907. [Google Scholar] [CrossRef]
- Salvia, R.; Rico, L.G.; Bradford, J.A.; Ward, M.D.; Olszowy, M.W.; Martínez, C.; Madrid-Aris, Á.D.; Grífols, J.R.; Ancochea, Á.; Gomez-Muñoz, L.; et al. Fast-Screening Flow Cytometry Method for Detecting Nanoplastics in Human Peripheral Blood. MethodsX 2023, 10, 102057. [Google Scholar] [CrossRef]
- Wu, J.; Shao, Y.; Hua, X.; Wang, Y.; Wang, D. Nanoplastic at Environmentally Relevant Concentrations Induces Toxicity across Multiple Generations Associated with Inhibition in Germline G Protein-Coupled Receptor CED-1 in Caenorhabditis Elegans. Chemosphere 2024, 364, 143011. [Google Scholar] [CrossRef]
- Deville, S.; Penjweini, R.; Smisdom, N.; Notelaers, K.; Nelissen, I.; Hooyberghs, J.; Ameloot, M. Intracellular Dynamics and Fate of Polystyrene Nanoparticles in A549 Lung Epithelial Cells Monitored by Image (Cross-) Correlation Spectroscopy and Single Particle Tracking. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2015, 1853, 2411–2419. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.-S.; Son, Y.; Kim, S.S.; Shin, D.-S.; Lim, S.H.; Yang, J.Y.; Jeong, H.N.; Lee, B.H.; Bae, M.A. Size-Dependent Effects of Polystyrene Nanoparticles (PS-NPs) on Behaviors and Endogenous Neurochemicals in Zebrafish Larvae. Int. J. Mol. Sci. 2022, 23, 10682. [Google Scholar] [CrossRef] [PubMed]
- Walczak, A.P.; Kramer, E.; Hendriksen, P.J.M.; Tromp, P.; Helsper, J.P.F.G.; Van Der Zande, M.; Rietjens, I.M.C.M.; Bouwmeester, H. Translocation of Differently Sized and Charged Polystyrene Nanoparticles in in Vitro Intestinal Cell Models of Increasing Complexity. Nanotoxicology 2015, 9, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Peng, C.; Xu, H.; Su, Z.; Yilihan, G.; Wei, X.; Shen, Y.; Jiang, C. Microplastics from Disposable Paper Cups Are Enriched in the Placenta and Fetus, Leading to Metabolic and Reproductive Toxicity during Pregnancy. bioRxiv 2024. [Google Scholar] [CrossRef]
- González-Caballero, M.C.; De Alba González, M.; Torres-Ruiz, M.; Iglesias-Hernández, P.; Zapata, V.; Terrón, M.C.; Sachse, M.; Morales, M.; Martin-Folgar, R.; Liste, I.; et al. Internalization and Toxicity of Polystyrene Nanoplastics on Inmortalized Human Neural Stem Cells. Chemosphere 2024, 355, 141815. [Google Scholar] [CrossRef]
- Shan, S.; Zhang, Y.; Zhao, H.; Zeng, T.; Zhao, X. Polystyrene Nanoplastics Penetrate across the Blood-Brain Barrier and Induce Activation of Microglia in the Brain of Mice. Chemosphere 2022, 298, 134261. [Google Scholar] [CrossRef]
- Mamun, A.A.; Prasetya, T.A.E.; Dewi, I.R.; Ahmad, M. Microplastics in Human Food Chains: Food Becoming a Threat to Health Safety. Sci. Total Environ. 2023, 858, 159834. [Google Scholar] [CrossRef]
- Chen, G.; Shan, H.; Xiong, S.; Zhao, Y.; Van Gestel, C.A.M.; Qiu, H.; Wang, Y. Polystyrene Nanoparticle Exposure Accelerates Ovarian Cancer Development in Mice by Altering the Tumor Microenvironment. Sci. Total Environ. 2024, 906, 167592. [Google Scholar] [CrossRef]
- Zarus, G.M.; Muianga, C.; Brenner, S.; Stallings, K.; Casillas, G.; Pohl, H.R.; Mumtaz, M.M.; Gehle, K. Worker Studies Suggest Unique Liver Carcinogenicity Potential of Polyvinyl Chloride Microplastics. Am. J. Ind. Med. 2023, 66, 1033–1047. [Google Scholar] [CrossRef]
- Cetin, M.; Demirkaya Miloglu, F.; Kilic Baygutalp, N.; Ceylan, O.; Yildirim, S.; Eser, G.; Gul, H.İ. Higher Number of Microplastics in Tumoral Colon Tissues from Patients with Colorectal Adenocarcinoma. Environ. Chem. Lett. 2023, 21, 639–646. [Google Scholar] [CrossRef]
- Li, S.; Keenan, J.I.; Shaw, I.C.; Frizelle, F.A. Could Microplastics Be a Driver for Early Onset Colorectal Cancer? Cancers 2023, 15, 3323. [Google Scholar] [CrossRef] [PubMed]
- Domenech, J.; Annangi, B.; Marcos, R.; Hernández, A.; Catalán, J. Insights into the Potential Carcinogenicity of Micro- and Nano-Plastics. Mutat. Res. Rev. Mutat. Res. 2023, 791, 108453. [Google Scholar] [CrossRef] [PubMed]
- Brynzak-Schreiber, E.; Schögl, E.; Bapp, C.; Cseh, K.; Kopatz, V.; Jakupec, M.A.; Weber, A.; Lange, T.; Toca-Herrera, J.L.; Del Favero, G.; et al. Microplastics Role in Cell Migration and Distribution during Cancer Cell Division. Chemosphere 2024, 353, 141463. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Hou, B.; Wang, Z.; Yang, Y. Polystyrene Microplastics Induce Mitochondrial Damage in Mouse GC-2 Cells. Ecotoxicol. Environ. Saf. 2022, 237, 113520. [Google Scholar] [CrossRef]
- He, Y.; Li, Z.; Xu, T.; Luo, D.; Chi, Q.; Zhang, Y.; Li, S. Polystyrene Nanoplastics Deteriorate LPS-Modulated Duodenal Permeability and Inflammation in Mice via ROS Drived-NF-κB/NLRP3 Pathway. Chemosphere 2022, 307, 135662. [Google Scholar] [CrossRef]
- Zhang, Y.; Jia, Z.; Gao, X.; Zhao, J.; Zhang, H. Polystyrene Nanoparticles Induced Mammalian Intestine Damage Caused by Blockage of BNIP3/NIX-Mediated Mitophagy and Gut Microbiota Alteration. Sci. Total Environ. 2024, 907, 168064. [Google Scholar] [CrossRef]
- Ballesteros, S.; Domenech, J.; Barguilla, I.; Cortés, C.; Marcos, R.; Hernández, A. Genotoxic and Immunomodulatory Effects in Human White Blood Cells after Ex Vivo Exposure to Polystyrene Nanoplastics. Environ. Sci. Nano 2020, 7, 3431–3446. [Google Scholar] [CrossRef]
- Malinowska, K.; Bukowska, B.; Piwoński, I.; Foksiński, M.; Kisielewska, A.; Zarakowska, E.; Gackowski, D.; Sicińska, P. Polystyrene Nanoparticles: The Mechanism of Their Genotoxicity in Human Peripheral Blood Mononuclear Cells. Nanotoxicology 2022, 16, 791–811. [Google Scholar] [CrossRef]
- Rubio, L.; Barguilla, I.; Domenech, J.; Marcos, R.; Hernández, A. Biological Effects, Including Oxidative Stress and Genotoxic Damage, of Polystyrene Nanoparticles in Different Human Hematopoietic Cell Lines. J. Hazard. Mater. 2020, 398, 122900. [Google Scholar] [CrossRef]
- Hou, L.; Zhang, X.; Wang, D.; Baccarelli, A. Environmental Chemical Exposures and Human Epigenetics. Int. J. Epidemiol. 2012, 41, 79–105. [Google Scholar] [CrossRef]
- Riley, P.A. Epimutation and Cancer: Carcinogenesis Viewed as Error-Prone Inheritance of Epigenetic Information. J. Oncol. 2018, 2018, 2645095. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-M. Detecting DNA Hydroxymethylation: Exploring Its Role in Genome Regulation. BMB Rep. 2024, 57, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Nabel, C.S.; Jia, H.; Ye, Y.; Shen, L.; Goldschmidt, H.L.; Stivers, J.T.; Zhang, Y.; Kohli, R.M. AID/APOBEC Deaminases Disfavor Modified Cytosines Implicated in DNA Demethylation. Nat. Chem. Biol. 2012, 8, 751–758. [Google Scholar] [CrossRef] [PubMed]
- De Wispelaere, N.; Rico, S.D.; Bauer, M.; Luebke, A.M.; Kluth, M.; Büscheck, F.; Hube-Magg, C.; Höflmayer, D.; Gorbokon, N.; Weidemann, S.; et al. High Prevalence of P16 Staining in Malignant Tumors. PLoS ONE 2022, 17, e0262877. [Google Scholar] [CrossRef] [PubMed]
- Hatano, Y. The Pathology According to P53 Pathway. Pathobiology 2023, 91, 230–243. [Google Scholar] [CrossRef]
- Shamloo, B.; Usluer, S. P21 in Cancer Research. Cancers 2019, 11, 1178. [Google Scholar] [CrossRef]
- Kulaberoglu, Y.; Gundogdu, R.; Hergovich, A. The Role of P53/P21/P16 in DNA-Damage Signaling and DNA Repair. In Genome Stability; Elsevier: Amsterdam, The Netherlands, 2016; pp. 243–256. ISBN 978-0-12-803309-8. [Google Scholar]
- Szybińska, A.; Leśniakx, L. P53 Dysfunction in Neurodegenerative Diseases—The Cause or Effect of Pathological Changes? Aging Dis. 2017, 8, 506. [Google Scholar] [CrossRef]
- Kooijman, R.; Devos, S.; Hooghe-Peters, E. Inhibition of in Vitro Cytokine Production by Human Peripheral Blood Mononuclear Cells Treated with Xenobiotics: Implications for the Prediction of General Toxicity and Immunotoxicity. Toxicol. Vitr. 2010, 24, 1782–1789. [Google Scholar] [CrossRef]
- Chen, R.; Curran, J.; Pu, F.; Zhuola, Z.; Bayon, Y.; Hunt, J. In Vitro Response of Human Peripheral Blood Mononuclear Cells (PBMC) to Collagen Films Treated with Cold Plasma. Polymers 2017, 9, 254. [Google Scholar] [CrossRef]
- Sen, P.; Kemppainen, E.; Orešič, M. Perspectives on Systems Modeling of Human Peripheral Blood Mononuclear Cells. Front. Mol. Biosci. 2018, 4, 96. [Google Scholar] [CrossRef]
- Kwiatkowska, M.; Reszka, E.; Woźniak, K.; Jabłońska, E.; Michałowicz, J.; Bukowska, B. DNA Damage and Methylation Induced by Glyphosate in Human Peripheral Blood Mononuclear Cells (In Vitro Study). Food Chem. Toxicol. 2017, 105, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, E.; Reszka, E.; Jabłońska, E.; Balcerczyk, A.; Broncel, M.; Bukowska, B. Glyphosate Affects Methylation in the Promoter Regions of Selected Tumor Suppressors as Well as Expression of Major Cell Cycle and Apoptosis Drivers in PBMCs (In Vitro Study). Toxicol. Vitr. 2020, 63, 104736. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, E.; Reszka, E.; Jabłońska, E.; Michałowicz, J.; Huras, B.; Bukowska, B. Glyphosate and AMPA Induce Alterations in Expression of Genes Involved in Chromatin Architecture in Human Peripheral Blood Mononuclear Cells (In Vitro). Int. J. Mol. Sci. 2021, 22, 2966. [Google Scholar] [CrossRef] [PubMed]
- Sicińska, P.; Jabłońska, E.; Bukowska, B.; Balcerczyk, A.; Reszka, E. The Selected Epigenetic Effects of Phthalates: DBP, BBP and Their Metabolites: MBP, MBzP on Human Peripheral Blood Mononuclear Cells (In Vitro). Toxicol. Vitr. 2022, 82, 105369. [Google Scholar] [CrossRef] [PubMed]
- Kik, K.; Bukowska, B.; Krokosz, A.; Sicińska, P. Oxidative Properties of Polystyrene Nanoparticles with Different Diameters in Human Peripheral Blood Mononuclear Cells (In Vitro Study). Int. J. Mol. Sci. 2021, 22, 4406. [Google Scholar] [CrossRef]
- Woźniak, E.; Reszka, E.; Jabłońska, E.; Mokra, K.; Balcerczyk, A.; Huras, B.; Zakrzewski, J.; Bukowska, B. The Selected Epigenetic Effects of Aminomethylphosphonic Acid, a Primary Metabolite of Glyphosate on Human Peripheral Blood Mononuclear Cells (In Vitro). Toxicol. Vitr. 2020, 66, 104878. [Google Scholar] [CrossRef]
- Ramsperger, A.F.R.M.; Bergamaschi, E.; Panizzolo, M.; Fenoglio, I.; Barbero, F.; Peters, R.; Undas, A.; Purker, S.; Giese, B.; Lalyer, C.R.; et al. Nano- and Microplastics: A Comprehensive Review on Their Exposure Routes, Translocation, and Fate in Humans. NanoImpact 2023, 29, 100441. [Google Scholar] [CrossRef]
- Zhao, Y.; Hua, X.; Bian, Q.; Wang, D. Nanoplastic Exposure at Predicted Environmental Concentrations Induces Activation of Germline Ephrin Signal Associated with Toxicity Formation in the Caenorhabditis Elegans Offspring. Toxics 2022, 10, 699. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kim, M.-S.; Lee, Y.; Kim, D.-H.; Lee, J.-S. Nanoplastics Induce Epigenetic Signatures of Transgenerational Impairments Associated with Reproduction in Copepods under Ocean Acidification. J. Hazard. Mater. 2023, 449, 131037. [Google Scholar] [CrossRef]
- Stojkovic, M.; Ortuño Guzmán, F.M.; Han, D.; Stojkovic, P.; Dopazo, J.; Stankovic, K.M. Polystyrene Nanoplastics Affect Transcriptomic and Epigenomic Signatures of Human Fibroblasts and Derived Induced Pluripotent Stem Cells: Implications for Human Health. Environ. Pollut. 2023, 320, 120849. [Google Scholar] [CrossRef]
- Malinowska, K.; Sicińska, P.; Michałowicz, J.; Bukowska, B. The Effects of Non-Functionalized Polystyrene Nanoparticles of Different Diameters on the Induction of Apoptosis and mTOR Level in Human Peripheral Blood Mononuclear Cells. Chemosphere 2023, 335, 139137. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, M.; Zhang, Z.; Halimu, G.; Li, Y.; Li, Y.; Gu, W.; Zhang, B.; Wang, X. In Vitro Study on the Toxicity of Nanoplastics with Different Charges to Murine Splenic Lymphocytes. J. Hazard. Mater. 2022, 424, 127508. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.; O’Leary, C.; O’Byrne, K.J.; Burgess, J.; Richard, D.J.; Suraweera, A. Epigenetic Mechanisms in DNA Double Strand Break Repair: A Clinical Review. Front. Mol. Biosci. 2021, 8, 685440. [Google Scholar] [CrossRef] [PubMed]
- Brzóska, K.; Grądzka, I.; Kruszewski, M. Silver, Gold, and Iron Oxide Nanoparticles Alter miRNA Expression but Do Not Affect DNA Methylation in HepG2 Cells. Materials 2019, 12, 1038. [Google Scholar] [CrossRef]
- Pfaffeneder, T.; Spada, F.; Wagner, M.; Brandmayr, C.; Laube, S.K.; Eisen, D.; Truss, M.; Steinbacher, J.; Hackner, B.; Kotljarova, O.; et al. Tet Oxidizes Thymine to 5-Hydroxymethyluracil in Mouse Embryonic Stem Cell DNA. Nat. Chem. Biol. 2014, 10, 574–581. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Kumar, D.; Singh, D.; Singh, R.K. Modulation of the Epigenome by Xenobiotics in Cancer. In Xenobiotics in Chemical Carcinogenesis; Elsevier: Amsterdam, The Netherlands, 2022; pp. 127–153. ISBN 978-0-323-90560-2. [Google Scholar]
- Li, Y.; Fan, Z.; Meng, Y.; Liu, S.; Zhan, H. Blood-Based DNA Methylation Signatures in Cancer: A Systematic Review. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2023, 1869, 166583. [Google Scholar] [CrossRef]
- Raut, J.R.; Guan, Z.; Schrotz-King, P.; Brenner, H. Whole-Blood DNA Methylation Markers for Risk Stratification in Colorectal Cancer Screening: A Systematic Review. Cancers 2019, 11, 912. [Google Scholar] [CrossRef]
- Terry, M.B.; Delgado-Cruzata, L.; Vin-Raviv, N.; Wu, H.C.; Santella, R.M. DNA Methylation in White Blood Cells: Association with Risk Factors in Epidemiologic Studies. Epigenetics 2011, 6, 828–837. [Google Scholar] [CrossRef]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why Are There Hotspot Mutations in the TP53 Gene in Human Cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef]
- Alaraby, M.; Abass, D.; Domenech, J.; Hernández, A.; Marcos, R. Hazard Assessment of Ingested Polystyrene Nanoplastics in Drosophila Larvae. Environ. Sci. Nano 2022, 9, 1845–1857. [Google Scholar] [CrossRef]
- Banerjee, A.; Billey, L.O.; Shelver, W.L. Uptake and Toxicity of Polystyrene Micro/Nanoplastics in Gastric Cells: Effects of Particle Size and Surface Functionalization. PLoS ONE 2021, 16, e0260803. [Google Scholar] [CrossRef] [PubMed]
- Forte, M.; Iachetta, G.; Tussellino, M.; Carotenuto, R.; Prisco, M.; De Falco, M.; Laforgia, V.; Valiante, S. Polystyrene Nanoparticles Internalization in Human Gastric Adenocarcinoma Cells. Toxicol. Vitr. 2016, 31, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Tian, X.; Lin, X.; Chen, R.; Hameed, S.; Wang, L.; Yu, Y.-L.; Li, B.; Li, Y.-F. Nanoplastic-Induced Biological Effects In Vivo and In Vitro: An Overview. Rev. Environ. Contam. Toxicol. 2023, 261, 2. [Google Scholar] [CrossRef]
- Schmidt, A.; Da Silva Brito, W.A.; Singer, D.; Mühl, M.; Berner, J.; Saadati, F.; Wolff, C.; Miebach, L.; Wende, K.; Bekeschus, S. Short- and Long-Term Polystyrene Nano- and Microplastic Exposure Promotes Oxidative Stress and Divergently Affects Skin Cell Architecture and Wnt/Beta-Catenin Signaling. Part. Fibre Toxicol. 2023, 20, 3. [Google Scholar] [CrossRef] [PubMed]
- Chortarea, S.; Gupta, G.; Saarimäki, L.A.; Netkueakul, W.; Manser, P.; Aengenheister, L.; Wichser, A.; Fortino, V.; Wick, P.; Greco, D.; et al. Transcriptomic Profiling Reveals Differential Cellular Response to Copper Oxide Nanoparticles and Polystyrene Nanoplastics in Perfused Human Placenta. Environ. Int. 2023, 177, 108015. [Google Scholar] [CrossRef]
- Martin-Folgar, R.; González-Caballero, M.C.; Torres-Ruiz, M.; Cañas-Portilla, A.I.; De Alba González, M.; Liste, I.; Morales, M. Molecular Effects of Polystyrene Nanoplastics on Human Neural Stem Cells. PLoS ONE 2024, 19, e0295816. [Google Scholar] [CrossRef]
- Liang, Y.; Yang, Y.; Lu, C.; Cheng, Y.; Jiang, X.; Yang, B.; Li, Y.; Chen, Q.; Ao, L.; Cao, J.; et al. Polystyrene Nanoplastics Exposure Triggers Spermatogenic Cell Senescence via the Sirt1/ROS Axis. Ecotoxicol. Environ. Saf. 2024, 279, 116461. [Google Scholar] [CrossRef]
- Płuciennik, K.; Sicińska, P.; Duchnowicz, P.; Bonarska-Kujawa, D.; Męczarska, K.; Solarska-Ściuk, K.; Miłowska, K.; Bukowska, B. The Effects of Non-Functionalized Polystyrene Nanoparticles with Different Diameters on Human Erythrocyte Membrane and Morphology. Toxicol. Vitr. 2023, 91, 105634. [Google Scholar] [CrossRef]
- Stapleton, P. 1 Department of Pharmacology and Toxicology, Ernest Mario School of Pharmacy, Rutgers University, 160 Frelinghuysen Rd., Piscataway, NJ 08854, USA; 2 Environmental and Occupational Health Sciences Institute, 170 Frelinghuysen Rd., Piscataway, NJ 08854, USA Toxicological Considerations of Nano-Sized Plastics. AIMS Environ. Sci. 2019, 6, 367–378. [Google Scholar] [CrossRef]
- Divakar, K.J.; Reese, C.B. 4-(1,2,4-Triazol-1-Yl)- and 4-(3-Nitro-1,2,4-Triazol-1-Yl)-1-(β-D-2,3,5-Tri-O-Acetylarabinofuranosyl)Pyrimidin-2(1H)-Ones. Valuable Intermediates in the Synthesis of Derivatives of 1-(β-D-Arabinofuranosyl)Cytosine (Ara-C). J. Chem. Soc. Perkin Trans. 1 1982, 1, 1171–1176. [Google Scholar] [CrossRef]
- Abdel Rahman, A.A.-H.; Wada, T.; Saigo, K. Facile Methods for the Synthesis of 5-Formylcytidine. Tetrahedron Lett. 2001, 42, 1061–1063. [Google Scholar] [CrossRef]
- Woźniak, E.; Sicińska, P.; Michałowicz, J.; Woźniak, K.; Reszka, E.; Huras, B.; Zakrzewski, J.; Bukowska, B. The Mechanism of DNA Damage Induced by Roundup 360 PLUS, Glyphosate and AMPA in Human Peripheral Blood Mononuclear Cells—Genotoxic Risk Assessement. Food Chem. Toxicol. 2018, 120, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Sicińska, P.; Mokra, K.; Wozniak, K.; Michałowicz, J.; Bukowska, B. Genotoxic Risk Assessment and Mechanism of DNA Damage Induced by Phthalates and Their Metabolites in Human Peripheral Blood Mononuclear Cells. Sci. Rep. 2021, 11, 1658. [Google Scholar] [CrossRef] [PubMed]
- Mokra, K.; Woźniak, K.; Bukowska, B.; Sicińska, P.; Michałowicz, J. Low-Concentration Exposure to BPA, BPF and BPAF Induces Oxidative DNA Bases Lesions in Human Peripheral Blood Mononuclear Cells. Chemosphere 2018, 201, 119–126. [Google Scholar] [CrossRef]
- Guz, J.; Zarakowska, E.; Mijewski, P. Unprecedentedly High Level of Intracellular Vitamin C and DNA Epigenetic Marks in Prostate: Relevant for Male Fertility? Cell. Physiol. Biochem. 2023, 57, 200–211. [Google Scholar] [CrossRef]
- Gackowski, D.; Starczak, M.; Zarakowska, E.; Modrzejewska, M.; Szpila, A.; Banaszkiewicz, Z.; Olinski, R. Accurate, Direct, and High-Throughput Analyses of a Broad Spectrum of Endogenously Generated DNA Base Modifications with Isotope-Dilution Two-Dimensional Ultraperformance Liquid Chromatography with Tandem Mass Spectrometry: Possible Clinical Implication. Anal. Chem. 2016, 88, 12128–12136. [Google Scholar] [CrossRef]
- Starczak, M.; Gawronski, M.; Olinski, R.; Gackowski, D. Quantification of DNA Modifications Using Two-Dimensional Ultraperformance Liquid Chromatography Tandem Mass Spectrometry (2D-UPLC-MS/MS). In DNA Modifications; Ruzov, A., Gering, M., Eds.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2021; Volume 2198, pp. 91–108. ISBN 978-1-07-160875-3. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malinowska, K.; Tarhonska, K.; Foksiński, M.; Sicińska, P.; Jabłońska, E.; Reszka, E.; Zarakowska, E.; Gackowski, D.; Górecka, K.; Balcerczyk, A.; et al. Impact of Short-Term Exposure to Non-Functionalized Polystyrene Nanoparticles on DNA Methylation and Gene Expression in Human Peripheral Blood Mononuclear Cells. Int. J. Mol. Sci. 2024, 25, 12786. https://doi.org/10.3390/ijms252312786
Malinowska K, Tarhonska K, Foksiński M, Sicińska P, Jabłońska E, Reszka E, Zarakowska E, Gackowski D, Górecka K, Balcerczyk A, et al. Impact of Short-Term Exposure to Non-Functionalized Polystyrene Nanoparticles on DNA Methylation and Gene Expression in Human Peripheral Blood Mononuclear Cells. International Journal of Molecular Sciences. 2024; 25(23):12786. https://doi.org/10.3390/ijms252312786
Chicago/Turabian StyleMalinowska, Kinga, Kateryna Tarhonska, Marek Foksiński, Paulina Sicińska, Ewa Jabłońska, Edyta Reszka, Ewelina Zarakowska, Daniel Gackowski, Karolina Górecka, Aneta Balcerczyk, and et al. 2024. "Impact of Short-Term Exposure to Non-Functionalized Polystyrene Nanoparticles on DNA Methylation and Gene Expression in Human Peripheral Blood Mononuclear Cells" International Journal of Molecular Sciences 25, no. 23: 12786. https://doi.org/10.3390/ijms252312786
APA StyleMalinowska, K., Tarhonska, K., Foksiński, M., Sicińska, P., Jabłońska, E., Reszka, E., Zarakowska, E., Gackowski, D., Górecka, K., Balcerczyk, A., & Bukowska, B. (2024). Impact of Short-Term Exposure to Non-Functionalized Polystyrene Nanoparticles on DNA Methylation and Gene Expression in Human Peripheral Blood Mononuclear Cells. International Journal of Molecular Sciences, 25(23), 12786. https://doi.org/10.3390/ijms252312786





