Root Microbiome and Metabolome Traits Associated with Improved Post-Harvest Root Storage for Sugar Beet Breeding Lines Under Southern Idaho Conditions
Abstract
1. Introduction
2. Results
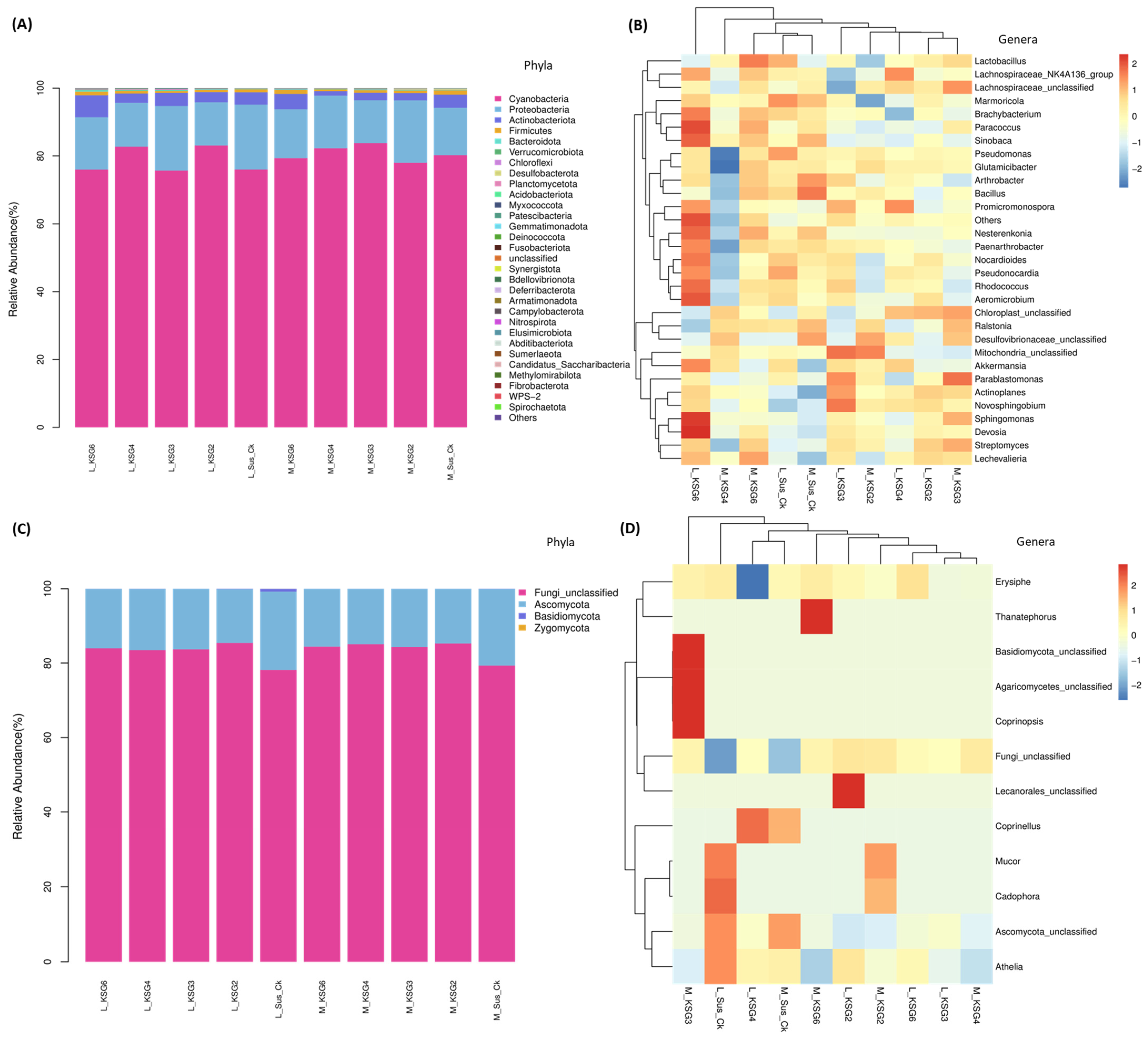
2.1. Bacterial and Fungal Phyla and Genera Showed Significant Changes Between the Resistant and Susceptible Lines and with Storage Times
2.2. Bacterial and Fungal Diversity in the Roots Varied Depending upon Genotype and Storage Stages
2.2.1. Alpha Diversity
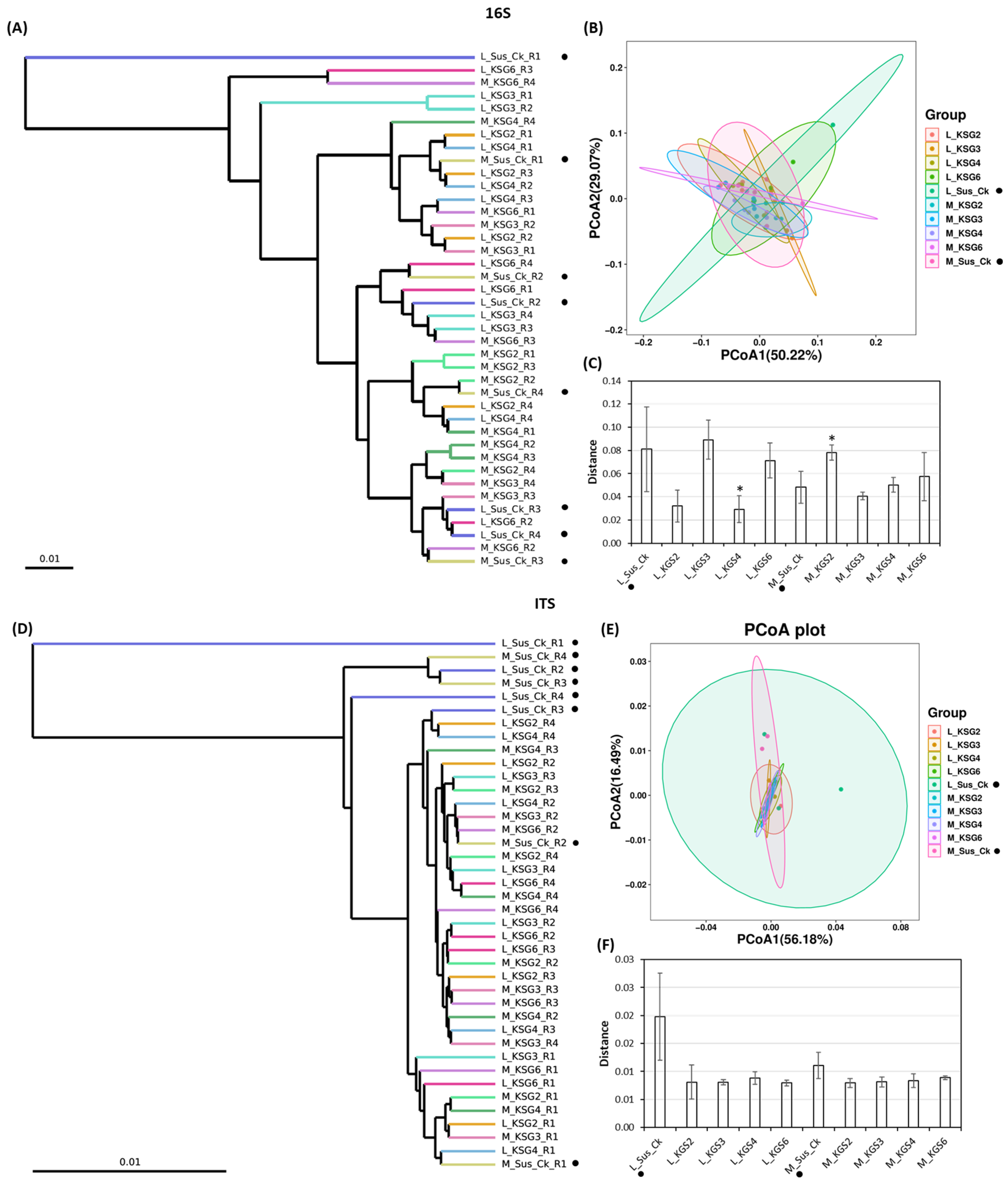
2.2.2. Beta Diversity
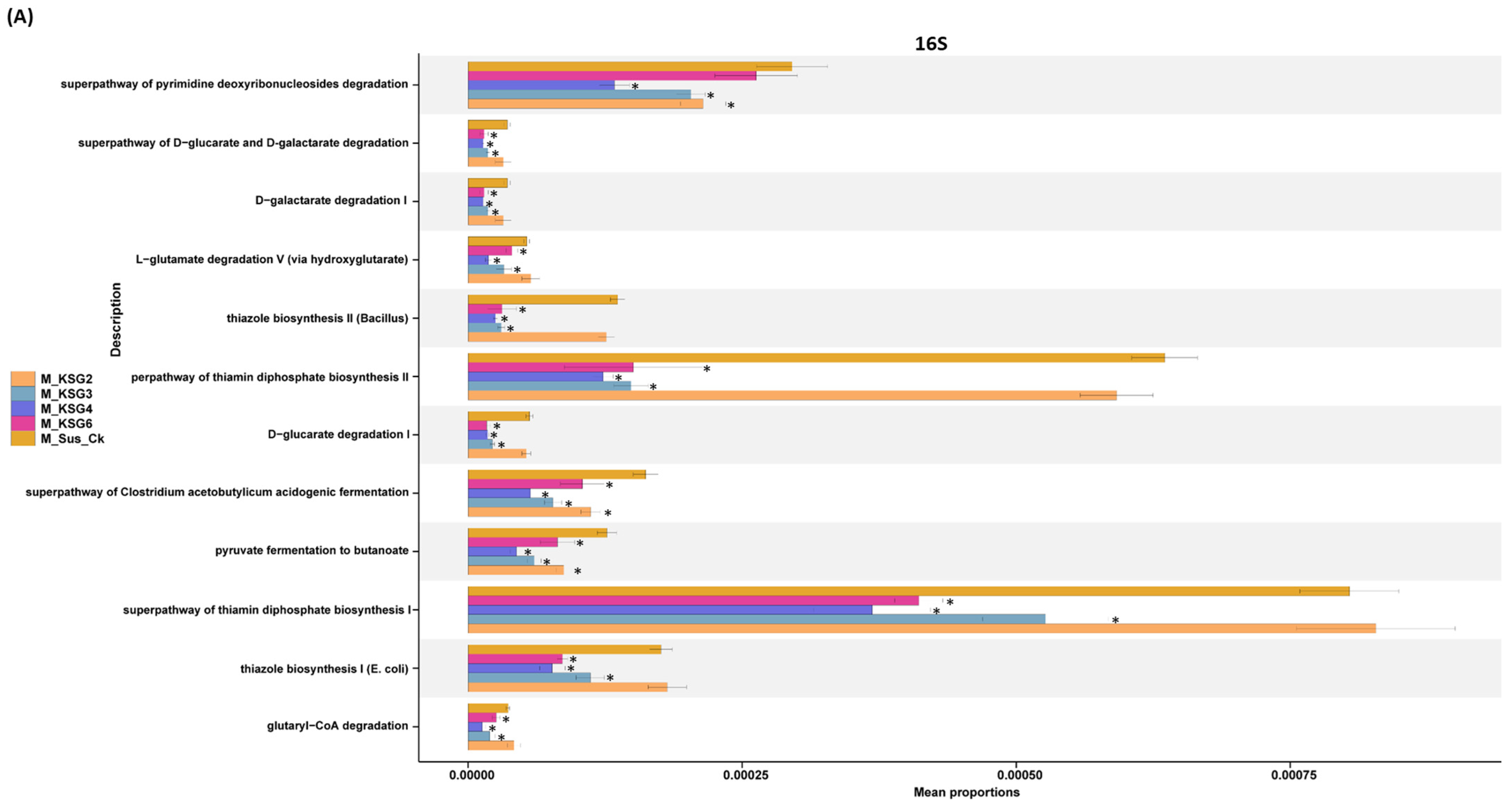
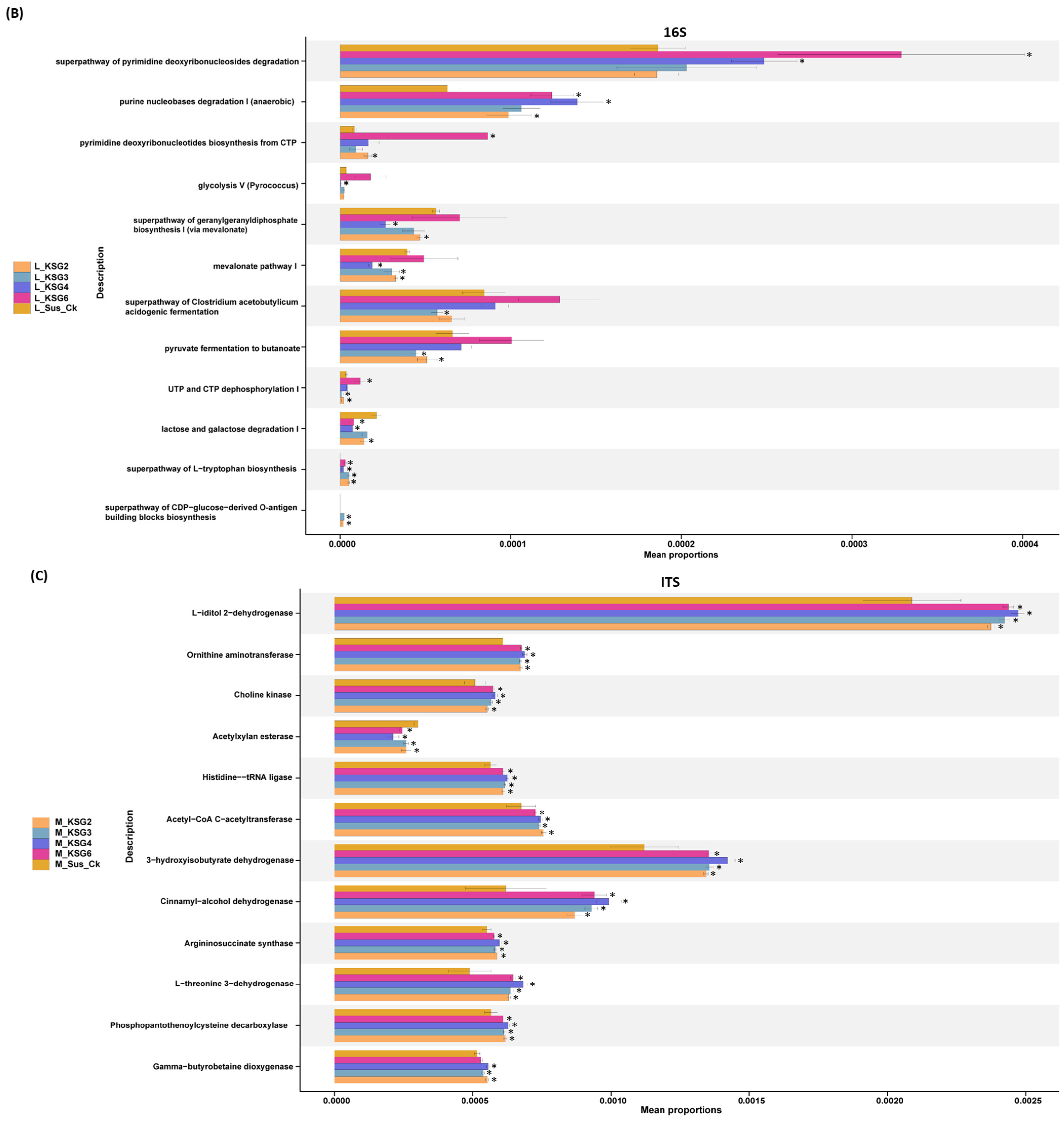
2.3. Pathway Enrichment Analysis of the Microbiome Data Showed Differential Regulation in the Resistant vs. Susceptible Lines
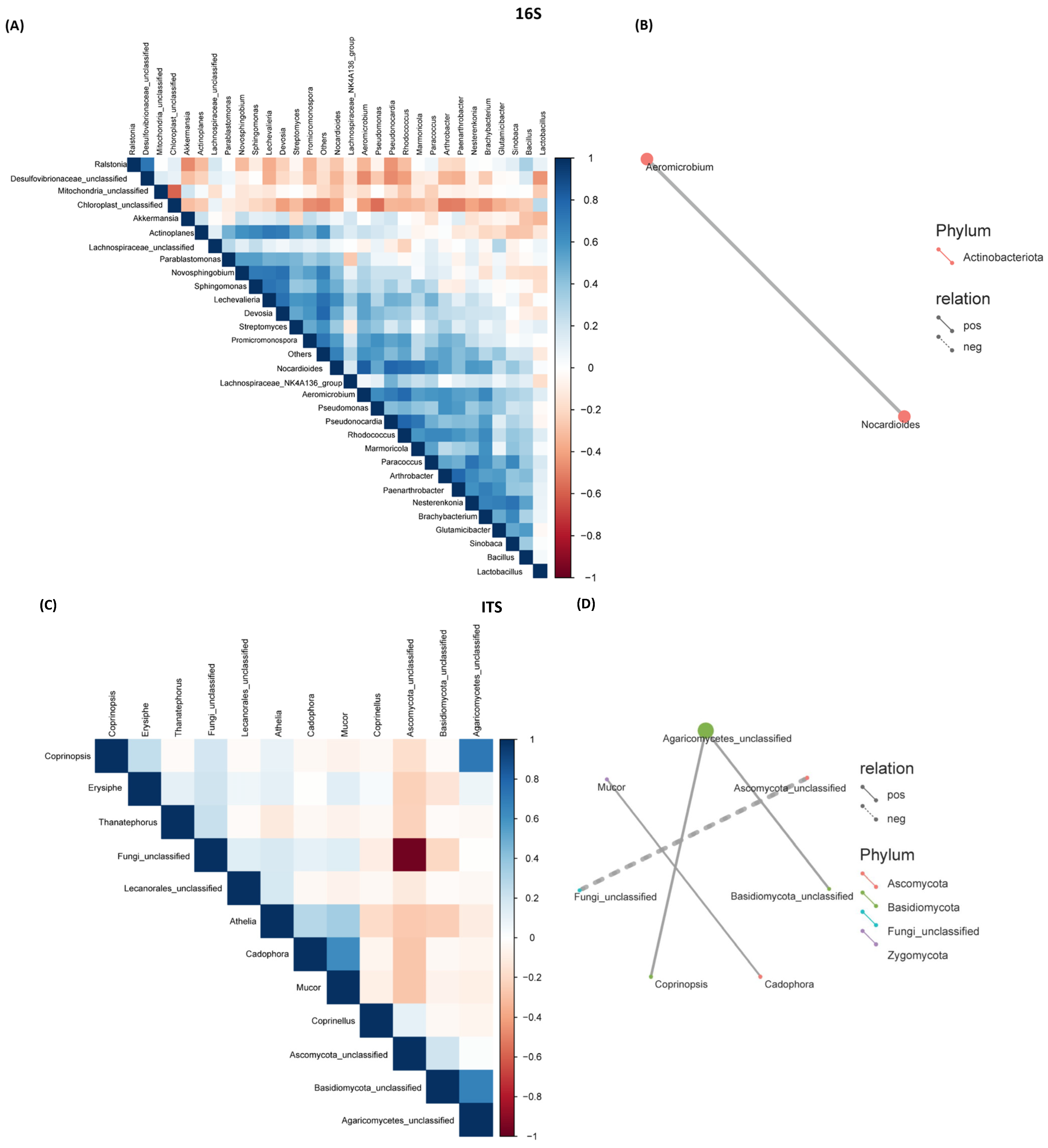
2.4. Correlation Between Bacterial and Fungal Communities Across Genotypes
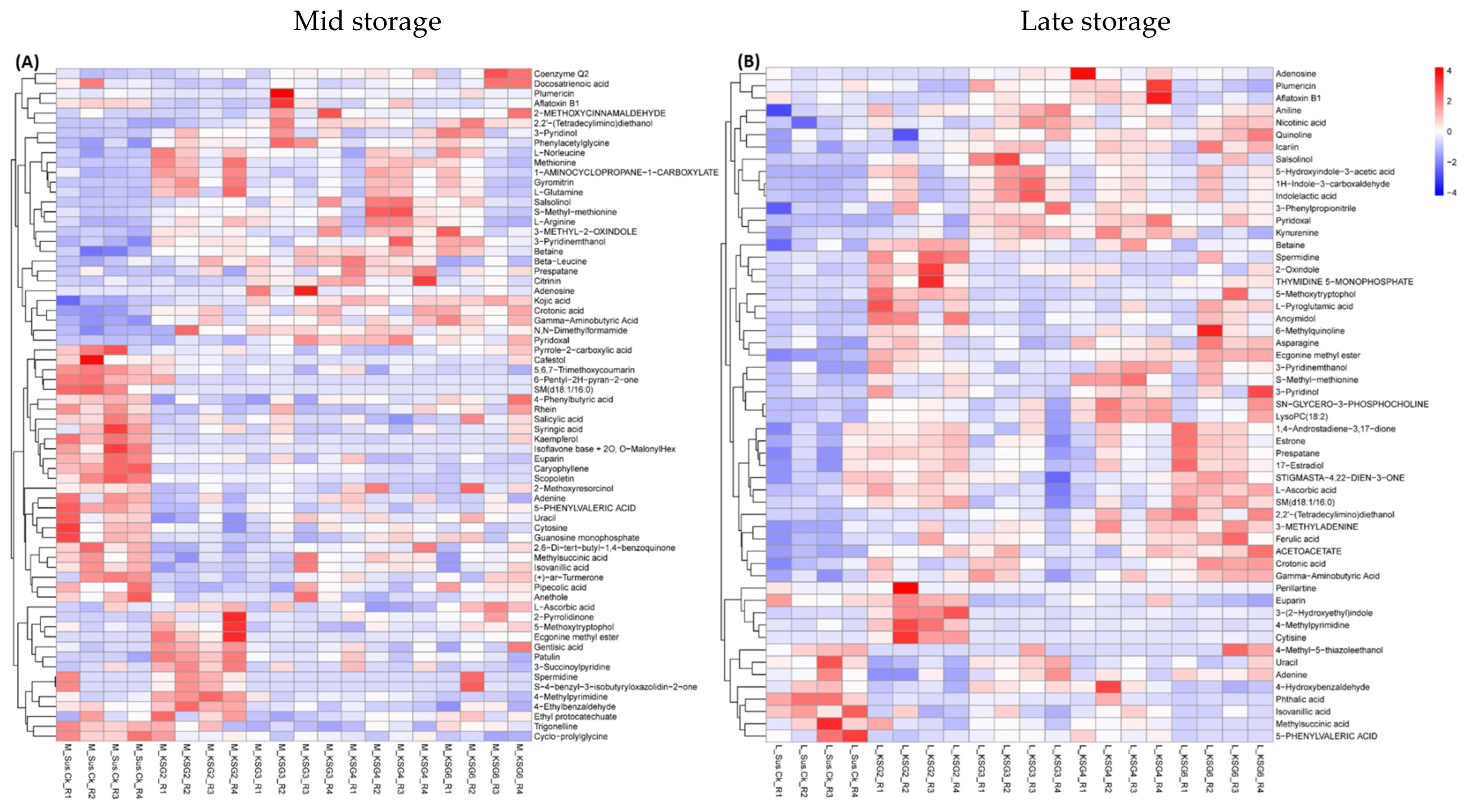
2.5. Untargeted Metabolome Analysis of the Roots of Susceptible (S) and Resistant (R) Lines
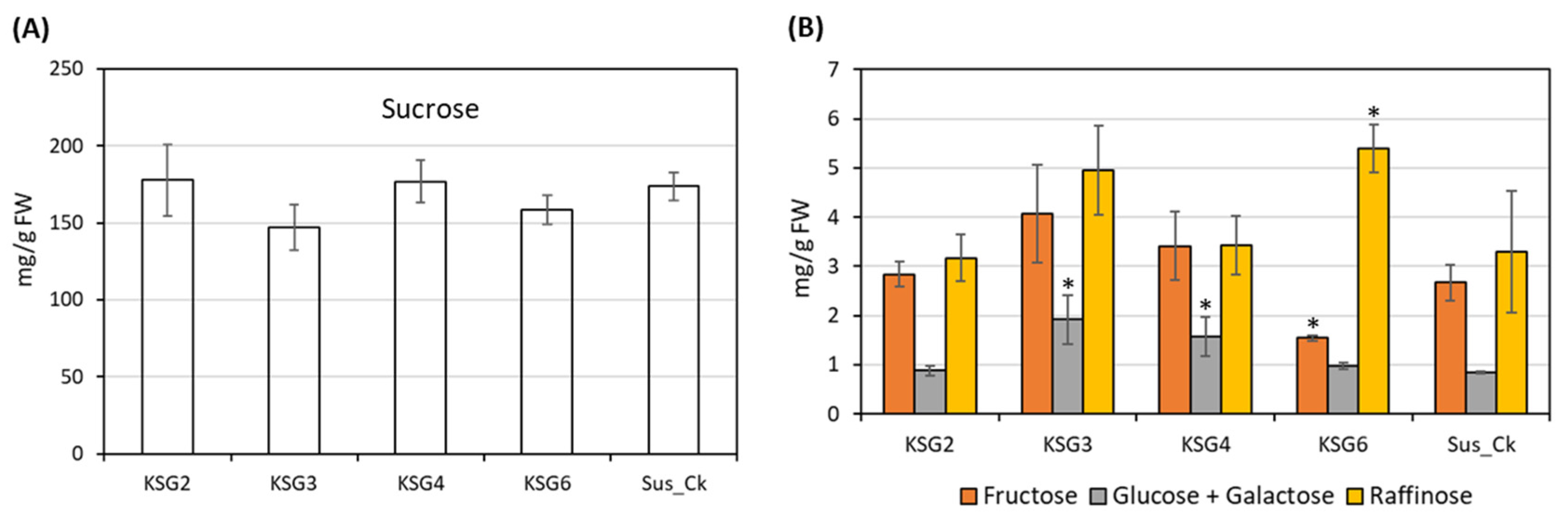
2.6. Carbohydrate Content in the Roots of Susceptible (S) and Resistant (R) Lines
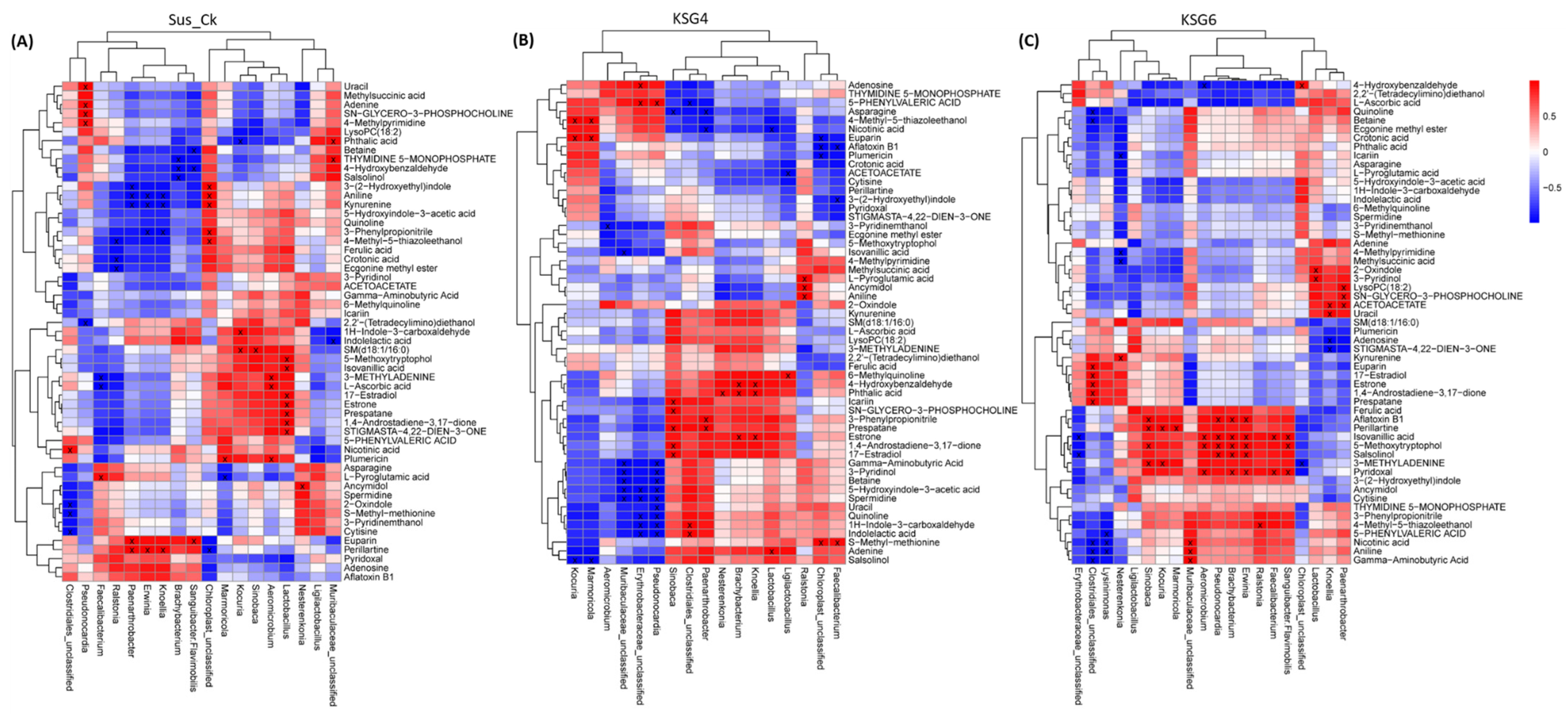
2.7. Correlation Analysis Between the Root Microbiome and Metabolome
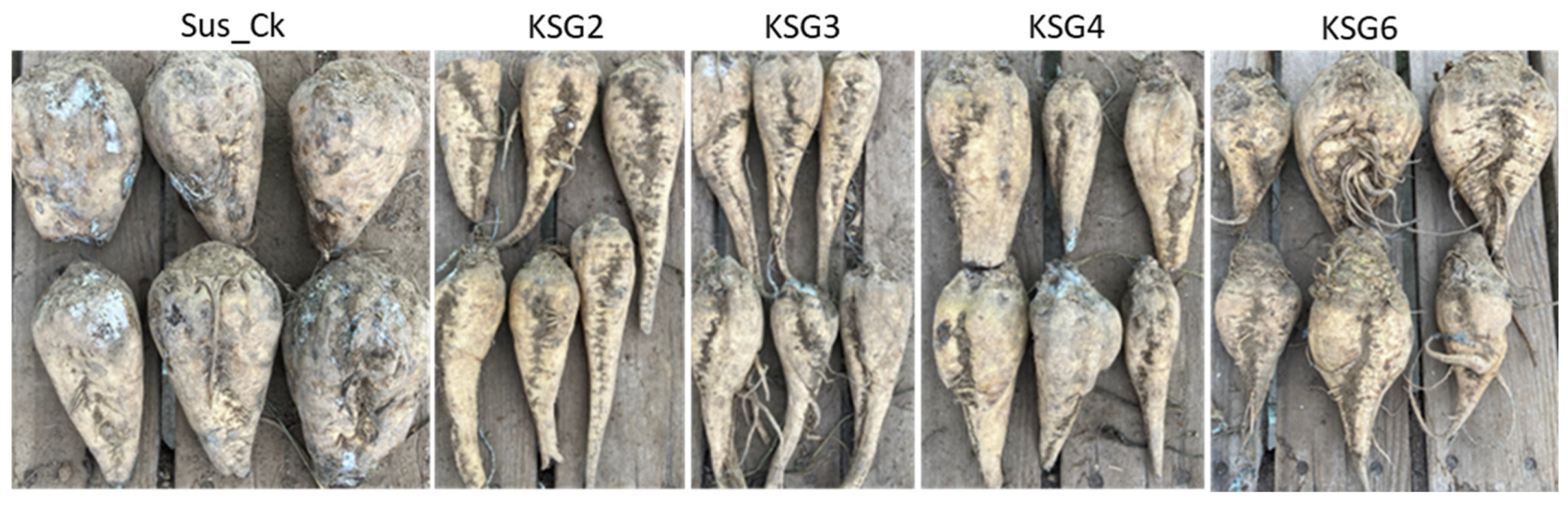
2.8. Resistant Lines Exhibited Lower Disease Symptoms vs. the Susceptible Line
3. Discussion
3.1. Microbial Diversity in the Resistant Lines and Their Putative Roles in Post-Harvest Disease Resistance
3.2. Metabolic Signatures of Resistant Lines Contributing to Resistance
3.3. Correlation Between Root Metabolites and the Microbiome Indicates the Role of Host Genotype in Resistance
4. Materials and Methods
4.1. Storage Conditions of Sugar Beet Roots, Evaluation of Roots, and Sample Collection During Prolonged Indoor Storage
4.2. Genomic DNA Extraction, PCR Amplification, and 16S rRNA and ITS Sequencing
4.3. Data Analysis
4.4. Untargeted Metabolome Analysis
4.5. Carbohydrate Analysis
4.6. Statistical Analysis
4.7. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bugbee, W.M. Storage. In The Sugar Beet Crop; Cooke, D.A., Scott, R.K., Eds.; Springer: Dordrecht, The Netherlands, 1993; pp. 551–570. [Google Scholar]
- Strausbaugh, C.A.; Neher, O.; Rearick, E.; Eujayl, I.A. Influence of Harvest Timing, Fungicides, and Beet necrotic yellow vein virus on Sugar Beet Storage. Plant Dis. 2015, 99, 1296–1309. [Google Scholar] [CrossRef] [PubMed]
- Liebe, S.; Varrelmann, M. Impact of root rot pathogens on storage of sugar beets and control measures. Zuckerind. Sugar Ind. 2014, 139, 443–452. [Google Scholar] [CrossRef]
- Strausbaugh, C.A. Incidence, Distribution, and Pathogenicity of Fungi Causing Root Rot in Idaho Long-Term Sugar Beet Storage Piles. Plant Dis. 2018, 102, 2296–2307. [Google Scholar] [CrossRef]
- Bernhardson, D. Sugarbeet storage techniques developed. Int. Sugar J. 2009, 111, 628–631. [Google Scholar]
- Bugbee, W.M. Storage Rot of Sugar Beet. Plant Dis. 1982, 66, 871–873. [Google Scholar] [CrossRef]
- Strausbaugh, C.A.; Eujayl, I.A. Influence of Beet necrotic yellow vein virus and Freezing Temperatures on Sugar Beet Roots in Storage. Plant Dis. 2018, 102, 932–937. [Google Scholar] [CrossRef]
- Peterson, C.L.; Traveller, D.J.; Hall, M.C. Loss of sucrose during controlled and conventional storage. J. Am. Soc. Sugar Beet Technol. 1980, 20, 517–530. [Google Scholar] [CrossRef]
- Van Eerd, L.L.; Congreves, K.A.; Zandstra, J.W. Sugar beet (Beta vulgaris L.) storage quality in large outdoor piles is impacted by pile management but not by nitrogen fertilizer or cultivar. Can. J. Plant Sci. 2012, 92, 129–139. [Google Scholar] [CrossRef]
- Akeson, W.R. Effect of Chemicals on Sucrose Loss in Sugarbeets During Storage. J. Am. Soc. Sugar Beet Technol. 1978, 20, 255–268. [Google Scholar] [CrossRef]
- Bugbee, W.; Cole, D. Comparison of thiabendazole and genetic resistance for control of sugar beet storage rot. Phytopathology 1979, 69, 1230–1232. [Google Scholar] [CrossRef]
- Fugate, K.; Ferrareze, J.; Bolton, M.; Deckard, E.; Campbell, L. Postharvest jasmonic acid treatment of sugarbeet roots reduces rot due to Botrytis cinerea, Penicillium claviforme, and Phoma betae. Postharvest Biol. Technol. 2012, 65, 1–4. [Google Scholar] [CrossRef]
- Miles, W.G.; Shaker, F.M.; Nielson, A.K.; Ames, R.R. A laboratory study on the ability of fungicides to control beet rotting fungi. J. Am. Soc. Sugar Beet Technol. 1977, 19, 288–293. [Google Scholar] [CrossRef]
- Mumford, D.L.; Wyse, R.E. Effect of Fungus Infection on Respiration and Reducing Sugar Accumulation of Sugarbeet Roots and Use of Fungicides to Reduce Infection. J. Am. Soc. Sugar Beet Technol. 1976, 19, 157–162. [Google Scholar] [CrossRef]
- Wu, M.; Singh, B.; Olson, L.; Salunkhe, K. Control of sucrose loss in sugarbeet during storage by chemicals and modified atmosphere and certain associated physiological changes. J. Am. Soc. Sugar Beet Technol. 1970, 16, 117–127. [Google Scholar] [CrossRef]
- Bugbee, W.M.; Campbell, L.G. Combined resistance in sugar beet to Rhizoctonia solani, Phoma betae, and Botrytis cinerea. Plant Dis. 1990, 74, 353–355. [Google Scholar] [CrossRef]
- Campbell, L.G.; Bugbee, W.M. Selection for Improved Sugarbeet Storability. Crop Sci. 1988, 28, 33–36. [Google Scholar] [CrossRef]
- Strausbaugh, C.A.; Rearick, E.; Camp, S. Influence of Curly Top and Poncho Beta on Storability of Sugarbeet. J. Sugarbeet Res. 2008, 45, 31–47. [Google Scholar] [CrossRef]
- Strausbaugh, C.A.; Rearick, E.; Camp, S.; Gallian, J.J.; Dyer, A.T. Influence of Beet necrotic yellow vein virus on Sugar Beet Storability. Plant Dis. 2008, 92, 581–587. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Strausbaugh, C.A.; Eujayl, I.; Rearick, E.; Foote, P.; Elison, D. Sugar Beet Cultivar Evaluation for Storability and Rhizomania Resistance. Plant Dis. 2009, 93, 632–638. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Vannier, N.; Agler, M.; Hacquard, S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019, 15, e1007740. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Brettell, L.E.; Qiu, Z.; Singh, B.K. Microbiome-Mediated Stress Resistance in Plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cook, J.; Nearing, J.T.; Zhang, J.; Raudonis, R.; Glick, B.R.; Langille, M.G.I.; Cheng, Z. Harnessing the plant microbiome to promote the growth of agricultural crops. Microbiol. Res. 2021, 245, 126690. [Google Scholar] [CrossRef] [PubMed]
- González, R.; Elena, S.F. The Interplay between the Host Microbiome and Pathogenic Viral Infections. mBio 2021, 12, e02496-21. [Google Scholar] [CrossRef]
- Wassermann, B.; Abdelfattah, A.; Cernava, T.; Wicaksono, W.; Berg, G. Microbiome-based biotechnology for reducing food loss post harvest. Curr. Opin. Biotechnol. 2022, 78, 102808. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Fan, X.; Wang, Y.; Kusstatscher, P.; Duan, J.; Wu, S.; Chen, S.; Qiao, K.; Wang, Y.; Ma, B.; et al. Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 2021, 7, 60–72. [Google Scholar] [CrossRef]
- Zachow, C.; Muller, H.; Tilcher, R.; Berg, G. Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima-ancestor of all beet crops-and modern sugar beets. Front. Microbiol. 2014, 5, 415. [Google Scholar] [CrossRef]
- Kusstatscher, P.; Cernava, T.; Harms, K.; Maier, J.; Eigner, H.; Berg, G.; Zachow, C. Disease Incidence in Sugar Beet Fields Is Correlated with Microbial Diversity and Distinct Biological Markers. Phytobiomes J. 2019, 3, 22–30. [Google Scholar] [CrossRef]
- Liebe, S.; Wibberg, D.; Winkler, A.; Puhler, A.; Schluter, A.; Varrelmann, M. Taxonomic analysis of the microbial community in stored sugar beets using high-throughput sequencing of different marker genes. FEMS Microbiol. Ecol. 2016, 92, fiw004. [Google Scholar] [CrossRef]
- Kusstatscher, P.; Zachow, C.; Harms, K.; Maier, J.; Eigner, H.; Berg, G.; Cernava, T. Microbiome-driven identification of microbial indicators for postharvest diseases of sugar beets. Microbiome 2019, 7, 112. [Google Scholar] [CrossRef]
- Fugate, K.K.; Campbell, L.G.; Lafta, A.M.; Eide, J.D.; Khan, M.F.R.; Chu, C.; Finger, F.L. Newly developed sugarbeet lines with altered postharvest respiration rates differ in transcription factor and glycolytic enzyme expression. Crop Sci. 2022, 62, 1251–1263. [Google Scholar] [CrossRef]
- Wyse, R.E.; Theurer, J.C.; Doney, D.L. Genetic Variability in Post-harvest Respiration Rates of Sugarbeet Roots. Crop Sci. 1978, 18, 264–266. [Google Scholar] [CrossRef]
- Eujayl, I.A.; Strausbaugh, C.A. Kimberly germplasm evaluated for rhizomania and storage rot resistance in Idaho, 2015. Plant Dis. Manag. Rep. 2016, 10, FC185. [Google Scholar]
- Eujayl, I.A.; Strausbaugh, C.A. Kimberly sugar beet germplasm evaluated for rhizomania and storage rot resistance in Idaho, 2017. Plant Dis. Manag. Rep. 2018, 12, CF153. [Google Scholar]
- Eujayl, I.A.; Strausbaugh, C.A. Kimberly sugar beet germplasm evaluated for rhizomania and storage rot resistance in Idaho, 2019. Plant Dis. Manag. Rep. 2020, 14, V139. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Fugate, K.K.; Eide, J.D.; Lafta, A.M.; Tehseen, M.M.; Chu, C.; Khan, M.F.R.; Finger, F.L. Transcriptomic and metabolomic changes in postharvest sugarbeet roots reveal widespread metabolic changes in storage and identify genes potentially responsible for respiratory sucrose loss. Front. Plant Sci. 2024, 15, 1320705. [Google Scholar] [CrossRef]
- Gippert, A.L.; Madritsch, S.; Woryna, P.; Otte, S.; Mayrhofer, M.; Eigner, H.; Garibay-Hernandez, A.; D’Auria, J.C.; Molin, E.M.; Mock, H.P. Unraveling metabolic patterns and molecular mechanisms underlying storability in sugar beet. BMC Plant Biol. 2022, 22, 430. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Bae, H. Plant Microbiome: An Ocean of Possibilities for Improving Disease Resistance in Plants. Microorganisms 2023, 11, 392. [Google Scholar] [CrossRef]
- Strausbaugh, C.A. Leuconostoc spp. Associated with Root Rot in Sugar Beet and Their Interaction with Rhizoctonia solani. Phytopathology 2016, 106, 432–441. [Google Scholar] [CrossRef]
- Strausbaugh, C.A.; Gillen, A.M. Bacteria and Yeast Associated with Sugar Beet Root Rot at Harvest in the Intermountain West. Plant Dis. 2008, 92, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Bruni, G. Draft genomes of 17 bacterial isolates from Louisiana raw sugarcane factory juices and biofilms. Microbiol. Resourc. Announc. 2023, 12, e0041623. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Du, C.; Gao, Y.; Zhou, X.; Ejaz, I.; Guo, J.; Chen, K.; Ma, J.; Zhang, Y.; Wang, Z.; et al. Watering Shapes a Robust and Stable Microbial Community under Fusarium Crown Rot Infection. Agronomy 2023, 13, 1356. [Google Scholar] [CrossRef]
- Lin, M.; Zhou, Y.; Xu, R.; Du, C.; Wang, R.; Lu, W.; Abudukadier, K.; Sun, Z. Contrasting Key Bacteria and Fungi Related to Sugar Beet (Beta vulgaris L.) with Different Resistances to Beet Rot under Two Farming Modes. Agronomy 2023, 13, 825. [Google Scholar] [CrossRef]
- Lazcano, C.; Boyd, E.; Holmes, G.; Hewavitharana, S.; Pasulka, A.; Ivors, K. The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions. Sci. Rep. 2021, 11, 3188. [Google Scholar] [CrossRef]
- Schäfer, M.; Vogel, C.M.; Bortfeld-Miller, M.; Mittelviefhaus, M.; Vorholt, J.A. Mapping phyllosphere microbiota interactions in planta to establish genotype-phenotype relationships. Nat. Microbiol. 2022, 7, 856–867. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, D.; Liu, Q. Connections Between Amino Acid Metabolisms in Plants: Lysine as an Example. Front. Plant Sci. 2020, 11, 928. [Google Scholar] [CrossRef]
- Trovato, M.; Funck, D.; Forlani, G.; Okumoto, S.; Amir, R. Editorial: Amino Acids in Plants: Regulation and Functions in Development and Stress Defense. Front. Plant Sci. 2021, 12, 772810. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Seifi, H.S.; Van Bockhaven, J.; Angenon, G.; Höfte, M. Glutamate Metabolism in Plant Disease and Defense: Friend or Foe? Mol. Plant-Microbe Interact. 2013, 26, 475–485. [Google Scholar] [CrossRef]
- Liao, H.S.; Lee, K.T.; Chung, Y.H.; Chen, S.Z.; Hung, Y.J.; Hsieh, M.H. Glutamine induces lateral root initiation, stress responses, and disease resistance in Arabidopsis. Plant Physiol. 2024, 195, 2289–2308. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.; Barchi, B.; Turlapati, S.A.; Gagne, M.; Minocha, R.; Long, S.; Minocha, S.C. Glutamate, Ornithine, Arginine, Proline, and Polyamine Metabolic Interactions: The Pathway Is Regulated at the Post-Transcriptional Level. Front. Plant Sci. 2016, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Aghdam, M.S.; Flaherty, E.J.; Shelp, B.J. γ-Aminobutyrate improves the postharvest marketability of horticultural commodities: Advances and prospects. Front. Plant Sci. 2022, 13, 884572. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.; Wani, K.I.; Naeem, M.; Aftab, T. From Neurotransmitter to Plant Protector: The Intricate World of GABA Signaling and its Diverse Functions in Stress Mitigation. J. Plant Growth Regul. 2024. [Google Scholar] [CrossRef]
- Majumdar, R.; Minocha, R.; Lebar, M.D.; Rajasekaran, K.; Long, S.; Carter-Wientjes, C.; Minocha, S.; Cary, J.W. Contribution of Maize Polyamine and Amino Acid Metabolism Toward Resistance Against Aspergillus flavus Infection and Aflatoxin Production. Front. Plant Sci. 2019, 10, 692. [Google Scholar] [CrossRef]
- Roy, T.; Pal, N.; Das, N. Regulation of the polyamine pool in plants: Metabolic implications and stress mitigation, with emphasis on microbial influence. Physiol. Mol. Plant Pathol. 2024, 132, 102317. [Google Scholar] [CrossRef]
- Kumar, N.; Pruthi, V. Potential applications of ferulic acid from natural sources. Biotechnol. Rep. 2014, 4, 86–93. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, X.; Liu, Y.; Han, Y.; Zuo, J.; Deng, J.; Yuan, L.; Gao, L.; Bai, W. Mixed oligosaccharides-induced changes in bacterial assembly during cucumber (Cucumis sativus L.) growth. Front. Microbiol. 2023, 14, 1195096. [Google Scholar] [CrossRef]
- Guo, M.; Li, C.; Huang, R.; Qu, L.; Liu, J.; Zhang, C.; Ge, Y. Ferulic acid enhanced resistance against blue mold of Malus domestica by regulating reactive oxygen species and phenylpropanoid metabolism. Postharvest Biol. Technol. 2023, 202, 112378. [Google Scholar] [CrossRef]
- Davies, C.R.; Wohlgemuth, F.; Young, T.; Violet, J.; Dickinson, M.; Sanders, J.W.; Vallieres, C.; Avery, S.V. Evolving challenges and strategies for fungal control in the food supply chain. Fungal Biol. Rev. 2021, 36, 15–26. [Google Scholar] [CrossRef]
- Hoffmann, C.; Märländer, B. Composition of harmful nitrogen in sugar beet (Beta vulgaris L.)—Amino acids, betaine, nitrate—As affected by genotype and environment. Eur. J. Agron. 2005, 22, 255–265. [Google Scholar] [CrossRef]
- Kumar, A.; Bachhawat, A.K. Pyroglutamic acid: Throwing light on a lightly studied metabolite. Curr. Sci. 2012, 102, 288–297. [Google Scholar]
- Bilska, K.; Stuper-Szablewska, K.; Kulik, T.; Buśko, M.; Załuski, D.; Perkowski, J. Resistance-Related l-Pyroglutamic Acid Affects the Biosynthesis of Trichothecenes and Phenylpropanoids by F. graminearum Sensu Stricto. Toxins 2018, 10, 492. [Google Scholar] [CrossRef] [PubMed]
- Mejri, S.; Ghinet, A.; Magnin-Robert, M.; Randoux, B.; Abuhaie, C.-M.; Tisserant, B.; Gautret, P.; Benoit, R.; Halama, P.; Reignault, P.; et al. New plant immunity elicitors from a sugar beet byproduct protect wheat against Zymoseptoria tritici. Sci. Rep. 2023, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, Q.; Zhao, J.; Chen, J.; Wang, S.; Ma, M.; Liu, H.; Zhang, Q.; Zhao, H.; Zhou, D.; et al. Metabolites from Bacillus subtilis J-15 Affect Seedling Growth of Arabidopsis thaliana and Cotton Plants. Plants 2022, 11, 3205. [Google Scholar] [CrossRef] [PubMed]
- Marszalek-Grabska, M.; Walczak, K.; Gawel, K.; Wicha-Komsta, K.; Wnorowska, S.; Wnorowski, A.; Turski, W.A. Kynurenine emerges from the shadows—Current knowledge on its fate and function. Pharmacol. Ther. 2021, 225, 107845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, Q.; Qin, Y.; Si, W.; Zhang, H.; Zhang, J. The Effect of Epimedium Isopentenyl Flavonoids on the Broiler Gut Health Using Microbiomic and Metabolomic Analyses. Int. J. Mol. Sci. 2023, 24, 7646. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Koprivova, A.; Kopriva, S. Pinpointing secondary metabolites that shape the composition and function of the plant microbiome. J. Exp. Bot. 2020, 72, 57–69. [Google Scholar] [CrossRef]
- Drewes, J.L.; White, J.R.; Dejea, C.M.; Fathi, P.; Iyadorai, T.; Vadivelu, J.; Roslani, A.C.; Wick, E.C.; Mongodin, E.F.; Loke, M.F.; et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. npj Biofilms Microbiomes 2017, 3, 34. [Google Scholar] [CrossRef]
- Shinohara, N.; Woo, C.; Yamamoto, N.; Hashimoto, K.; Yoshida-Ohuchi, H.; Kawakami, Y. Comparison of DNA sequencing and morphological identification techniques to characterize environmental fungal communities. Sci. Rep. 2021, 11, 2633. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps: R Package; Version 1.0.10; R Core Team: Vienna, Austria, 2023. [Google Scholar]
- Majumdar, R.; Strausbaugh, C.A.; Galewski, P.J.; Minocha, R.; Rogers, C.W. Cell-Wall-Degrading Enzymes-Related Genes Originating from Rhizoctonia solani Increase Sugar Beet Root Damage in the Presence of Leuconostoc mesenteroides. Int. J. Mol. Sci. 2022, 23, 1366. [Google Scholar] [CrossRef]
- Pismennoi, D.; Kiritsenko, V.; Marhivka, J.; Kutt, M.L.; Vilu, R. Development and Optimization of HILIC-LC-MS Method for Determination of Carbohydrates in Fermentation Samples. Molecules 2021, 26, 3669. [Google Scholar] [CrossRef]


| (A) | ||
|---|---|---|
| Treatment | Observed_OTUs | Shannon |
| M_KSG2 | 186.25 ± 23.84 | 1.26 ± 0.09 |
| M_KSG3 | 221.25 ± 17.50 | 1.17 ± 0.09 |
| M_KSG4 | 133.5 * ± 28.12 | 1.04 ± 0.10 |
| M_KSG6 | 257.75 ± 50.80 | 1.48 ± 0.25 |
| M_Sus_Ck | 214 ± 19.55 | 1.35 ± 0.13 |
| L_KSG2 | 231 ± 26.54 | 1.17 ± 0.09 |
| L_KSG3 | 243.75 ± 48.02 | 1.49 ± 0.13 |
| L_KSG4 | 235.50 ± 26.22 | 1.21 ± 0.05 |
| L_KSG6 | 273 ± 38.94 | 1.84 ± 0.28 |
| L_Sus_Ck | 185.50 ± 33.28 | 1.60 ± 0.32 |
| (B) | ||
| M_KSG2 | 227.75 ± 17 | 7.27 ± 0.11 |
| M_KSG3 | 233.25 ± 19.75 | 7.26 ± 0.16 |
| M_KSG4 | 278.25 ± 56.81 | 7.48 ± 0.29 |
| M_KSG6 | 297.75 * ± 27.75 | 7.59 * ± 0.08 |
| M_Sus_Ck | 210 ± 83.85 | 7.08 ± 2.34 |
| L_KSG2 | 180.75 ± 6.14 | 6.86 ± 0.13 |
| L_KSG3 | 200 ± 10.40 | 7.04 ± 0.15 |
| L_KSG4 | 223.75 ± 14.05 | 7.21 ± 0.09 |
| L_KSG6 | 179.75 ± 7.34 | 6.93 ± 0.09 |
| L_Sus_Ck | 199 ± 30.11 | 7.04 ± 0.25 |
| Time (min) | Flow Rate (mL/min) | A (%) | B (%) |
|---|---|---|---|
| 0.00 | 0.30 | 95 | 5 |
| 1.00 | 0.30 | 95 | 5 |
| 12.50 | 0.30 | 5 | 95 |
| 13.50 | 0.30 | 5 | 95 |
| 13.60 | 0.30 | 95 | 5 |
| 16.00 | 0.30 | 95 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majumdar, R.; Kandel, S.L.; Strausbaugh, C.A.; Singh, A.; Pokhrel, S.; Bill, M. Root Microbiome and Metabolome Traits Associated with Improved Post-Harvest Root Storage for Sugar Beet Breeding Lines Under Southern Idaho Conditions. Int. J. Mol. Sci. 2024, 25, 12681. https://doi.org/10.3390/ijms252312681
Majumdar R, Kandel SL, Strausbaugh CA, Singh A, Pokhrel S, Bill M. Root Microbiome and Metabolome Traits Associated with Improved Post-Harvest Root Storage for Sugar Beet Breeding Lines Under Southern Idaho Conditions. International Journal of Molecular Sciences. 2024; 25(23):12681. https://doi.org/10.3390/ijms252312681
Chicago/Turabian StyleMajumdar, Rajtilak, Shyam L. Kandel, Carl A. Strausbaugh, Anuradha Singh, Suresh Pokhrel, and Malick Bill. 2024. "Root Microbiome and Metabolome Traits Associated with Improved Post-Harvest Root Storage for Sugar Beet Breeding Lines Under Southern Idaho Conditions" International Journal of Molecular Sciences 25, no. 23: 12681. https://doi.org/10.3390/ijms252312681
APA StyleMajumdar, R., Kandel, S. L., Strausbaugh, C. A., Singh, A., Pokhrel, S., & Bill, M. (2024). Root Microbiome and Metabolome Traits Associated with Improved Post-Harvest Root Storage for Sugar Beet Breeding Lines Under Southern Idaho Conditions. International Journal of Molecular Sciences, 25(23), 12681. https://doi.org/10.3390/ijms252312681






