Abstract
The Chinese fir (Cunninghamia lanceolata) is a significant species utilized in afforestation efforts in southern China. It is distinguished by its rapid growth and adaptability to diverse environmental conditions. The GRAS gene family comprises a group of plant-specific transcription factors that play a pivotal role in plant growth and development, response to adversity, and hormone regulatory networks. However, the exploration of the GRAS family in gymnosperm Chinese fir has not yet begun. In this study, a total of 43 GRAS genes were identified in the whole genome of Chinese fir, and a phylogenetic analysis classified them into nine distinct subfamilies. Gene structure analysis revealed that the majority of ClGRAS genes lacked introns. It is notable that among these proteins, both ClGAI and ClGRA possess distinctive DELLA structural domains. Cis-acting element analysis revealed that nearly all ClGRAS genes contained light-responsive elements, while hormone-responsive elements, environmental-responsive elements (low-temperature- or defense-responsive elements), and meristem-organization-related elements were also identified. Based on transcriptome data and RT-qPCR expression patterns, we analyzed the expression of ClGAI and ClRGA genes across different developmental stages, hormones, and three abiotic stresses. Subcellular localization analysis demonstrated that ClGAI and ClRGA were localized to the nucleus. Transcriptional activation assays showed that both genes have self-activating activity. In conclusion, the results of this study indicate that the ClGRAS gene family is involved in the response of Chinese fir to environmental stress. Further research on the ClDELLA genes provides valuable information for exploring the potential regulatory network of DELLA proteins in Chinese fir.
1. Introduction
The Chinese fir is a fast-growing coniferous species renowned for its exceptional wood properties and high wood productivity in southern China [1]. It is the most extensively cultivated tree species in southern China, with a history spanning over a thousand years. However, adverse environmental conditions, such as elevated temperatures and drought, can affect the growth and development of Chinese fir [2]. The GRAS transcription factors play a role in plant development and response to a variety of stresses, but the GRAS gene family has been rarely studied in Chinese fir.
Plant transcription factors (TFs) play a significant role in a multitude of biological processes [3]. TFs are not only integral components of plant signaling pathways but are also capable of participating in plant responses to biotic and abiotic stresses [4]. The GRAS family is named after three transcription factors: gibberellin insensitive factor (GAI), repressor of GA (RGA), and scarecrow (SCR) [5]. These TFs are typically distinguished by a highly conserved C-terminal structural domain, which belongs to the GRAS family of structural domains, and a variable N-terminal structural domain [6]. Based on the N-terminal length and sequence variation, GRAS is typically classified into 8–17 subfamilies, including LS, DELLA, SCL9, SCR, PAT1, SHR, HAM, SCL3, and others [7].
The advent of next-generation sequencing technologies has resulted in the generation of a substantial amount of data pertaining to genomic, transcriptomic, proteomic, and metabolomic processes, which has in turn led to the development of new analytical techniques capable of processing and interpreting these data. Consequently, GRAS gene families have been identified and analyzed in numerous plant species, including Arabidopsis thaliana [8], Nicotiana tabacum [9], Populus tomentosa [10], Oryza sativa [11], Triticum aestivum [12], Actinidia chinensis [13], Eucalyptus megacephalus [14], and Manihot esculenta [15]. As a consequence of the divergence in the N-terminal structural domains, the functions exhibit variation among subfamilies. It has been demonstrated that the PAT1 subfamily plays a multifaceted role in light signaling and transcriptional regulation [16]. In A. thaliana, three genes within the PAT1 subfamily, namely PAT1, SCL5, and SCL21, have been identified as positive regulators of light signaling [17]. The grape (Vitis vinifera) VaPAT1 transcription factor has been demonstrated to regulate jasmonic acid (JA) biosynthesis in response to grape cold stress [18]. GhSCL13-2A, a member of the Gossypium hirsutum PAT1 family, has been demonstrated to positively regulate resistance to Botrytis cinerea in cotton [19]. This is accomplished by modulating JA and salicylic acid (SA) signaling pathways, in addition to the accumulation of reactive oxygen species. The SCR and SHR form the SCR/SHR complex, which plays an important role in the radial organization of roots and stems [20]. It is worthy of note that the SCL3 subfamily and the DELLA subfamily exert opposing effects on GA signaling in plants [21]. Following gibberellin A3 (GA3) treatment, nine of the LkGRAS genes in Larix kaempferi exhibited upregulation [16].
The DELLA subfamily occupies a distinctive position within the GRAS family. All TFs in the subfamily possess a distinctive DELLA structural domain at their N-terminus [22]. Interestingly, the DELLA family derives its name from a short-chain amino acid, D-E-L-L-A [23], which is present in this conserved structural domain. It is noteworthy that DELLA proteins lack the typical DNA-binding structural domains observed in other proteins. Nevertheless, they have been demonstrated to interact with a variety of transcription factors, thereby influencing plant growth and development, as well as plant responses to diverse environmental stresses [24]. It is well established that DELLA proteins act as negative regulators of the gibberellin signaling pathway [25]. The gibberellin hormone plays a pivotal role in this process, whereby it triggers the polyubiquitination and subsequent degradation of DELLA proteins via the 26S proteasome. DELLA and JAZ proteins act synergistically to repress the transcriptional functions of MYB21 and MYB24, thereby inhibiting fiber elongation [26]. DELLA proteins interact with FLC to inhibit perforation [27]. Moreover, DELLA proteins have been demonstrated to activate the JA defense pathway, and they bind to the MYC2 transcription factor [28]. In poplar, however, DELLA has been demonstrated to inhibit the promotion of vascular cambium by ARK2 and WOX4 [29]. In this study, we employed all the genomic data from the Chinese fir species to identify and analyze GRAS genes. Our analysis yielded a total of 43 GRAS genes. The phylogenetic relationships, chromosomal locations, gene structures, structural domains and conserved motifs, and cis-acting elements of the ClDELLA genes were also analyzed. Additionally, the expression patterns of ClDELLAs were investigated in different tissues, hormones, and abiotic stresses using transcriptome data and qRT-PCR. The findings presented here will prove invaluable for future research into the multifaceted roles of Chinese fir DELLA proteins.
2. Results
2.1. Identification and Physicochemical Characterization of Chinese Fir GRAS Gene Family
We screened and analyzed the whole genome of Chinese fir by BLAST comparison and HMM search. In total, 43 ClGRAS genes were identified by manual screening and named ClGRAS1~ClGRAS41, among which two DELLA genes were named ClGAI and ClRGA individually. In addition, the physicochemical properties of ClGRAS proteins were analyzed using the online tool Expasy (https://web.expasy.org/protparam/, accessed on 8 May 2024) (Table 1). According to the results in the table, the number of amino acids in ClGRAS proteins varied widely among them, with lengths ranging from 390 aa (ClGRAS29) to 1316 aa (ClGRAS1), with an average of 605 aa. The molecular weights (MW) of 43 genes ranged from 43.72 kDa (ClGRAS29) to 146.94 kDa (ClGRAS1), with an average of 67.55 kDa. The isoelectric point (pI) ranged from 4.46 (ClGRAS35) to 8.88 (ClGRAS25), with a total of 41 ClGRAS genes exhibiting pI values less than 7.0, indicative of acidic properties. Conversely, two genes, ClGRAS24 and ClGRAS25, demonstrated pI values greater than 7.0, suggesting alkali characteristics. The aliphatic index of the amino acid composition of the protein was found to be 81.83 on average, which is conducive to the thermal stability of the spherical protein. The instability index ranged from 35.08 (ClGRAS31) to 62.62 (ClGRAS25), most of which were associated with unstable proteins; it is noteworthy that the instability indices of ClGRAS5, ClGRAS20, ClGRAS31, ClGRAS37 and ClGRAS39 were less than 40, which would indicate that these five proteins were stable. The average hydrophilicity (GRAVY)value of all 43 genes was less than 0, indicating that they are all hydrophilic proteins. Finally, we predicted the subcellular localization of the 43 ClGRAS genes using the Plant-mPLoc online tool (version 2.0), which showed that all of them were localized in the nucleus (Table 1).

Table 1.
Detailed information and physicochemical properties of ClGRASs.
2.2. Phylogenetic Analysis of Chinese Fir GRAS Gene Family
In order to gain insight into the evolutionary relationships and classification of the members of the Chinese fir GRAS gene family, we constructed a phylogenetic evolutionary tree containing 43 ClGRAS members and 32 A. thaliana AtGRAS members (Supplementary Material, Table S1) using the NJ method in MEGA software. As shown, we classified the GRAS genes into nine subfamilies based on the subfamily classification of GRAS gene families in A. thaliana, namely SCR, SCL3, DELLA, LAS, HAM, SCL26, SCL9, SHR, and PAT1 (Figure 1). Of these, the SCL26 subfamily was the most numerous, with 12 members. This was followed by the PAT1 and SCL3 subfamilies with nine and six members, respectively. The DELLA and SCL9 subfamilies were the smallest, comprising only two ClGRAS genes each.
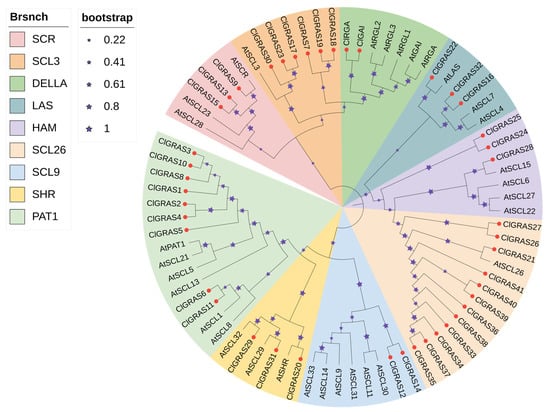
Figure 1.
Phylogenetic evolutionary tree of A. thaliana and Chinese fir GRAS gene families. Nine colors represent different subfamilies, red dots indicate ClGRAS genes, and purple stars represent bootstrap.
2.3. Analysis of Gene Structure and Conserved Motifs in ClGRAS Family of Chinese Fir
To gain further insight into the sequence characterization of ClGRAS proteins, we conducted an in-depth analysis of the motif composition of these proteins via the MEME website. The results demonstrated that among the ten identified motifs, the conserved motifs 1, 2, 4, 5, 6, 8, and 9 exhibited high conservation and were present in 95% of the protein sequences (Figure 2A). It is notable that the number of motifs present in the C-terminal region exceeds those observed in the N-terminal region. The characteristics of the 10 motifs are listed in Figure S1. Moreover, even members of the same subfamily exhibit some variation in the range and frequency of motif distribution. For instance, within the PAT1 subfamily, six genes contain motif 10, while the remaining three (ClGRAS5, ClGRAS6, and ClGRAS11) lack this motif. Additionally, motif 3 is observed only once in ClGRAS11, but twice in each of the other seven genes (Figure 1 and Figure 2A). This indicates that the ClGRAS11 gene may be implicated in particular functions within the PAT1 subfamily. Subsequently, the conserved structural domains of 43 protein sequences were analyzed using the Batch-CDD tool (version 3.21) from the NCBI website. As illustrated in Figure 2B, all ClGRAS proteins possess GRAS or GRAS superfamily structural domains. It is noteworthy that the DELLA subfamily is the sole one to possess a distinctive DELLA structural domain (Figure 2B). The different structural domains all play crucial roles in the signaling process [30]. Given the pivotal role of gene structural diversity in the evolution of gene families, we undertook an analysis of the intron–exon distribution of ClGRAS genes. It was observed that 28 ClGRAS genes exhibited no introns, while the number of introns in the remaining 15 genes ranged from two to four (Figure 2C).
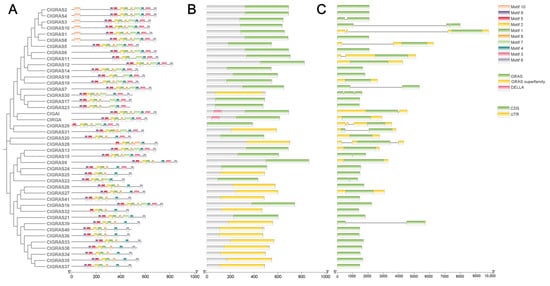
Figure 2.
Motif and gene structure analysis of the ClGRAS gene family. (A) Conserved motifs of ClGRAS genes. The phylogenetic evolutionary tree of the 43 ClGRAS genes is shown on the left. Different colored boxes on the right represent different motifs and their positions in the GRAS protein sequences. (B) Structural domain analysis of ClGRAS proteins. Green is the GRAS family structural domain; yellow is the GRAS superfamily structural domain; pink is the DELLA family structural domain. (C) The gene structure of ClGRAS genes. Exons are represented as green rectangles. Yellow color represents introns.
2.4. Analysis of Cis-Acting Elements and Chromosomal Localization in the ClGRAS Family of Chinese Fir
In order to identify potential cis-acting elements and to further investigate the regulatory functions of GRAS genes, the 2000-base pair promoter region upstream of 43 ClGRAS genes was analyzed using the PlantCARE online tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, access on 15 May 2024). As illustrated in Figure 3, light-responsive elements were identified in the majority of genes. Hormone-associated cis-acting elements include abscisic acid (ABA), GA, Methyl jasmonate (MeJA), SA, and indole acetic acid (IAA). It is noteworthy that, with the exception of the DELLA subfamily, where GA-responsive elements are present in the promoters of both genes, ClGAI and ClRGA, the majority of genes are only present in the other eight subfamilies. To illustrate, in the PATI family, six out of the eight genes were found to contain this element, while it was absent from the promoters of ClGRAS5 and ClGRAS6 (Figure 1 and Figure 3). Nevertheless, the IAA response element is present only in ClGRAS2 in this subfamily, with the other genes lacking this response element. In contrast, the promoters of the majority of ClGRAS genes exhibited the presence of ABA- and MeJA-responsive elements, while SA-responsive elements were identified in only a limited number of promoters. Furthermore, some environment-responsive elements, such as low-temperature-responsive elements, were identified. This type of element is predominantly observed in the SCL3 and HAM families, which may be attributed to the capacity of the genes within this subfamily to engage in temperature-related biological processes. Additionally, meristematic organization-related elements are present, particularly in the promoters of SCL9 and SCL26 subfamily genes. In light of these findings, it can be posited that ClGRAS genes may serve a pivotal function in the growth and development of Chinese fir, as well as in its stress response.
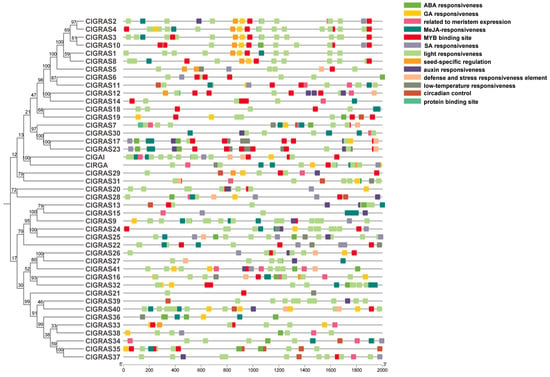
Figure 3.
The distribution of cis-acting elements in the 2000 bp promoter upstream of the ClGRAS genes. The phylogenetic evolutionary tree of ClGRASs is shown on the left, and different colored rectangles in the figure represent different cis-acting elements.
The genes on the chromosomes were visualized using TBtools software (version 2.119) based on the annotation files of the Chinese fir genome, which facilitated our understanding of the distribution of ClGRAS genes on the chromosomes and the genome-wide density (Figure 4). The results demonstrated that 42 genes were not uniformly distributed across the 11 chromosomes of the Chinese fir genome. Additionally, ClGRAS40 was not localized to any specific chromosome. Of the ClGRAS genes, chromosome 05 exhibited the greatest distribution, with 11 genes identified. Subsequently, chromosomes 10 and 11 exhibited a distribution of six and five genes, respectively. The remaining eight chromosomes demonstrated a range of one to four genes. In particular, gene clusters are formed on chromosomes 1, 4, 5, 8, and 10, and functional similarities may exist between genes belonging to the same cluster.
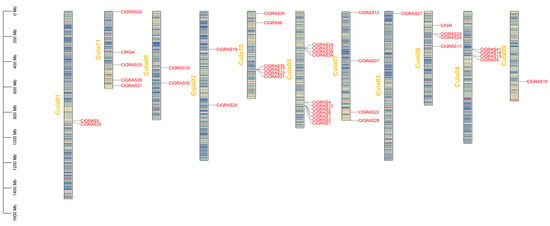
Figure 4.
Illustrates the chromosomal distribution of members of the ClGRAS gene family. The vertical bars indicate chromosomes, with the chromosome number in yellow on the left side of each bar, the gene name in red on the right side, and the chromosome density in the center. The scale on the left indicates chromosome length.
2.5. Collinearity Analysis in the ClGRAS Family of Chinese Fir
The analysis of collinearity among species can provide further insight into phylogenetic mechanisms. Consequently, we conducted an analysis of the collinearity of GRAS genes in Chinese fir, Cryptomeria japonica, and Populus trichocarpa. The results showed that among the woody plants, Chinese fir had a higher rate of GRAS gene pairing with Japanese cedar, which is also a Cupressaceae, with a total of 29 pairs of genes (Figure 5). Conversely, no pairing was observed between the P. trichocarpa GRAS genes and the Chinese fir. This indicates that the Chinese fir and C. japonica are more closely related in evolutionary terms.
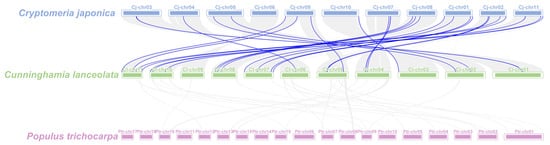
Figure 5.
Collinearity analysis of GRAS genes between C. japonica (blue), Chinese fir (green), and P. trichocarpa (purple). Gray lines indicate collinear blocks in genomes of Chinese fir and other plants, and blue lines indicate syntenic GRAS gene pairs.
2.6. Sequence Comparison, Conserved Motifs, and Structural Domains Analysis of Chinese Fir DELLA Proteins
The Chinese fir ClGRAS gene family comprises nine subfamilies, exhibiting functional variability among each subfamily. In the present study, we elected to explore the functions of two genes within the DELLA subfamily: ClGAI and ClRGA. A comparative analysis of the protein sequences of two Chinese fir DELLA proteins and five A. thaliana DELLA proteins was conducted. As illustrated in Figure 6A, there was a notable degree of similarity between the two species. To gain further insight into the DELLA protein sequences, we also undertook a detailed characterization of their conserved motifs and structural domains. A total of 10 conserved motifs were identified (Figure 6B), and the results demonstrated a high degree of similarity between the conserved motifs of the ClDELLA protein and those of the A. thaliana protein sequences. Both exhibited the presence of the GRAS and DELLA structural domains (Figure 6A,C), indicating that the functions of the DELLA proteins of the two may exhibit some degree of similarity.
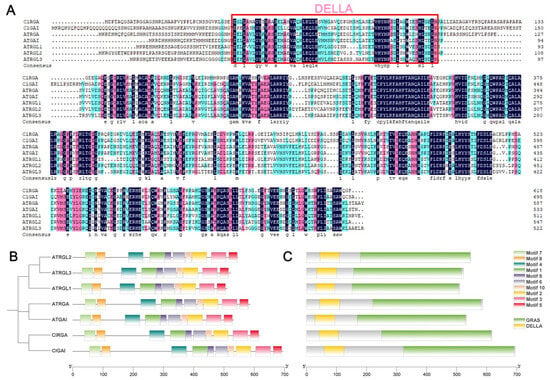
Figure 6.
(A) A sequence comparison of ClDELLAs and AtDELLAs. The pink box indicates the DELLA structural domain. (B) The distribution of conserved motifs of ClDELLAs and AtDELLAs. The phylogenetic evolutionary tree of the two is shown on the left. A total of 10 conserved motifs were identified, which are indicated by different colors. (C) Conserved structural domains of ClDELLAs and AtDELLAs. A yellow color indicates DELLA family structural domains and a green color indicates GRAS family structural domains.
2.7. Subcellular Localization and Transcriptional Activation Activity of ClDELLA Protein
We explored the fundamental properties of transcription factors through nuclear localization and transcriptional activation. The results of subcellular localization are shown in Figure 7A, where ClGAI and ClRGA are localized in the nucleus, consistent with the predicted results from bioinformatics analysis of ClGAI and ClRGA proteins (Table 1). To investigate the transcriptional activation activity of the two genes, the full-length CDS was fused to the DNA-binding domain of GAL4 (GAL4-BD) within the pGBKT7 vector, and the resulting construct was introduced to the yeast strain AH109. The AH109 strain was obtained by screening on tryptophan-free SD medium (SD/-Trp). Transformants containing the pBD-ClDELLA construct exhibited a blue coloration on SD medium containing X-α-gal but not Trp, His, and Ade (SD/-Trp-His-Ade), whereas the negative control displayed no coloration (Figure 7D). This suggests that both ClGAI and ClRGA possess transcriptional activation activity.
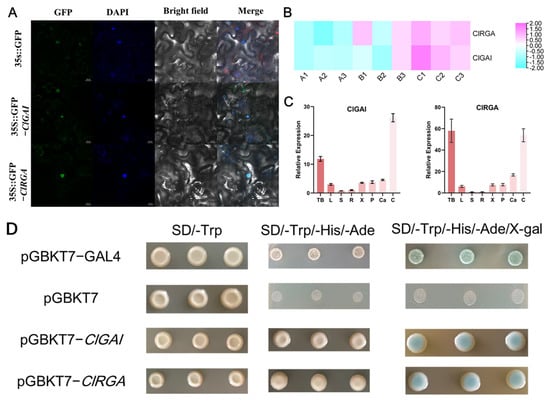
Figure 7.
Expression patterns and transient expression analysis of ClGAI and ClRGA. (A). Subcellular localization of ClDELLAs in tobacco leaves. The expression of ClGAI and ClRGA, which have been fused with GFP tags, was observed in tobacco leaves. The image is composed of four columns, each representing a different set of data: GFP, DAPI, bright field, and these images have been merged to create a single, composite representation. Bar = 25 μm. (B) Transcriptional profiles of Chinese fir DELLA genes under different developmental stages. A1-A3, B1-B3, and C1-C3 represent the fir growth reactivation, active, and dormant phases, respectively. The generation of heatmaps was based on log2 (FPKM + 1) values, which were subsequently normalized to a line scale. The color scales indicate the relative expression levels. (C) The relative expression levels of ClDELLAs in different tissues of the fir are presented. tb, terminal bud; L, leaf; S, stem; R, root; X, xylem; P, phloem; Ca, cambium; C, cones. (D) The positive control, negative control, pGBKT7-ClGAI, and pGBKT7-ClRGA yeast cells were incubated in SD/-Trp, SD/-Trp-His-Ade, and SD/-Trp-His-Ade + X-α-gal medium, respectively.
2.8. Expression Pattern of ClGAI and ClRGA in Different Tissues
It has been shown that in A. thaliana and populus, DELLA proteins can directly or indirectly affect the stem vascular cambium [31]. Therefore, to further understand the function of ClDELLA genes, we first analyzed two DELLA genes by real-time quantitative PCR (qRT-PCR). The results demonstrated that both genes were expressed in a majority of organs and tissues, including terminal buds, leaves, stems, roots, phloem, xylem, the vascular cambium, and cones. However, the expression levels exhibited considerable variation (Figure 7C). Both genes were highly expressed in terminal buds and cones, followed by the vascular cambium, while lower expression was found in stems and roots. This suggests that the expression of ClGAI and ClRGA is spatiotemporally specific during the growth and development of Chinese fir. However, the results of the transcriptomic analysis of the vascular cambium of Chinese fir at different developmental stages (reactivation (April), active (July), and dormant (September)) demonstrated that the expression levels of the two genes exhibited a gradual increase over time (Figure 6B). This finding is in accordance with the results presented in Figure 7C.
2.9. Expression Pattern Analysis of ClDELLAs Under Different Hormonal and Abiotic Stresses
To gain further insight into the functions of ClGAI and ClRGA, we conducted RT-qPCR analysis to examine the expression patterns of ClDELLAs in two-year-old Chinese fir seedlings subjected to various hormonal and abiotic stresses (Figure 8). It is noteworthy that under 200 μM GA3 treatment, the expression of both genes showed a gradual decrease with increasing time. This demonstrates the response of the DELLA genes to GA stress. Under 20 μM strigolactone (GR24) treatment, the expression of both ClGAI and ClRGA significantly increased at 6 h, and then gradually increased at 24 h. This phenomenon may be attributed to a gradual decline in GR24 hormone levels over time. In addition, both genes could be induced by IAA at different time points. Under 15% (w/v) PEG stress treatment, the expression of both genes was significantly decreased at 6 h, and both were repressed. In contrast, under 200 mM NaCl stress, the two genes showed different expression trends, with ClGAI expression increasing while ClRGA expression was suppressed. After the application of heat stress, the expression of both genes was found to be significantly decreased, but the expression of ClGAI increased at 8 h, and we speculate that it might be induced by other genes.
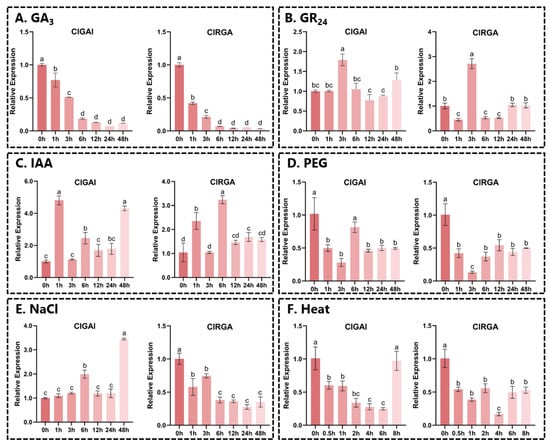
Figure 8.
Relative expression of ClDELLA genes under different treatments. (A) GA3, (B) GR24, (C) IAA, (D) PEG, (E) NaCl, and (F) heat. Vertical coordinates are relative expressions and horizontal coordinates indicate different sampling times. Same lowercase letters between different columns indicate that differences are not significant. Highest column is marked with “a”, then “b”, and so on. Completely different lowercase letter between columns indicates significant difference; p < 0.0001.
3. Discussion
The GRAS family plays a pivotal role in the signaling pathways of plants [32], as well as in their growth and development. However, the characterization and expression patterns of GRAS genes involved in abiotic stress responses in Chinese fir have not been previously elucidated. Consequently, we identified and analyzed GRAS genes in Chinese fir, and two DELLA genes were selected for functional analysis under different abiotic stress conditions.
In the present study, a total of 43 GRAS genes were identified in Chinese fir based on a whole genome thereof, and they were classified into nine subfamilies based on the A. thaliana subfamily classification. These subfamilies were as follows: SCR, SCL3, DELLA, LAS, HAM, SCL26, SCL9, SHR, and PAT1 (Figure 1). The number of GRAS family members in Chinese fir (43), similar to that of Betula platyphylla (40) [33], is higher than that of A. thaliana (33) [8], but less than that of Cyclocarya paliurus (51) [34], Castanea mollissima (48) [35], and Malus domestica (127) [36]. Variations in the number of gene families may be linked to genome size or gene duplication events during evolutionary processes [34]. Furthermore, the combination of conserved motifs, structural domains, and gene structures demonstrated similarities among genes within the same subfamily and differences among those in different subfamilies. It is noteworthy that the PAT1 family was the only one to contain motif 10 (Figure 2A), a result that aligns with the motifs of Glycine max [37] and Symplocos tanakana [38], but differs from those of cassava [13], E. grandis [12], and Cymbidium goeringii [39]. For instance, an examination of the GRAS family of E. grandis [12] reveals that the PAT1 subfamily exhibits a comparable motif composition and distribution. However, within the Chinese fir GRAS family, PAT1 is observed to occur with a motif 10 that is not present in other families. The presence of specific motifs in proteins indicates that they have unique functions. This indicates that the genes within the Chinese fir PATI subfamily may possess distinctive functions. It is noteworthy that the DELLA structural domain is exclusive to the DELLA family, while other subfamilies encompass the GRAS family or the GRAS superfamily. This may contribute to the diversification of the GRAS gene family and influence its functional specialization. An analysis of exon–intron patterns can provide further insights into the evolution of gene families. The gene structure of the ClGRAS family revealed that 28 genes lacked introns, while the remaining 15 genes had introns ranging from two to four (Figure 2C). The presence of introns serves to mitigate the deleterious effects of mutations on coding sequences. It has been demonstrated that the primary cause of intron deletion in gene families is rapid replication at the bacterial level following horizontal gene transfer [40].
Cis-acting elements are transcription factor binding sites and other regulatory motifs present in the paralogous sequences of genes with specific functions [41]. These elements are of significant importance with regard to the processes of gene transcription and regulation. In this study, we have identified cis-acting elements within the 2000 bp promoter region upstream of the ClGRAS genes. (Figure 3). The results of our investigation demonstrated the pervasive occurrence of phytohormone-responsive elements (SA, GA, MeJA, etc.) and abiotic stress elements (defense, low temperature, drought, etc.). Additionally, we observed the occurrence of elements related to plant growth and development, including meristematic tissue expression. These components were also commonly found with GRAS family genes in woody plants such as B. platyphylla [33] and Larix kaempferi [42], as well as in herbaceous plants such as C. macrorrhizum [39] and soybean [37]. This suggests that there is a similarity in the function of the GRAS gene family across species. It is noteworthy that the Chinese fir GRAS family contains some differences in the elements among the different subfamilies. To illustrate, the DELLA subfamily functions as a gibberellin response factor and comprises genes that all contain GA elements; the low-temperature response elements are predominantly found in the SCL3 and HAM families, indicating that they may be involved in the regulation of responses to temperature changes; and the majority of meristem-organization-related elements are concentrated in the SCL9 and SCL26 subfamilies, which suggests that they may have a regulatory function with regard to growth. To gain further insight into the characteristics of the ClGRAS family of genes, we conducted a chromosomal localization analysis on 43 genes. The results indicated that 42 of the genes were unevenly distributed across 11 chromosomes, while ClGRAS40 was not localized on any chromosomes.
Due to its rapid growth and superior wood properties, Chinese fir has become the most widely cultivated gymnosperm [43] in southern China. Consequently, a comprehensive understanding of its growth and development mechanisms is of paramount importance. In the GRAS family, the DELLA protein inhibits the mitotic activity of poplar cambium cells by interacting with ARK2 and WOX4, key regulators of the vascular cambium, and affecting the binding of ARK2 and WOX4 to downstream target genes [27]. Additionally, the DELLA protein is responsible for the formation of the SCR-SHR-DELLA complex, which plays a pivotal role in the circumferential morphogenesis of roots and stems [44]. Accordingly, an investigation of the DELLA gene in Chinese fir was deemed appropriate. The initial step involved an examination of the expression levels of ClGAI and ClRGA in various tissues. The findings indicated that these genes were implicated in the diverse stages of growth and development of Chinese fir. The elevated expression of the two genes in the vascular cambium indicates the potential involvement of these genes in the secondary growth of fir. Furthermore, the transcript levels of DELLA genes were analyzed in the vascular cambium at different developmental stages. The expression levels of DELLA genes demonstrated a gradual increase over time, with the highest transcript levels observed during the dormant stage. This indicates the potential involvement of DELLA genes in the response to adversity. However, further analyses and experiments are necessary to substantiate this hypothesis.
It has been demonstrated that DELLA proteins play a significant role in plant signaling pathways and stress response [20]. The expression of DELLA genes varies in response to abiotic stress in plants [45]. To illustrate this, salt stress or low-temperature stress has been observed to regulate the transcript abundance of specific DELLA genes in Cucurbita moschata [46]. Similarly, our findings indicated that both genes were repressed in response to GA treatment, drought stress, and high-temperature stress. Conversely, they were induced in IAA treatment. However, the two genes exhibited disparate trends in GR and NaCl treatments [47]. The aforementioned results indicate that ClGAI and ClRGA are integral to the response to abiotic stresses. For the proteins to perform their functions, they must be localized in appropriate regions. The results of subcellular localization showed that ClGAI and ClRGA play a regulatory role in the nucleus, which is consistent with the predicted results and with the localization of DELLA genes in A. thaliana [8], Vaccinium darrowii [48], poplar [14], and Paulownia fortune [49]. Moreover, both genes demonstrated transcriptional activation activity. Further investigation of their downstream regulatory genes may provide insights into the molecular mechanisms of growth and development and stress resistance in Chinese fir. As GA-negative regulators [50], they exhibited repression under GA treatment and demonstrated disparate responses to GR and IAA. Both genes exhibited repression under conditions of drought and high-temperature stress; however, they demonstrated disparate trends under salt treatment, indicating that they may serve as promising candidates for further investigation into their abiotic resistance. It is noteworthy that this study represents the inaugural report of GRAS genes in Chinese fir, has augmented the genetic data pertaining to the ClGRAS gene family, and has established a foundation for future investigations into the functional roles of GRAS genes, particularly the DELLA protein. Furthermore, this study offers a theoretical foundation for the selection and improvement of forest trees.
4. Materials and Methods
4.1. Identification and Physicochemical Characterization of Chinese Fir GRAS Gene Family
The Hidden Markov Model (HMM) profile for the GRAS domain (PF03514) was obtained from the Pfam database (Pfam is now hosted by InterPro (xfam.org)). The Chinese fir genome files were sourced from the NCBI website (National Center for Biotechnology Information (nih.gov)). The GRAS protein sequences from Arabidopsis were obtained from the TAIR website (TAIR–Home (arabidopsis.org)). The Hidden Markov Model (HMM) was employed as a query in the TBtools software to identify potential ClGRAS genes within the Chinese fir genome. A total of 43 candidate ClGRAS genes were identified and their protein sequences were extracted. The physicochemical properties of the candidate proteins were subsequently analyzed and plotted in a table using the online tool ExPasy (SIB Swiss Institute of Bioinformatics|Expasy) (Table 1). Furthermore, the subcellular localization of the candidate genes was predicted using the Plant-mPLoc (Plant-mPLoc server (sjtu.edu.cn) online tool).
4.2. Phylogenetic Analysis of the Chinese Fir GRAS Gene Family
Phylogenetic trees were constructed using Arabidopsis and Chinese fir GRAS protein sequences in MEGA 7.0 software, employing the neighbor-joining (NJ) method and 1000 bootstrap test replicates.
4.3. Analysis of Gene Structure and Conserved Motifs in the ClGRAS Family of Chinese Fir
The gene structure was analyzed using TBtools software, based on the Chinese fir genome annotation file (GFF3). The structural domains of 43 ClGRAS genes were then predicted by the Batch-CDD (Welcome to NCBI Batch CD-search (nih.gov)) online tool of the NCBI database, utilizing the default parameters. The conserved motifs of the GRAS proteins were analyzed using the MEME (Introduction–MEME Suite (meme-suite.org)) online tool (version 5.5.7), which revealed significant differences between members of the ClGRAS family. In conclusion, the aforementioned results were visualized using TBtools software (version 2.119).
4.4. Analysis of Cis-Acting Elements and Chromosomal Localization in the ClGRAS Family of Chinese Fir
In total, 2000 base pair sequences located upstream of each gene were extracted from the Chinese fir genome annotation file (GFF3) using TBtools software. These sequences were subsequently analyzed for promoter cis-acting elements using the online analysis software PlantCARE (PlantCARE, a database of plant promoters and their cis-acting regulatory elements (ugent.be)). The results were then filtered and simplified, and a visual representation was generated using TBtools software. Chromosome position data were received from the genome annotation file (GFF3) and mapped using TBtools software.
4.5. Expression Pattern Analysis of ClGAI and ClRGA
Thirty-year-old fir trees exhibiting robust growth were selected as sample materials for vascular cambium RNA-seq. The vascular cambium bands were obtained by scraping the stems. Samples were collected at three distinct growth phases: reactivation, active, and dormant. These samples were designated as A, B, and C, respectively. The samples were promptly placed in a freezer set to −80 °C for the purpose of RNA isolation. In addition, the two-year-old Chinese fir seedlings utilized in this study were sourced from Yangkou Forestry in Fujian, China, and subsequently transplanted to the greenhouse of Nanjing Forestry University for a two-month period of acclimatization. We collected RNA from organs such as terminal buds, leaves, stems, and roots of Chinese fir in April and tissues such as phloem, xylem, and vascular cambium as well as cones during the period of vigorous growth (July) (AG21019, ACCURATE BIOTECHNOLOGY (HUNAN), Co., Ltd., Changsha, China).
Healthy Chinese fir seedlings of equivalent height were selected and subjected to distinct treatments in a separate manner. For the purposes of this study, seedlings were selected for individual treatment with a solution of 200 μM GA3, 50 mg/L IAA, and 20 μM GR24 until the needle surfaces were completely wetted [51]. To simulate drought stress and salt stress, 15% (w/v) PEG-6000 and 200 mM NaCl were used as osmotic stressors, while a continuous 40 °C treatment was employed to imitate high-temperature stress [52]. Samples were collected at 0, 1, 3, 6, 12, 24, and 48 h, with the exception of those subjected to high-temperature stress, which were immediately stored at −80 °C for RNA isolation. Sampling was conducted at 0, 0.5, 1, 2, 4, and 8 h for the high-temperature stress treatment [53]. Three biological and three technical replicates were conducted for each treatment. Following the extraction of RNA from the samples, cDNA was generated through reverse transcription (SPARKscriptⅡRT Plus Kit (With gDNA Eraser) (Shandong Sparkjade Biotechnology Co., Ltd., Jinan, China)). Real-time quantitative PCR primers (Supplementary Material, Table S2) were then designed and tested using the Primer 6.0 software, The reaction was performed through the 2x Realab Green PCR Fast mixture (Beijing LABLEAD Inc., Beijing, China) in a 96-Well PCR plate (2040101, SAINING Biotechnology, Suzhou, China) with the quality of the PCR reactions evaluated from the melting curves. Three independent biological replicates and three technical replicates were tested for each biological replicate. A comparative cycle threshold (Ct) was employed for the quantification of gene expression levels, which were calculated as 2 (−∆Ct) [∆Ct = Ct Target − Ct TUA]. The results were then visualized and marked for significance using GraphPad software (version 10.1.2).
4.6. Subcellular Localization of the ClDELLA Protein
The cDNA sequence of the ClDELLA gene was extracted from the Chinese fir genome file using TBtools software. The coding sequences (CDSs) of the candidate genes, designated ClGAI and ClRGA, were cloned using the rapid amplification of cDNA ends (RACE) technique (Cat#10154; Yeasen, Shanghai, China). The enzymatic cleavage sites were selected to be BamHI and EcoRI, and the complete coding sequence was integrated into a modified vector, PCAMBIA1302, which contains a green fluorescent protein (GFP) reporter gene, via homologous recombination (CloneUFO® II One Step Cloning Kit (ATG Biotechnology, Nanjing, China)). Subsequently, the construct was transformed into strain GV3101, which was then used to infest tobacco epidermal cells. The gene localization was determined by observing the fluorescence signal.
4.7. Transcriptional Activation Activity of the ClDELLA Protein
The enzyme cleavage sites were selected as BamHI and EcoRI, the intact CDS was inserted into the pGBKT7 vector by homologous recombination, and the construct was introduced into Dh5α (#CC96102, Tolo Biotech Co., Ltd., Shanghai, China) and yeast strain AH109. Screening on tryptophan-free SD medium (SD/-Trp) yielded the AH109-positive strain (28 °C, 48–96 h). Subsequently, yeast cells were cultured on SD/-Trp-His-Ade and SD/-Trp-His-Ade+X-α-gal (28 °C, 48–96 h). The transcriptional activation activity of the proteins was determined by observing the growth of the strains on the medium and the resulting discoloration.
5. Conclusions
In the present study, a total of 43 ClGRAS genes were identified in the Chinese fir genome, and phylogenetic analyses revealed the existence of nine distinct clades. A gene structure analysis demonstrated that the majority of ClGRAS genes lacked introns, indicating that they are highly structurally conserved. The members of the same clade exhibit extensive similarity in conserved motifs and structural domains, which suggests that they have parallel gene functions. Furthermore, an investigation was conducted into the functions of the DELLA subfamily genes. The two genes demonstrated differential expression in discrete tissues, suggesting that they are spatio-temporally specific with regard to their involvement in plant growth and development. Based on the transcriptome data of the vascular cambium of Chinese fir at different developmental stages and RT-qPCR analysis after different treatments, it was found that they could be candidate genes for further study of their abiotic resistance. Importantly, this study reported the GRAS genes in Chinese fir for the first time, enriched the genetic information of the ClGRAS gene family, and established a foundation for the investigation of the function of GRAS genes, particularly the DELLA protein. Furthermore, it offers a theoretical foundation for the selection and improvement of forest trees.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms252212262/s1.
Author Contributions
Conceptualization, Y.L. and J.Y.; methodology, Y.Y., H.L. and T.G.; performed the experiments: Y.L., J.Y. and M.J.; analyzed the data: Y.L., J.Y., M.J. and R.G.; writing—original draft preparation, Y.L.; Funding acquisition, J.X. All authors have read and agreed to the published version of the manuscript.
Funding
This search was supported by the National Key Research and Development Program of China (2023YFD2200105), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We appreciate the reviewers’ and editors’ diligent reading and constructive criticism of this work.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hui, X.; Lin, Z.Z.; Su, S.D.; Jiang, X.L.; Chen, H.Q.; Wu, W.; Luo, S.J.; Pan, L.Y.; Zhen, R.H. Genetic variation analysis and ultra-early selection of nursery-determined traits in Chinese fir asexual lines. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2024, 48, 63–70. [Google Scholar]
- Lu, X.D.; Dong, Y.R.; Li, G.; Mao, L.F. Mechanisms of community construction in different stand development stages of subtropical Chinese fir plantation forests in China. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2024, 48, 67–73. [Google Scholar]
- Song, L.; Li, W.; Chen, X. Transcription factor is not just a transcription factor. Trends Plant. 2022, 27, 1087–1089. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, J.; Wang, W.; Gao, Y.; Xian, X.; Li, C.; Wang, Y. Transcription factors dealing with Iron-deficiency stress in plants: Focus on the bHLH transcription factor family. Physiol. Plantarum. 2023, 175, e14091. [Google Scholar] [CrossRef]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef]
- Zeng, X.; Ling, H.; Chen, X.; Guo, S. Genome-wide identification, phylogeny and function analysis of GRAS gene family in Dendrobium catenatum (Orchidaceae). Gene 2019, 705, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Cenci, A.; Rouard, M. Evolutionary Analyses of GRAS Transcription Factors in Angiosperms. Front. Plant Sci. 2017, 8, 273. [Google Scholar] [CrossRef]
- Lee, H.; Kim, B.; Song, S.; Heo, J.O.; Yu, N.L.; Lee, S.A.; Kim, M.; Sohn, S.O.; Lim, C.E.; Chang, K.; et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant. Mol. Biol. 2008, 67, 659–670. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Tai, S.; Wang, D.W.; Ding, A.M.; Sun, T.; Wang, W.F.; Sun, Y.H. Homology-based analysis of the GRAS gene family in tobacco. Genet. Mol. Res. 2015, 14, 15188–15200. [Google Scholar] [CrossRef]
- Liu, X.Y.; Widmer, A. Genome-wide Comparative Analysis of the GRAS Gene Family in Populus, Arabidopsis and Rice. Plant Mol. Bio. Rep. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Tian, C.G.; Wan, P.; Sun, S.H.; Li, J.Y.; Chen, M.S. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Bhalothia, P. Evolutionary analysis of GRAS gene family for functional and structural insights into hexaploid bread wheat (Triticum aestivum). J. Biosci. 2021, 46, 15. [Google Scholar] [CrossRef]
- Zhu, L.; Yin, T.; Zhang, M.J.; Yang, X.Y.; Wu, J.X.; Cai, H.B.; Yang, N.; Li, X.L.; Wen, K.; Chen, D.M.; et al. Genome-wide identification and expression pattern analysis of the kiwifruit GRAS transcription factor family in response to salt stress. BMC Genom. 2024, 25, 12. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xu, J.; Li, G.; Zhong, T.; Chen, D.; Lv, J. Genome-wide identification and expression analysis of GRAS gene family in Eucalyptus grandis. BMC Plant Biol. 2024, 24, 573. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Luo, X.; Wu, M.Y.; Wei, L.M.; Fan, Z.P.; Zhu, Y.M. Genome-wide identification and expression of GRAS gene family members in cassava. BMC Plant Biol. 2020, 20, 46. [Google Scholar] [CrossRef]
- Cai, H.; Xuan, L.; Xu, L.A.; Huang, M.R.; Xu, M. Identification and characterization of nine PAT1 branch genes in poplar. Plant Growth Regul. 2016, 81, 355–364. [Google Scholar] [CrossRef]
- Bisht, A.; Eekhout, T.; Canher, B.; Lu, R.; Vercauteren, I.; De Jaeger, G.; Heyman, J.; De Veylder, L. PAT1-type GRAS-domain proteins control regeneration by activating DOF3.4 to drive cell proliferation in Arabidopsis roots. Plant Cell 2023, 35, 1513–1531. [Google Scholar] [CrossRef]
- Wang, Z.; Wong, D.; Wang, Y.; Xu, G.Z.; Ren, C.; Liu, Y.F.; Kuang, Y.F.; Fan, P.G.; Li, S.H.; Xin, H.P.; et al. GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 2021, 186, 1660–1678. [Google Scholar] [CrossRef]
- Chen, C.; Lu, L.L.; Ma, S.Y.; Zhao, Y.P.; Wu, N.; Li, W.J.; Ma, L.; Kong, X.H.; Xie, Z.M.; Hou, Y.X. Analysis of PAT1 subfamily members in the GRAS family of upland cotton and functional characterization of GhSCL13-2A in Verticillium dahliae resistance. Plant Cell Rep. 2023, 42, 487–504. [Google Scholar] [CrossRef]
- Clark, N.M.; Hinde, E.; Winter, C.M.; Fisher, A.P.; Crosti, G.; Blilou, I.; Gratton, E.; Benfey, P.N.; Sozzani, R. Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. eLife 2016, 5, 25. [Google Scholar] [CrossRef]
- Heo, J.O.; Chang, K.S.; Kim, I.A.; Lee, M.H.; Lee, S.A.; Song, S.K.; Lee, M.; Lim, J. Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2011, 108, 2166–2171. [Google Scholar] [CrossRef]
- Blanco-Tourin, N.; Serrano-Mislata, A.; Alabad, D. Regulation of DELLA Proteins by Post-translational Modifications. Plant Cell Physiol. 2020, 61, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.F.; Zhang, Q.Q.; Chen, Y.C.; Zhao, Y.X.; Ren, F.S.; Shi, H.M.; Wu, X.Y. Comprehensive identification and analysis of DELLA genes throughout the plant kingdom. BMC Plant Biol. 2020, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Hirano, K.; Sato, T.; Mistuda, N.; Nomoto, M.; Maeo, K.; Koketsu, E.; Mitani, R.; Kawamura, M.; Ishiguro, S.; et al. DELLA protein functions as a transcriptional activator through the DNA binding of the INDETERMINATE DOMAIN family proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 7861–7866. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Hirano, Y.; Sun, T.P.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef]
- Huang, H.; Gong, Y.L.; Liu, B.; Wu, D.W.; Zhamg, M.; Xie, D.X.; Song, S.S. The DELLA proteins interact with MYB21 and MYB24 to regulate filament elongation in Arabidopsis. BMC Plant Biol. 2020, 20, 64. [Google Scholar] [CrossRef]
- Li, M.; An, F.Y.; Li, W.Y.; Ma, M.D.; Feng, Y.; Zhang, X.; Guo, H.W. DELLA proteins interact with FLC to repress flowering transition. J. Integr. Plant Biol. 2016, 58, 642–655. [Google Scholar] [CrossRef]
- Fukazawa, J.; Mori, K.; Ando, H.; Mori, R.; Kanno, Y.; Seo, M.; Takahashi, Y. Jasmonate inhibits plant growth and reduces gibberellin levels via microRNA5998 and transcription factor MYC2. Plant Physiol. 2023, 193, 2197–2214. [Google Scholar] [CrossRef]
- Hui, C. Protein Interactions Between DELLA, ARK2 and WOX4 Mediate the Molecular Mechanism of Gibberellin Regulation of Poplar Formative Layer Activity. Master’s Thesis, Southwest University, Chongqing, China, 2022. [Google Scholar]
- Tai, C.H.; Sam, V.; Gibrat, J.F.; Garnier, J.; Munson, P.; Lee, B. Protein domain assignment from the recurrence of locally similar structures. Proteins 2011, 79, 853–866. [Google Scholar] [CrossRef]
- Ben-Targem, M.; Ripper, D.; Bayer, M.; Ragni, L. Auxin and gibberellin signaling cross-talk promotes hypocotyl xylem expansion and cambium homeostasis. J. Exp. Bot. 2021, 72, 3647–3660. [Google Scholar] [CrossRef]
- Huang, M.X.; Zhang, G.F.; Gan, H.Y.; Liu, C.; Li, M.M.; Shu, Y.J. Genome-wide analysis of the GRAS gene family in white clover Trifolium repens L. provides insight into its critical role in response to cold stress. Biotechnol. Biotec. Eq. 2024, 38, 14. [Google Scholar] [CrossRef]
- He, Z.H.; Tian, Z.Z.; Zhang, Q.; Wang, Z.B.; Huang, R.K.; Xu, X.; Wang, Y.C.; Ji, X.Y. Genome-wide identification, expression and salt stress tolerance analysis of the GRAS transcription factor family in Betula platyphylla. Front. Plant Sci. 2022, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yang, Y.B.; Yu, Y.H.; Mei, D.; Mao, X.; Fu, X.X. Genome-Wide Characterization of the GRAS Gene Family in Cyclocarya paliurus and Its Involvement in Heterodichogamy. Agronomy 2024, 14, 2397. [Google Scholar] [CrossRef]
- Yu, L.; Hui, C.; Huang, R.; Wang, D.; Fei, C.; Guo, C.; Zhang, J. Genome-wide identification, evolution and transcriptome analysis of GRAS gene family in Chinese chestnut (Castanea mollissima). Front. Genet. 2023, 13, 1080759. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, D.; Gao, C.; Zhao, M.; Wu, H.; Li, Y.; Shen, Y.; Han, M. Identification, classification, and expression analysis of GRAS gene family in Malus domestica. Front. Physiol. 2017, 8, 253. [Google Scholar] [CrossRef]
- Wang, L.; Ding, X.L.; Gao, Y.Q.; Yang, S.P. Genome-wide identification and characterization of GRAS genes in soybean (Glycine max). BMC Plant Biol. 2020, 20, 415. [Google Scholar] [CrossRef]
- Wang, S.; Duan, Z.; Yan, Q.; Wu, F.; Zhou, P.; Zhang, J.Y. Genome–Wide Identification of the GRAS Family Genes in Melilotus albus and Expression Analysis under Various Tissues and Abiotic Stresses. Int. J. Mol. Sci. 2022, 23, 7403. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, Q.Y.; Zhang, M.M.; He, X.; Zhao, X.W.; Wang, L.Y.; Lan, S.R.; Liu, Z.J. Genome-Wide Identification and Expression Analysis of the GRAS Gene Family and Their Responses to Heat Stress in Cymbidium goeringii. Int. J. Mol. Sci. 2024, 25, 6363. [Google Scholar] [CrossRef]
- Yuan, W.; Yu, J.; Li, Z.C. Rapid functional activation of horizontally transferred eukaryotic intron-containing genes in the bacterial recipient. Nucleic Acids Res. 2024, 52, 8344–8355. [Google Scholar] [CrossRef]
- Rushton, P.J. What Have We Learned About Synthetic Promoter Construction? Methods Mol. Biol. 2016, 1482, 1–13. [Google Scholar]
- Ma, M.; Li, L.; Wang, X.H.; Zhang, C.Y.; Pak, S.; Li, C.H. Comprehensive Analysis of GRAS Gene Family and Their Expression under GA3, Drought Stress and ABA Treatment in Larix kaempferi. Forests 2022, 13, 1424. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Y.; Yang, Y. Inventory of gymnosperm species in China based on the latest gymnosperm taxonomic system. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2024, 48, 49–56. [Google Scholar]
- Aoyanagi, T.; Ilwya, S.; Kobayashi, A.; Kozaki, A. Gene Regulation via the Combination of Transcription Factors in the INDETERMINATE DOMAIN and GRAS Families. Genes 2020, 11, 18. [Google Scholar] [CrossRef]
- Briones-Moreno, A.; Hwrnandez-Garcia, J.; Vargas-Chavea, C.; Blanco-Tourinan, N.; Phokas, A.; Urbez, C.; Cerdan, P.D.; Coates, J.C.; Alabadi, D.; Blazquez, M.A. DELLA functions evolved by rewiring of associated transcriptional networks. Nat. Plants 2023, 9, 535–543. [Google Scholar] [CrossRef]
- Luo, W.R.; Zhao, Z.X.; Chen, H.Z.; Ao, W.H.; Lu, L.; Liu, J.J.; Li, X.Z.; Sun, Y.D. Genome-wide characterization and expression of DELLA genes in Cucurbita moschata reveal their potential roles under development and abiotic stress. Front. Plant Sci. 2023, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Wang, T.; Yang, K.; Li, L.B. Characterization of cireRNAs and its expression in germinating seeds of Phyllostachys edulis under PEG and NaCl stress. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2023, 47, 17–24. [Google Scholar]
- Zhou, H.; Wang, Y.W.; Wang, X.Y.; Cheng, R.; Zhang, H.X.; Yang, L. Genome-wide characterization of DELLA gene family in blueberry (Vaccinium darrowii) and their expression profiles in development and response to abiotic stress. BMC Genom. 2024, 25, 815. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Cao, Y.B.; Fan, Y.J.; Fan, G.Q. Comprehensive Analysis of the GRAS Gene Family in Paulownia fortunei and the Response of DELLA Proteins to Paulownia Witches’ Broom. Int. J. Mol. Sci. 2024, 25, 15. [Google Scholar] [CrossRef]
- Mäkilä, R.; Wybouw, B.; Smetana, O.; Vainio, L.; Sole-Gil, A.; Lyu, M.; Ye, L.L.; Wang, X.; Siligato, R.; Jeness, M.K.; et al. Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium. Nat. Plants 2023, 9, 631–644. [Google Scholar] [CrossRef]
- Wang, D.B.; Qiu, Z.M.; Xu, T.; Yao, S.; Zhang, M.Y.; Cheng, X.; Zhao, Y.L.; Ji, K.S. Identification and Expression Patterns of WOX Transcription Factors under Abiotic Stresses in Pinus massoniana. Int. J. Mol. Sci. 2024, 25, 1627. [Google Scholar] [CrossRef]
- Zhu, L.J.; Yang, J.J.; Zhang, Y.T.; Hu, H.L.; Cui, J.B.; Xue, J.Y.; Xu, J. Overexpression of CfICE1 from Cryptomeria fortunei Enhances Cold, Drought and Salt Stress in Poplar. Int. J. Mol. Sci. 2022, 23, 15214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.T.; Xue, J.Y.; Zhu, L.J.; Hu, H.L.; Yang, J.J.; Cui, J.B.; Xu, J. Selection and Optimization of Reference Genes for MicroRNA Expression Normalization by qRT-PCR in Chinese Cedar (Cryptomeria fortunei) under Multiple Stresses. Int. J. Mol. Sci. 2021, 22, 7246. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).