Metabolic Flux Analysis of Xanthomonas oryzae Treated with Bismerthiazol Revealed Glutathione Oxidoreductase in Glutathione Metabolism Serves as an Effective Target
Abstract
1. Introduction
2. Result and Discussion
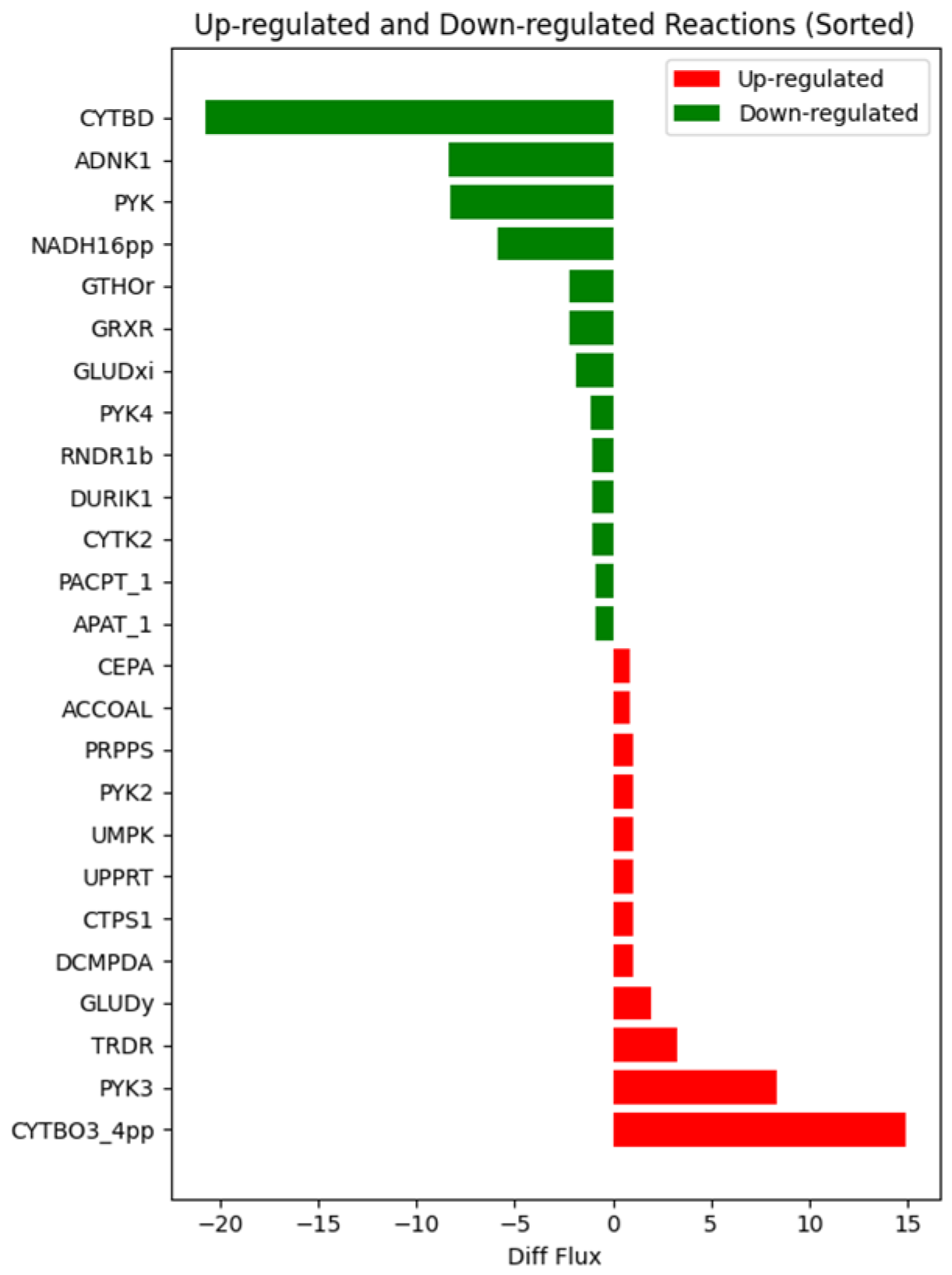
2.1. GSMM Reconstruction and FBA Analysis
2.2. Mechanism Analysis and Potential Target Discovery
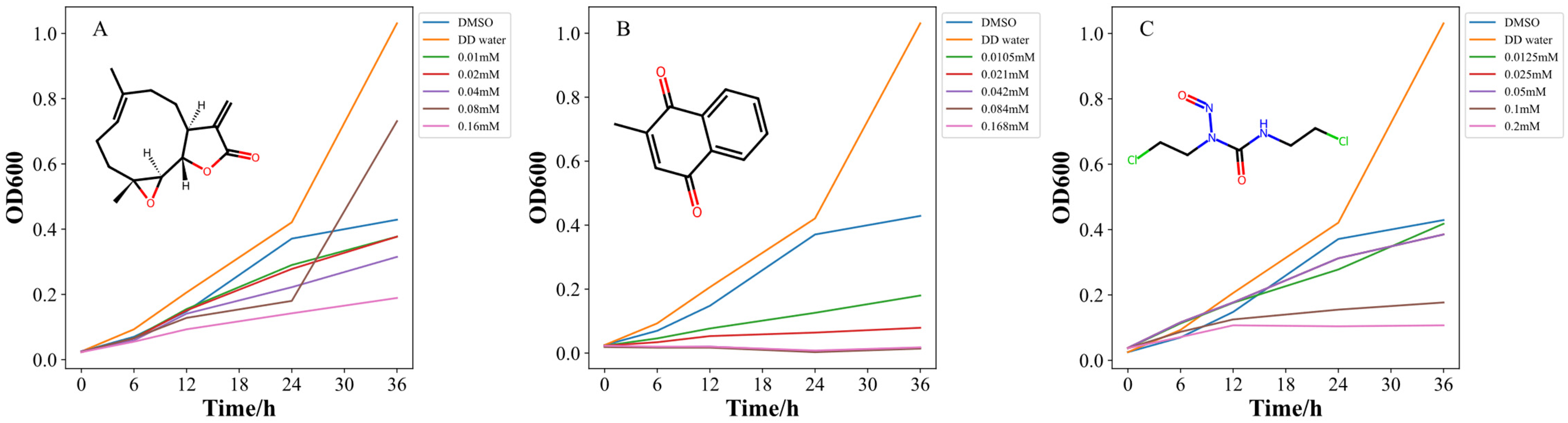
2.3. Effectiveness Test of Target GSR
2.4. Inhibitor Screening Based on Xoo-GSR
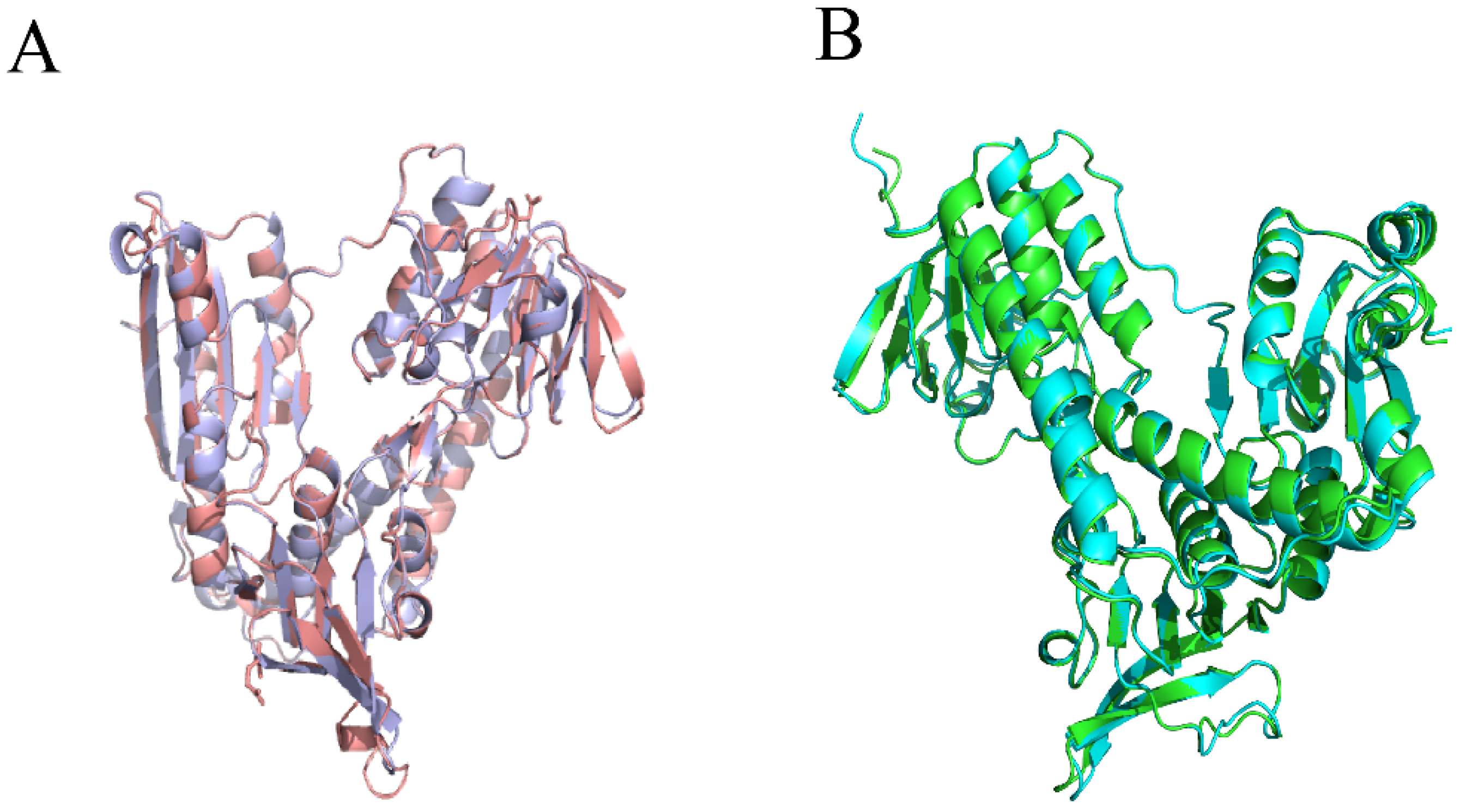
2.4.1. Homology Modeling of Xoo-GSR
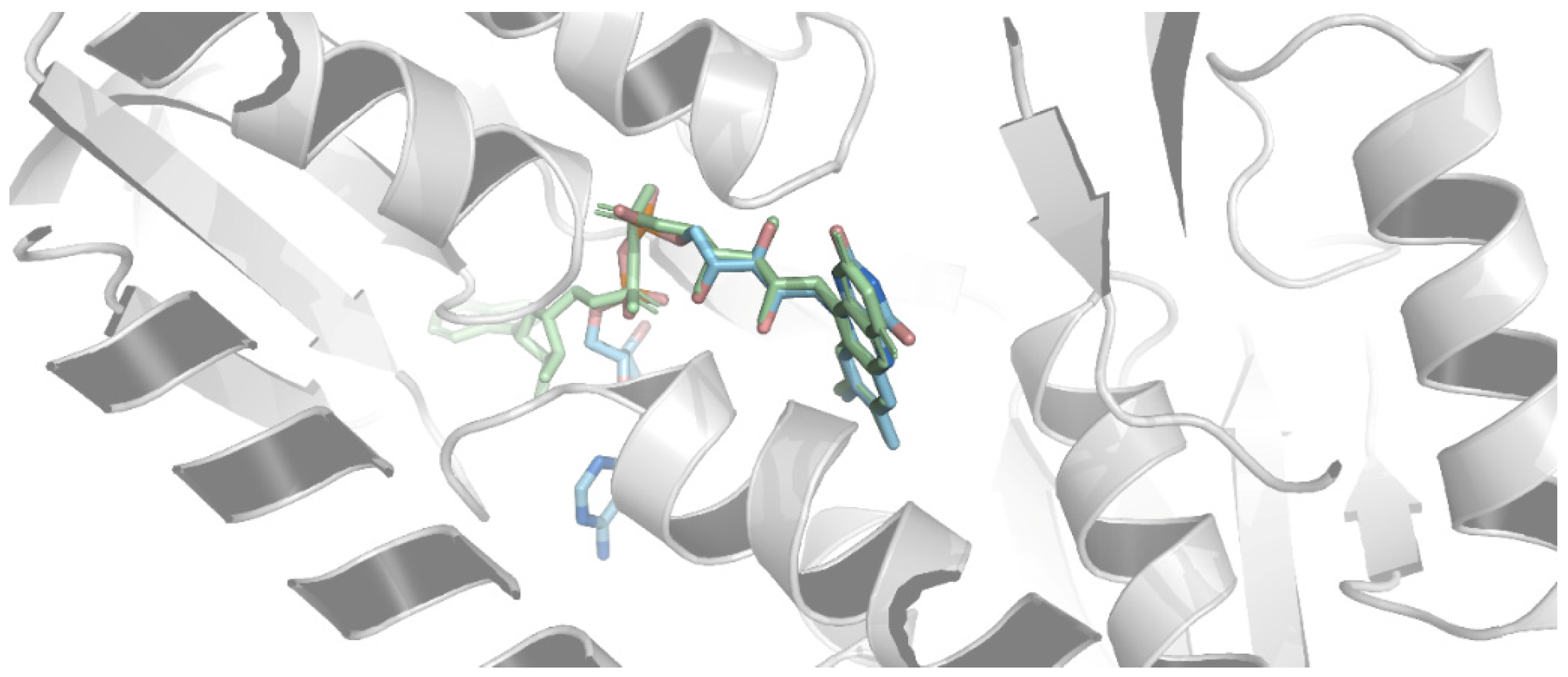
2.4.2. Docking Analysis
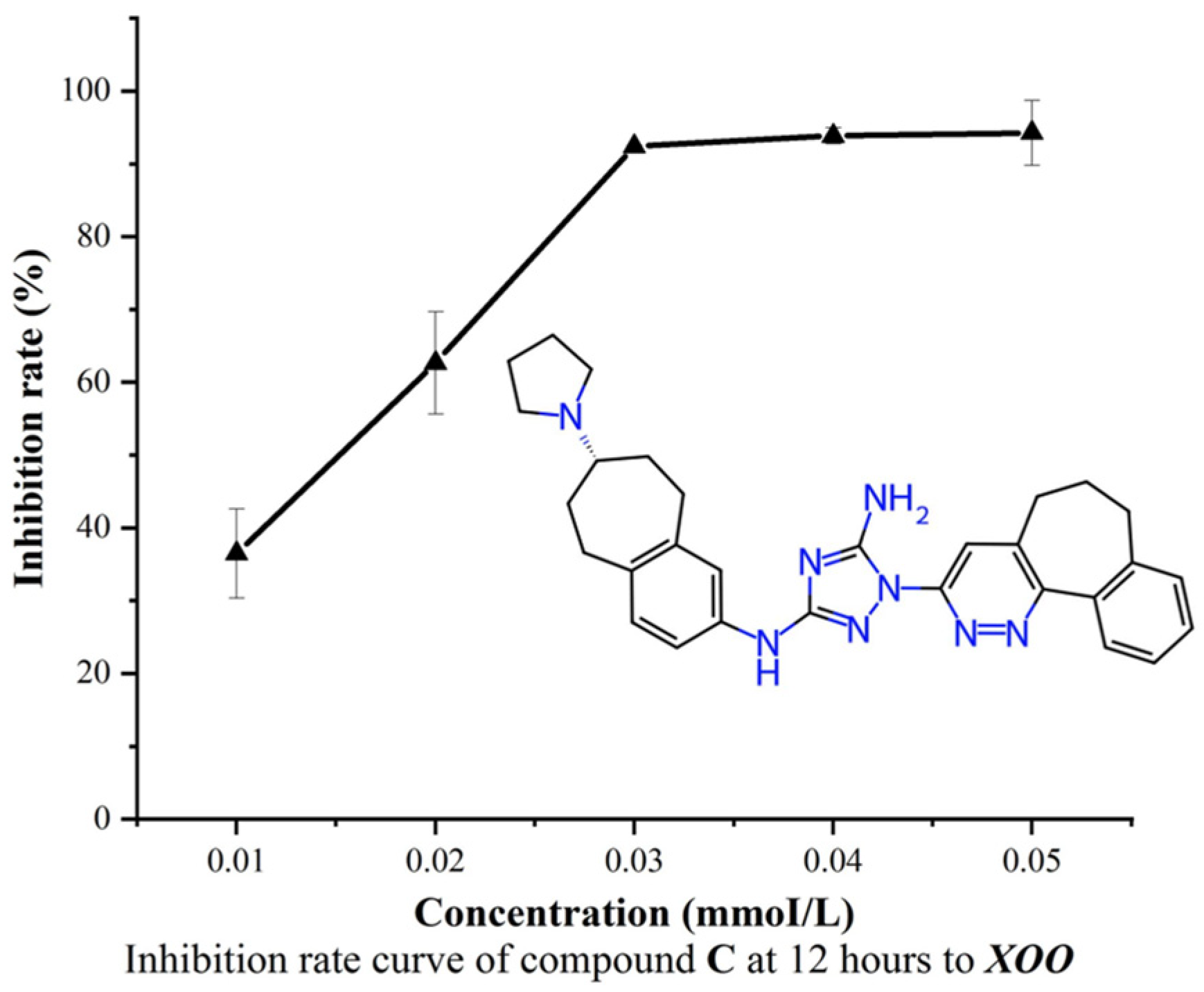
2.4.3. Experimental Evaluation
3. Materials and Methods
3.1. RNA-Seq Data Preprocessing
3.2. Automated Reconstruction and Analysis of GSMM
3.3. Antibacterial Experiment
3.4. Virtual Screening of Xoo-GSR Targets
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Chukwu, S.; Rafii, M.; Ramlee, S.; Ismail, S.; Hasan, M.; Oladosu, Y.; Magaji, U.; Akos, I.; Olalekan, K. Bacterial leaf blight resistance in rice: A review of conventional breeding to molecular approach. Mol. Biol. Rep. 2019, 46, 1519–1532. [Google Scholar] [CrossRef] [PubMed]
- NIÑO-LIU, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wu, Z.; Zhang, L.; Ma, Q.; Cai, D.; Zhang, J.; Hu, D. Design, synthesis, antibacterial activity, and mechanism of novel mesoionic compounds based on natural pyrazole isolated from an endophytic fungus Colletotrichum gloeosporioides. J. Agric. Food Chem. 2023, 71, 10018–10027. [Google Scholar] [CrossRef]
- Huang, X.; Liu, H.-W.; Long, Z.-Q.; Li, Z.-X.; Zhu, J.-J.; Wang, P.-Y.; Qi, P.-Y.; Liu, L.-W.; Yang, S. Rational optimization of 1, 2, 3-triazole-tailored carbazoles as prospective antibacterial alternatives with significant in vivo control efficiency and unique mode of action. J. Agric. Food Chem. 2021, 69, 4615–4627. [Google Scholar] [CrossRef]
- Nelson, R.J.; Baraoidan, M.R.; Cruz, C.M.V.; Yap, I.V.; Leach, J.E.; Mew, T.W.; Leung, H. Relationship between phylogeny and pathotype for the bacterial blight pathogen of rice. Appl. Environ. Microbiol. 1994, 60, 3275–3283. [Google Scholar] [CrossRef]
- Lee, S.-W.; Han, M.; Park, C.-J.; Seo, Y.-S.; Bartley, L.E.; Jeon, J.-S. The molecular mechanisms of rice resistance to the bacterial blight pathogen, Xanthomonas oryzae pathovar oryzae. Adv. Bot. Res. 2011, 60, 51–87. [Google Scholar]
- Teng, K.; Liu, Q.; Zhang, M.; Naz, H.; Zheng, P.; Wu, X.; Chi, Y.R. Design and Enantioselective Synthesis of Chiral Pyranone Fused Indole Derivatives with Antibacterial Activities against Xanthomonas oryzae pv oryzae for Protection of Rice. J. Agric. Food Chem. 2024, 72, 4622–4629. [Google Scholar] [CrossRef]
- Liang, X.; Duan, Y.; Yu, X.; Wang, J.; Zhou, M. Photochemical degradation of bismerthiazol: Structural characterisation of the photoproducts and their inhibitory activities against Xanthomonas oryzae pv. oryzae. Pest Manag. Sci. 2016, 72, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Mo, X.; Wang, W.; Chen, X.; Lou, Y. The commonly used bactericide bismerthiazol promotes rice defenses against herbivores. Int. J. Mol. Sci. 2018, 19, 1271. [Google Scholar] [CrossRef]
- Yahong, Z.; Xiaofang, C.; Shuo, W.; Congfeng, S. Functional analysis of tal4 of Xanthomonas oryzae pv. oryzae strain PXO99 A in resistance to bismerthiazol. J. Nanjing Agric. Univ./Nanjuing Nongye Daxue Xuebao 2014, 37, 57. [Google Scholar]
- Zhu, X.-F.; Xu, Y.; Peng, D.; Zhang, Y.; Huang, T.-T.; Wang, J.-X.; Zhou, M.-G. Detection and characterization of bismerthiazol-resistance of Xanthomonas oryzae pv. oryzae. Crop Prot. 2013, 47, 24–29. [Google Scholar] [CrossRef]
- Liang, X.; Yu, X.; Pan, X.; Wu, J.; Duan, Y.; Wang, J.; Zhou, M. A thiadiazole reduces the virulence of Xanthomonas oryzae pv. oryzae by inhibiting the histidine utilization pathway and quorum sensing. Mol. Plant Pathol. 2018, 19, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Duarte, N.C.; Becker, S.A.; Jamshidi, N.; Thiele, I.; Mo, M.L.; Vo, T.D.; Srivas, R.; Palsson, B.Ø. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. USA 2007, 104, 1777–1782. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.; Swainston, N.; Fleming, R.M.; Hoppe, A.; Sahoo, S.; Aurich, M.K.; Haraldsdottir, H.; Mo, M.L.; Rolfsson, O.; Stobbe, M.D. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013, 31, 419–425. [Google Scholar] [CrossRef]
- O’Brien, E.J.; Monk, J.M.; Palsson, B.O. Using genome-scale models to predict biological capabilities. Cell 2015, 161, 971–987. [Google Scholar] [CrossRef]
- Ye, C.; Wei, X.; Shi, T.; Sun, X.; Xu, N.; Gao, C.; Zou, W. Genome-scale metabolic network models: From first-generation to next-generation. Appl. Microbiol. Biotechnol. 2022, 106, 4907–4920. [Google Scholar] [CrossRef]
- Gu, C.; Kim, G.B.; Kim, W.J.; Kim, H.U.; Lee, S.Y. Current status and applications of genome-scale metabolic models. Genome Biol. 2019, 20, 121. [Google Scholar] [CrossRef]
- Machado, D.; Andrejev, S.; Tramontano, M.; Patil, K.R. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018, 46, 7542–7553. [Google Scholar] [CrossRef]
- Molina Ortiz, J.P.; Read, M.N.; McClure, D.D.; Holmes, A.; Dehghani, F.; Shanahan, E.R. High throughput genome scale modeling predicts microbial vitamin requirements contribute to gut microbiome community structure. Gut Microbes 2022, 14, 2118831. [Google Scholar] [CrossRef]
- Wang, H.; Marcišauskas, S.; Sánchez, B.J.; Domenzain, I.; Hermansson, D.; Agren, R.; Nielsen, J.; Kerkhoven, E.J. RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput. Biol. 2018, 14, e1006541. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S. KBase: The United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Mendoza, S.N.; Olivier, B.G.; Molenaar, D.; Teusink, B. A systematic assessment of current genome-scale metabolic reconstruction tools. Genome Biol. 2019, 20, 158. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, R.; Schilling, C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003, 5, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Schellenberger, J.; Palsson, B.Ø. Use of randomized sampling for analysis of metabolic networks. J. Biol. Chem. 2009, 284, 5457–5461. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.E.; Hixson, K.K.; Conrad, T.M.; Lerman, J.A.; Charusanti, P.; Polpitiya, A.D.; Adkins, J.N.; Schramm, G.; Purvine, S.O.; Lopez-Ferrer, D. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Biol. 2010, 6, 390. [Google Scholar] [CrossRef]
- Córdoba, S.C.; Tong, H.; Burgos, A.; Zhu, F.; Alseekh, S.; Fernie, A.R.; Nikoloski, Z. Identification of gene function based on models capturing natural variability of Arabidopsis thaliana lipid metabolism. Nat. Commun. 2023, 14, 4897. [Google Scholar] [CrossRef]
- Paul, A.; Anand, R.; Karmakar, S.P.; Rawat, S.; Bairagi, N.; Chatterjee, S. Exploring gene knockout strategies to identify potential drug targets using genome-scale metabolic models. Sci. Rep. 2021, 11, 213. [Google Scholar] [CrossRef]
- Becker, S.A.; Palsson, B.O. Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 2008, 4, e1000082. [Google Scholar] [CrossRef] [PubMed]
- Blazier, A.S.; Papin, J.A. Integration of expression data in genome-scale metabolic network reconstructions. Front. Physiol. 2012, 3, 299. [Google Scholar] [CrossRef]
- Jamialahmadi, O.; Hashemi-Najafabadi, S.; Motamedian, E.; Romeo, S.; Bagheri, F. A benchmark-driven approach to reconstruct metabolic networks for studying cancer metabolism. PLoS Comput. Biol. 2019, 15, e1006936. [Google Scholar] [CrossRef]
- Kim, M.K.; Lane, A.; Kelley, J.J.; Lun, D.S. E-Flux2 and SPOT: Validated methods for inferring intracellular metabolic flux distributions from transcriptomic data. PLoS ONE 2016, 11, e0157101. [Google Scholar] [CrossRef]
- Palsson, B. In silico biology through “omics”. Nat. Biotechnol. 2002, 20, 649–650. [Google Scholar] [CrossRef]
- Zur, H.; Ruppin, E.; Shlomi, T. iMAT: An integrative metabolic analysis tool. Bioinformatics 2010, 26, 3140–3142. [Google Scholar]
- Zhang, A.; Zhang, H.; Wang, R.; He, H.; Song, B.; Song, R. Bactericidal bissulfone B7 targets bacterial pyruvate kinase to impair bacterial biology and pathogenicity in plants. Sci. China Life Sci. 2024, 67, 391–402. [Google Scholar]
- Lucarelli, A.P.; Buroni, S.; Pasca, M.R.; Rizzi, M.; Cavagnino, A.; Valentini, G.; Riccardi, G.; Chiarelli, L.R. Mycobacterium tuberculosis phosphoribosylpyrophosphate synthetase: Biochemical features of a crucial enzyme for mycobacterial cell wall biosynthesis. PLoS ONE 2010, 5, e15494. [Google Scholar]
- Villela, A.D.; Ducati, R.G.; Rosado, L.A.; Bloch, C.J.; Prates, M.V.; Goncalves, D.C.; Ramos, C.H.I.; Basso, L.A.; Santos, D.S. Biochemical characterization of uracil phosphoribosyltransferase from Mycobacterium tuberculosis. PLoS ONE 2013, 8, e56445. [Google Scholar]
- Torrents, E. Ribonucleotide reductases: Essential enzymes for bacterial life. Front. Cell. Infect. Microbiol. 2014, 4, 52. [Google Scholar]
- Zhu, Z.; Du, S.; Du, Y.; Ren, J.; Ying, G.; Yan, Z. Glutathione reductase mediates drug resistance in glioblastoma cells by regulating redox homeostasis. J. Neurochem. 2018, 144, 93–104. [Google Scholar]
- Fleming, A.B.; Saltzman, W.M. Pharmacokinetics of the carmustine implant. Clin. Pharmacokinet. 2002, 41, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Brittain, H.G. Profiles of drug substances, excipients, and related methodology. Analy Profiles Drug Subst Excip. 2002, 29, 1–5. [Google Scholar]
- Yamashita, M. Auranofin: Past to Present, and repurposing. Int. Immunopharmacol. 2021, 101, 108272. [Google Scholar] [CrossRef]
- Hoffman, D.W.; Wiebkin, P.; Rybak, L.P. Inhibition of glutathione-related enzymes and cytotoxicity of ethacrynic acid and cyclosporine. Biochem. Pharmacol. 1995, 49, 411–415. [Google Scholar] [CrossRef]
- Dalmizrak, O.; Teralı, K.; Asuquo, E.B.; Ogus, I.H.; Ozer, N. The relevance of glutathione reductase inhibition by fluoxetine to human health and disease: Insights derived from a combined kinetic and docking study. Protein J. 2019, 38, 515–524. [Google Scholar] [CrossRef]
- Martí-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sánchez, R.; Melo, F.; Šali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef]
- King, Z.A.; Lu, J.; Dräger, A.; Miller, P.; Federowicz, S.; Lerman, J.A.; Ebrahim, A.; Palsson, B.O.; Lewis, N.E. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016, 44, D515–D522. [Google Scholar] [CrossRef]
- Liang, X.-L.; Liang, Z.-M.; Wang, S.; Chen, X.-H.; Ruan, Y.; Zhang, Q.-Y.; Zhang, H.-Y. An analysis of the mechanism underlying photocatalytic disinfection based on integrated metabolic networks and transcriptional data. J. Environ. Sci. 2020, 92, 28–37. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Clark, M.; Cramer III, R.D.; Van Opdenbosch, N. Validation of the general purpose tripos 5.2 force field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Song, Y.-L.; Liu, S.-S.; Yang, J.; Xie, J.; Zhou, X.; Wu, Z.-B.; Liu, L.-W.; Wang, P.-Y.; Yang, S. Discovery of Epipodophyllotoxin-Derived B2 as Promising Xoo FtsZ Inhibitor for Controlling Bacterial Cell Division: Structure-Based Virtual Screening, Synthesis, and SAR Study. Int. J. Mol. Sci. 2022, 23, 9119. [Google Scholar] [CrossRef]
- Ya, Y. Laboratory Identification of Resistance to Pesticides and rpfC Gene Sequence Analysis of Xanthomonas oryzae pv. oryzae in japonica Rice from Yunnan Plateau. Chin. J. Rice Sci. 2014, 28, 665. [Google Scholar]
- Sirén, J.; Välimäki, N.; Mäkinen, V. HISAT2-fast and sensitive alignment against general human population. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 375–388. [Google Scholar]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Cao, M.-H.; Tang, B.-H.; Ruan, Y.; Liang, X.-L.; Chu, X.-Y.; Liang, Z.-M.; Zhang, Q.-Y.; Zhang, H.-Y. Development of specific and selective bactericide by introducing exogenous metabolite of pathogenic bacteria. Eur. J. Med. Chem. 2021, 225, 113808. [Google Scholar] [CrossRef]
- Aminov, R. Metabolomics in antimicrobial drug discovery. Expert Opin. Drug Discov. 2022, 17, 1047–1059. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, J.; Li, J. Genome-scale metabolic modeling in antimicrobial pharmacology. Eng. Microbiol. 2022, 2, 100021. [Google Scholar] [CrossRef]

| Model | Residues in Most Favoured Regions | Residues in Additional Allowed Regions | Residues in Generously Allowed Regions | Residues in Disallowed Regions |
|---|---|---|---|---|
| Xoo-GSR (378) | 348 (92.1%) | 24 (6.3%) | 6 (1.6%) | 0 (0.0%) |
| Molecular ID | Docking Score |
|---|---|
| GSR-DB12411 | −12.3 |
| FAD | −12.2 |
| GSR-DB15039 | −11.8 |
| GSR-DB04888 | −11.7 |
| GSR-DB11852 | −11.7 |
| GSR-DB12886 | −11.7 |
| Molecular | CAS | Molecular Weight | Inhibition Rate (6 h) | Inhibition Rate (12 h) |
|---|---|---|---|---|
| DB12411 | 1037624-75-1 | 506.64 | 97.08% | 99.23% |
| DB15039 | 1642303-38-5 | 605.56 | 45.20% | 27.33% |
| DB11852 | 1000787-75-6 | 517.4 | 29.51% | 13.74% |
| DMSO | 9.72% | 6.39% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.-L.; Liang, X.-L.; Ge, Z.-Y.; Zhang, Z.; Ruan, Y.; Tang, H.; Zhang, Q.-Y. Metabolic Flux Analysis of Xanthomonas oryzae Treated with Bismerthiazol Revealed Glutathione Oxidoreductase in Glutathione Metabolism Serves as an Effective Target. Int. J. Mol. Sci. 2024, 25, 12236. https://doi.org/10.3390/ijms252212236
Yu H-L, Liang X-L, Ge Z-Y, Zhang Z, Ruan Y, Tang H, Zhang Q-Y. Metabolic Flux Analysis of Xanthomonas oryzae Treated with Bismerthiazol Revealed Glutathione Oxidoreductase in Glutathione Metabolism Serves as an Effective Target. International Journal of Molecular Sciences. 2024; 25(22):12236. https://doi.org/10.3390/ijms252212236
Chicago/Turabian StyleYu, Hai-Long, Xiao-Long Liang, Zhen-Yang Ge, Zhi Zhang, Yao Ruan, Hao Tang, and Qing-Ye Zhang. 2024. "Metabolic Flux Analysis of Xanthomonas oryzae Treated with Bismerthiazol Revealed Glutathione Oxidoreductase in Glutathione Metabolism Serves as an Effective Target" International Journal of Molecular Sciences 25, no. 22: 12236. https://doi.org/10.3390/ijms252212236
APA StyleYu, H.-L., Liang, X.-L., Ge, Z.-Y., Zhang, Z., Ruan, Y., Tang, H., & Zhang, Q.-Y. (2024). Metabolic Flux Analysis of Xanthomonas oryzae Treated with Bismerthiazol Revealed Glutathione Oxidoreductase in Glutathione Metabolism Serves as an Effective Target. International Journal of Molecular Sciences, 25(22), 12236. https://doi.org/10.3390/ijms252212236





