Genome-Wide Identification of the ClpB Gene Family in Tomato and Expression Analysis Under Heat Stress
Abstract
1. Introduction
2. Results
2.1. Identification of SlClpB Genes
2.2. Systematic Profiles of SlClpB Genes
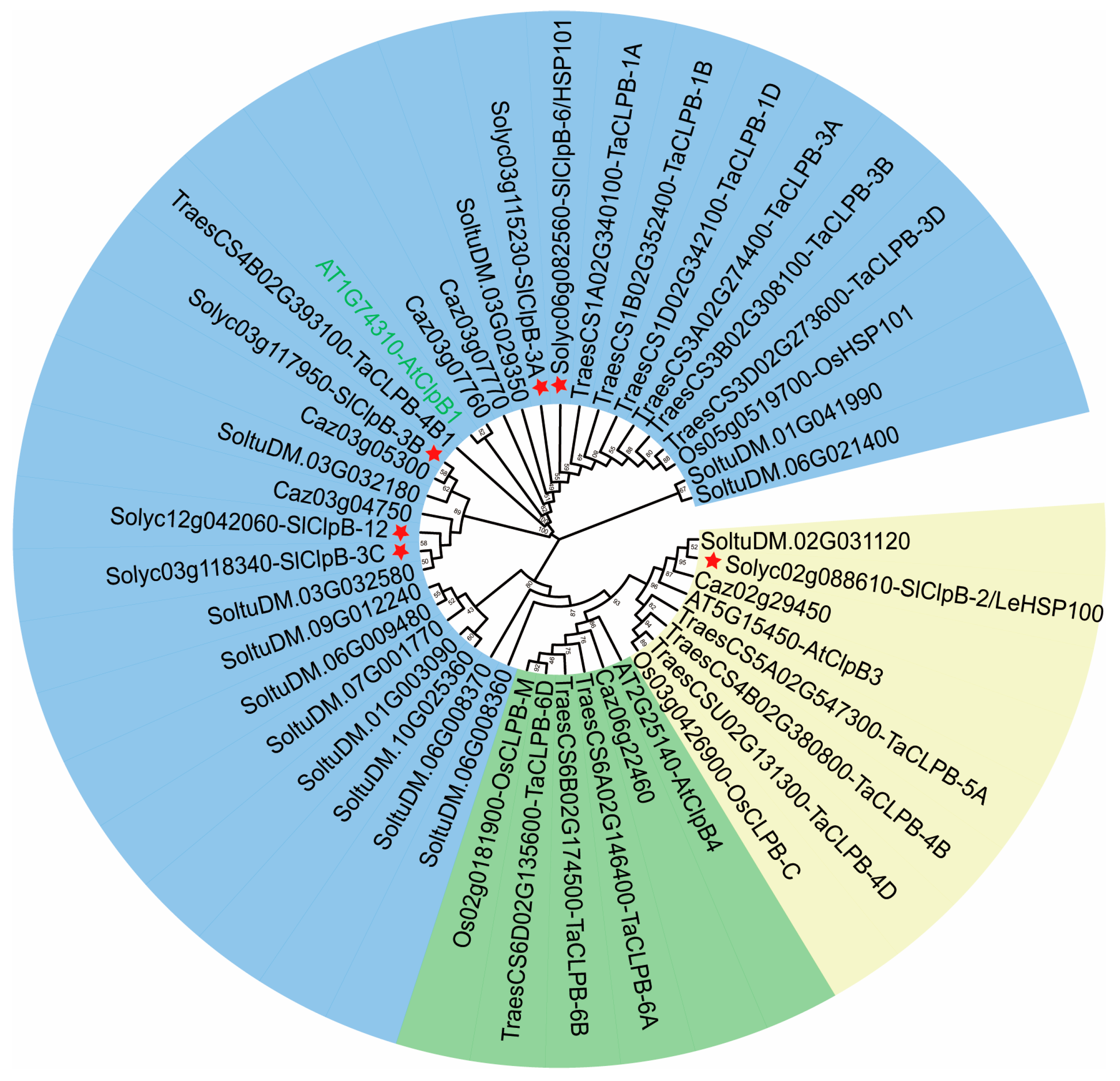
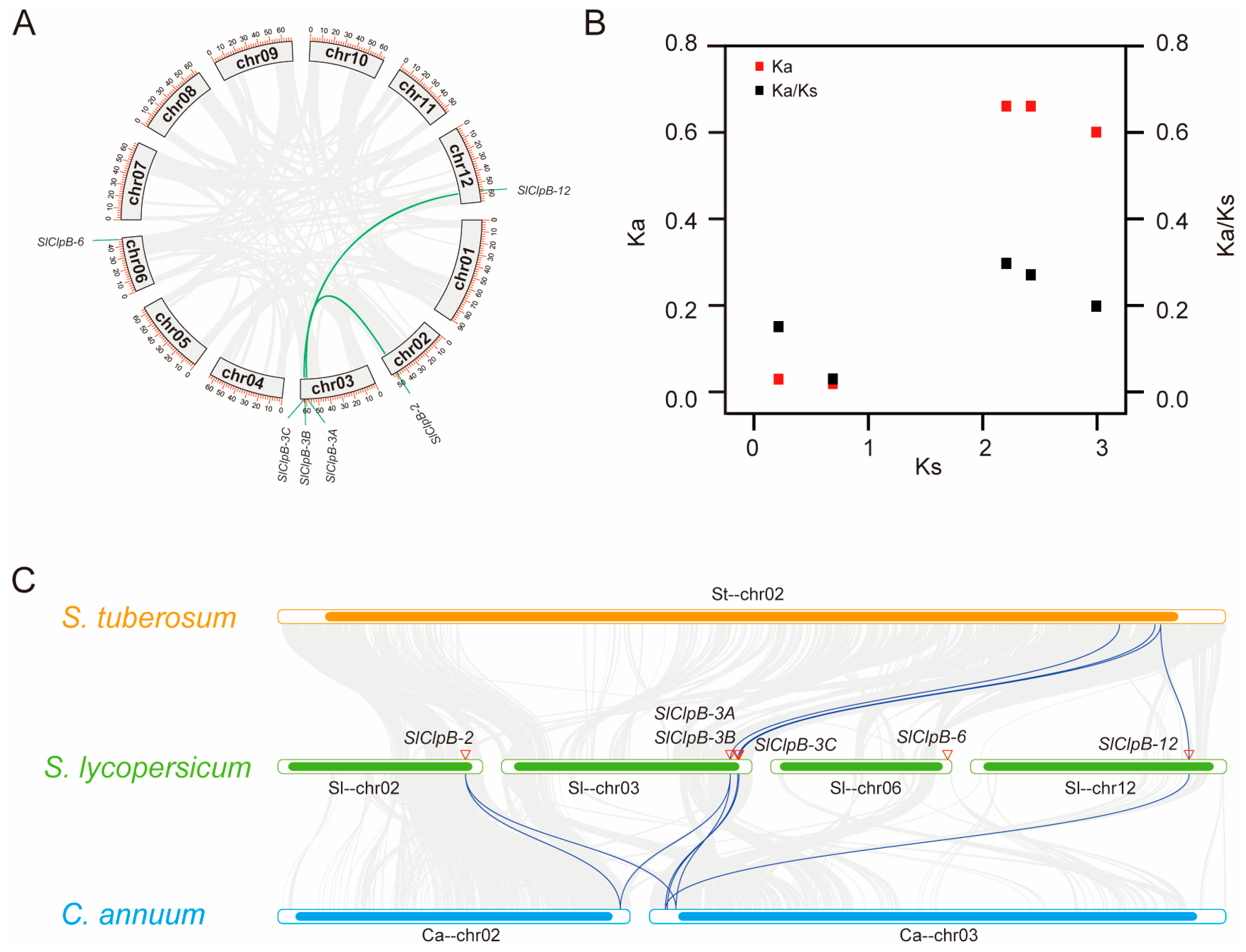
2.3. Evolutionary Investigation of SlClpB Genes
2.4. Conserved Motifs and Domains of SlClpB Genes
2.5. Identification of Cis-Acting Elements of SlClpB Promoters
2.6. Expression Pattern of SlClpB Genes
3. Discussion
4. Materials and Methods
4.1. Tomato Plant Cultivation and Heat Stress Treatment
4.2. Identification and Characterization of SlClpB Genes
4.3. Motif Analysis of SlClpB Genes
4.4. Phylogenetic Tree Construction of SlClpB Genes
4.5. Genome Localization and Synteny Analysis
4.6. Gene Expression Profile Using RNA-seq Data
4.7. RNA Extraction and qRT-PCR Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanchez, Y.; Lindquist, S.L. HSP104 required for induced thermotolerance. Science 1990, 248, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.G.; Baneyx, F. Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: Comparison with ClpA, ClpB, and HtpG in vivo. J. Bacteriol. 1998, 180, 5165–5172. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, D.W.; Lee, Y.; Mayer, U.; Stierhof, Y.D.; Lee, S.; Jurgens, G.; Hwang, I. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the Ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 2009, 21, 3984–4001. [Google Scholar] [CrossRef] [PubMed]
- Glover, J.R.; Lindquist, S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 1998, 94, 73–82. [Google Scholar] [CrossRef]
- Watanabe, Y.; Takano, M.; Yoshida, M. ATP binding to nucleotide binding domain (NBD) 1 of the ClpB chaperone induces motion of the long coiled-coil, stabilizes the hexamer, and activates NBD2. J. Biol. Chem. 2005, 280, 24562–24567. [Google Scholar] [CrossRef]
- Lee, S.; Sowa, M.E.; Watanabe, Y.; Sigler, P.B.; Chiu, W.; Yoshida, M.; Tsai, F.T. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell 2003, 115, 229–240. [Google Scholar] [CrossRef]
- Barnett, M.E.; Nagy, M.; Kedzierska, S.; Zolkiewski, M. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 2005, 280, 34940–34945. [Google Scholar] [CrossRef]
- Lee, S.; Choi, J.; Tsai, F.T. Visualizing the ATPase cycle in a protein disaggregating machine: Structural basis for substrate binding by ClpB. Mol. Cell 2007, 25, 261–271. [Google Scholar] [CrossRef]
- Mizuno, S.; Nakazaki, Y.; Yoshida, M.; Watanabe, Y.H. Orientation of the amino-terminal domain of ClpB affects the disaggregation of the protein. FEBS J. 2012, 279, 1474–1484. [Google Scholar] [CrossRef]
- Kim, K.I.; Cheong, G.; Park, S.; Ha, J.; Woo, K.M.; Choi, S.J.; Chung, C.H. Heptameric ring structure of the heat-shock protein ClpB, a protein-activated ATPase in Escherichia coli. J. Mol. Biol. 2000, 303, 655–666. [Google Scholar] [CrossRef]
- Watanabe, Y.; Motohashi, K.; Yoshida, M. Roles of the two ATP binding sites of ClpB from Thermus thermophilus. J. Biol. Chem. 2002, 277, 5804–5809. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Schlieker, C.; Strub, C.; Rist, W.; Weibezahn, J.; Bukau, B. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 2003, 278, 17615–17624. [Google Scholar] [CrossRef] [PubMed]
- Uchihashi, T.; Watanabe, Y.; Nakazaki, Y.; Yamasaki, T.; Watanabe, H.; Maruno, T.; Ishii, K.; Uchiyama, S.; Song, C.; Murata, K. Dynamic structural states of ClpB involved in its disaggregation function. Nat. Commun. 2018, 9, 2147. [Google Scholar] [CrossRef]
- Cashikar, A.G.; Schirmer, E.C.; Hattendorf, D.A.; Glover, J.R.; Ramakrishnan, M.S.; Ware, D.M.; Lindquist, S.L. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 2002, 9, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Barnett, M.E.; Zolkiewska, A.; Zolkiewski, M. Structure and activity of ClpB from Escherichia coli: Role of the amino-and carboxyl-terminal domains. J. Biol. Chem. 2000, 275, 37565–37571. [Google Scholar] [CrossRef]
- Lo, J.H.; Baker, T.A.; Sauer, R.T. Characterization of the N-terminal repeat domain of Escherichia coli ClpA-A class I Clp/HSP100 ATPase. Protein Sci. 2001, 10, 551–559. [Google Scholar] [CrossRef]
- Smith, C.K.; Baker, T.A.; Sauer, R.T. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc. Natl. Acad. Sci USA 1999, 96, 6678–6682. [Google Scholar] [CrossRef]
- Oguchi, Y.; Kummer, E.; Seyffer, F.; Berynskyy, M.; Anstett, B.; Zahn, R.; Wade, R.C.; Mogk, A.; Bukau, B. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat. Struct. Mol. Biol. 2012, 19, 1338–1346. [Google Scholar] [CrossRef]
- Mishra, R.C.; Grover, A. ClpB/Hsp100 proteins and heat stress tolerance in plants. Crit. Rev. Biotechnol. 2016, 36, 862–874. [Google Scholar] [CrossRef]
- Singh, A.; Singh, U.; Mittal, D.; Grover, A. Genome-wide analysis of rice ClpB/HSP100, ClpC and ClpD genes. BMC Genom. 2010, 11, 95. [Google Scholar] [CrossRef]
- Davoudi, M.; Chen, J.; Lou, Q. Genome-wide identification and expression analysis of heat shock protein 70 (HSP70) gene family in pumpkin (Cucurbita moschata) rootstock under drought stress suggested the potential role of these chaperones in stress How. Int. J. Mol. Sci. 2022, 23, 1918. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Mittler, R.; Balfagon, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, E.C.; Glover, J.R.; Singer, M.A.; Lindquist, S. HSP100/Clp proteins: A common mechanism explains diverse functions. Trends Biochem. Sci. 1996, 21, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Parsell, D.A.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef]
- Schirmer, E.C.; Lindquist, S.; Vierling, E. An Arabidopsis heat shock protein complements a thermotolerance defect in yeast. Plant Cell 1994, 6, 1899–1909. [Google Scholar] [CrossRef]
- Lee, Y.R.; Nagao, R.T.; Key, J.L. A soybean 101-kD heat shock protein complements a yeast HSP104 deletion mutant in acquiring thermotolerance. Plant Cell 1994, 6, 1889–1897. [Google Scholar] [CrossRef]
- Park, S.K.; Kim, K.I.; Woo, K.M.; Seol, J.H.; Tanaka, K.; Ichihara, A.; Ha, D.B.; Chung, C.H. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J. Biol. Chem. 1993, 268, 20170–20174. [Google Scholar] [CrossRef]
- Wells, D.R.; Tanguay, R.L.; Le, H.; Gallie, D.R. HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Gene Dev. 1998, 12, 3236–3251. [Google Scholar] [CrossRef]
- Hong, S.W.; Vierling, E. Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J. 2001, 27, 25–35. [Google Scholar] [CrossRef]
- Keeler, S.J.; Boettger, C.M.; Haynes, J.G.; Kuches, K.A.; Johnson, M.M.; Thureen, D.L.; Keeler Jr, C.L.; Kitto, S.L. Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiol. 2000, 123, 1121–1132. [Google Scholar] [CrossRef]
- Wu, T.; Juan, Y.; Hsu, Y.; Wu, S.; Liao, H.; Fung, R.W.; Charng, Y. Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol. 2013, 161, 2075–2084. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-H.; Tseng, T.-S.; Wu, C.-R.; Cho, S.-T.; Kuo, C.-H.; Huang, X.-J.; Cheng, J.-Y.; Hsu, K.-H.; Lin, K.F.; Liu, C.-C.; et al. Rice OsHsp16. 9A interacts with OsHsp101 to confer thermotolerance. Plant Sci. 2023, 330, 111634. [Google Scholar] [CrossRef]
- Gupta, S.; Shi, X.; Lindquist, I.E.; Devitt, N.; Mudge, J.; Rashotte, A.M. Transcriptome profiling of cytokinin and auxin regulation in tomato root. J. Exp. Bot. 2013, 64, 695–704. [Google Scholar] [CrossRef]
- Bonierbale, M.W.; Plaisted, R.L.; Tanksley, S.D. RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 1988, 120, 1095–1103. [Google Scholar] [CrossRef]
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar] [CrossRef]
- Sato, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Complex plant responses to drought and heat stress under climate change. Plant J. 2024, 117, 1873–1892. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Yang, J.; Sun, Y.; Sun, A.; Yi, S.; Qin, J.; Li, M.; Liu, J. The involvement of chloroplast HSP100/ClpB in the acquired thermotolerance in tomato. Plant Mol. Biol. 2006, 62, 385–395. [Google Scholar] [CrossRef]
- De Klerk, E.; AC T Hoen, P. Alternative mRNA transcription, processing, and translation: Insights from RNA sequencing. Trends Genet. 2015, 31, 128–139. [Google Scholar] [CrossRef]
- Graci, S.; Cigliano, R.A.; Barone, A. Exploring the gene expression network involved in the heat stress response of a thermotolerant tomato genotype. BMC Genom. 2024, 25, 509. [Google Scholar] [CrossRef]
- Cárdenas, P.D.; Sonawane, P.D.; Pollier, J.; Vanden Bossche, R.; Dewangan, V.; Weithorn, E.; Tal, L.; Meir, S.; Rogachev, I.; Malitsky, S. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat. Commun. 2016, 7, 10654. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Y.; Li, Y.; Yang, Q.; Yang, T.; Zhou, Z.; Li, Y.; Zhang, N.; Lyu, Y.; Zhu, Y. Heat shock transcription factors regulate thermotolerance gene networks in tomato (Solanum lycopersicum) flower buds. Hortic. Plant J. 2023; in press. [Google Scholar] [CrossRef]
- Almeida, J.; Perez Fons, L.; Fraser, P.D. A transcriptomic, metabolomic and cellular approach to the physiological adaptation of tomato fruit to high temperature. Plant Cell Environ. 2021, 44, 2211–2229. [Google Scholar] [CrossRef] [PubMed]
- Erdayani, E.; Nagarajan, R.; Grant, N.P.; Gill, K.S. Genome-wide analysis of the HSP101/CLPB gene family for heat tolerance in hexaploid wheat. Sci. Rep. 2020, 10, 3948. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Sotelo, J.; Kannan, K.B.; Martınez, L.M.; Segal, C. Characterization of a maize heat-shock protein 101 gene, HSP101, encoding a ClpB/Hsp100 protein homologue. Genes 1999, 230, 187–195. [Google Scholar] [CrossRef]
- Shen, J.; Yu, Q.; Chen, S.; Tan, Q.; Li, J.; Li, Y. Unbiased organism-agnostic and highly sensitive signal peptide predictor with deep protein language model. Nat. Comput. Sci. 2024, 4, 29–42. [Google Scholar] [CrossRef]
- Rozov, S.M.; Deineko, E.V. Increasing the efficiency of the accumulation of recombinant proteins in plant cells: The role of transport signal peptides. Plants 2022, 11, 2561. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, L.; Lin, X.; Wang, Y.; Li, Y.; Guo, Q.; Li, S.; Sun, Y.; Tao, X.; Zhang, D.; et al. A translocation pathway for vesicle-mediated unconventional protein secretion. Cell 2020, 181, 637–652. [Google Scholar] [CrossRef]
- Steringer, J.P.; Nickel, W. A direct gateway into the extracellular space: Unconventional secretion of FGF2 through self-sustained plasma membrane pores. Semin. Cell Dev. Biol. 2018, 83, 3–7. [Google Scholar] [CrossRef]
- Malhotra, V. Unconventional protein secretion: An evolving mechanism. EMBO J. 2013, 32, 1660–1664. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Strader, L.C. Auxin-abscisic acid interactions in plant growth and development. Biomolecules 2020, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.; Yoon, Y.; Choi, Y.D. Crosstalk with jasmonic acid integrates multiple responses in plant development. Int. J. Mol. Sci. 2020, 21, 305. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kesawat, M.S.; Ali, A.; Lee, S.; Gill, S.S.; Kim, H.U. Integration of abscisic acid signaling with other signaling pathways in plant stress responses and development. Plants 2019, 8, 592. [Google Scholar] [CrossRef] [PubMed]
- Küpers, J.J.; Oskam, L.; Pierik, R. Photoreceptors regulate plant developmental plasticity through auxin. Plants 2020, 9, 940. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Li, N.; Euring, D.; Cha, J.Y.; Lin, Z.; Lu, M.; Huang, L.; Kim, W.Y. Plant hormone-mediated regulation of heat tolerance in response to global climate change. Front. Plant Sci. 2021, 11, 627969. [Google Scholar] [CrossRef]
- Mani, N.; Ramakrishna, K.; Suguna, K. Characterization of rice small heat shock proteins targeted to different cellular organelles. Cell Stress Chaperones 2015, 20, 451–460. [Google Scholar] [CrossRef]
- Miot, M.; Reidy, M.; Doyle, S.M.; Hoskins, J.R.; Johnston, D.M.; Genest, O.; Vitery, M.; Masison, D.C.; Wickner, S. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc. Natl. Acad. Sci. USA 2011, 108, 6915–6920. [Google Scholar] [CrossRef]
- Carroni, M.; Kummer, E.; Oguchi, Y.; Wendler, P.; Clare, D.K.; Sinning, I.; Kopp, J.; Mogk, A.; Bukau, B.; Saibil, H.R. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. eLife 2014, 3, e02481. [Google Scholar] [CrossRef]
- Qin, F.; Yu, B.; Li, W. Heat shock protein 101 (HSP101) promotes flowering under nonstress conditions. Plant Physiol. 2021, 186, 407–419. [Google Scholar] [CrossRef]
- Wu, H.; Ren, Y.; Dong, H.; Xie, C.; Zhao, L.; Wang, X.; Zhang, F.; Zhang, B.; Jiang, X.; Huang, Y. FLOURY ENDOSPERM24, a heat shock protein 101 (HSP101), is required for starch biosynthesis and endosperm development in rice. New Phytol. 2024, 242, 2635–2651. [Google Scholar] [CrossRef] [PubMed]
- Gurley, W.B. HSP101: A key component for the acquisition of thermotolerance in plants. Plant Cell 2000, 12, 457–460. [Google Scholar] [CrossRef]
- Lee, U.; Rioflorido, I.; Hong, S.W.; Larkindale, J.; Waters, E.R.; Vierling, E. The Arabidopsis ClpB/Hsp100 family of proteins: Chaperones for stress and chloroplast development. Plant J. 2007, 49, 115–127. [Google Scholar] [CrossRef]
- Surekha Katiyar-Agarwal, S.K.; Manu Agarwal, M.A.; Anil Grover, A.G. Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol. Biol. 2003, 51, 677–686. [Google Scholar] [CrossRef]
- Queitsch, C.; Hong, S.; Vierling, E.; Lindquist, S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 2000, 12, 479–492. [Google Scholar] [CrossRef]
- Burke, J.J. Enhancement of Reproductive Heat Tolerance in Plants. PLoS ONE 2015, 10, e0122933. [Google Scholar] [CrossRef]
- Tonsor, S.J.; Scott, C.; Boumaza, I.; Liss, T.R.; Brodsky, J.L.; Vierling, E. Heat shock protein 101 effects in A. thaliana: Genetic variation, fitness and pleiotropy in controlled temperature conditions. Mol. Ecol. 2008, 17, 1614–1626. [Google Scholar] [CrossRef]
- Dhaliwal, A.K.; Mohan, A.; Gill, K.S. Comparative analysis of ABCB1 reveals novel structural and functional conservation between monocots and dicots. Front. Plant Sci. 2014, 5, 657. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Eidi, M.; Abdolalizadeh, S.; Nasirpour, M.H.; Zahiri, J.; Garshasbi, M. 123FASTQ: An intuitive and efficient tool for preprocessing Illumina FASTQ reads. bioRxiv 2024, 2023–2024. [Google Scholar] [CrossRef]
- Zhang, Y. High-Performance Software Development for Genomic Sequence Alignment and Analysis. Doctoral Dissertation, The University of Texas Southwestern Medical Center, Dallas, TX, USA, 2023. Available online: https://hdl.handle.net/2152.5/10071 (accessed on 1 May 2023).
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie, and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]





| Gene Name | Gene ID | Gene Position | CDS (bp) | AA (aa) | MW (kDa) | pI | Signal Peptide | Prediction of Subcellular Localization | |
|---|---|---|---|---|---|---|---|---|---|
| Start | End | ||||||||
| SlClpB-2/LeHSP100 | Solyc02g088610 | 48691924 | 48699037 | 3138 | 980 | 110.377 | 6.41 | NO | Chloroplast |
| SlClpB-3A | Solyc03g115230 | 59435365 | 59439567 | 3064 | 911 | 101.116 | 6.03 | NO | Cytoplasm |
| SlClpB-3B | Solyc03g117950 | 61374547 | 61381305 | 3119 | 964 | 105.713 | 6.88 | NO | Cytoplasm |
| SlClpB-3C | Solyc03g118340 | 67245569 | 61699244 | 3146 | 926 | 102.61 | 4.82 | NO | Cytoplasm |
| SlClpB-6/HSP101 | Solyc06g082560 | 45913978 | 45916542 | 2565 | 854 | 95.071 | 6.31 | NO | Cytoplasm |
| SlClpB-12 | Solyc12g042060 | 40789176 | 56728789 | 2772 | 923 | 102.21 | 5.99 | NO | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Yang, T.; Han, J.; Su, X.; Cong, Y.; Zhou, M.; Wang, Y.; Lin, T. Genome-Wide Identification of the ClpB Gene Family in Tomato and Expression Analysis Under Heat Stress. Int. J. Mol. Sci. 2024, 25, 12325. https://doi.org/10.3390/ijms252212325
Zhang Y, Yang T, Han J, Su X, Cong Y, Zhou M, Wang Y, Lin T. Genome-Wide Identification of the ClpB Gene Family in Tomato and Expression Analysis Under Heat Stress. International Journal of Molecular Sciences. 2024; 25(22):12325. https://doi.org/10.3390/ijms252212325
Chicago/Turabian StyleZhang, Yuemei, Tailai Yang, Jiaxi Han, Xiao Su, Yanqing Cong, Ming Zhou, Yan Wang, and Tao Lin. 2024. "Genome-Wide Identification of the ClpB Gene Family in Tomato and Expression Analysis Under Heat Stress" International Journal of Molecular Sciences 25, no. 22: 12325. https://doi.org/10.3390/ijms252212325
APA StyleZhang, Y., Yang, T., Han, J., Su, X., Cong, Y., Zhou, M., Wang, Y., & Lin, T. (2024). Genome-Wide Identification of the ClpB Gene Family in Tomato and Expression Analysis Under Heat Stress. International Journal of Molecular Sciences, 25(22), 12325. https://doi.org/10.3390/ijms252212325





