The Expression of HPV-16 E5 Oncoprotein Impacts the Transcript Profiles of FGFR2 and EMT-Related Genes in Preneoplastic Anal Epithelium Lesions
Abstract
1. Introduction
2. Results
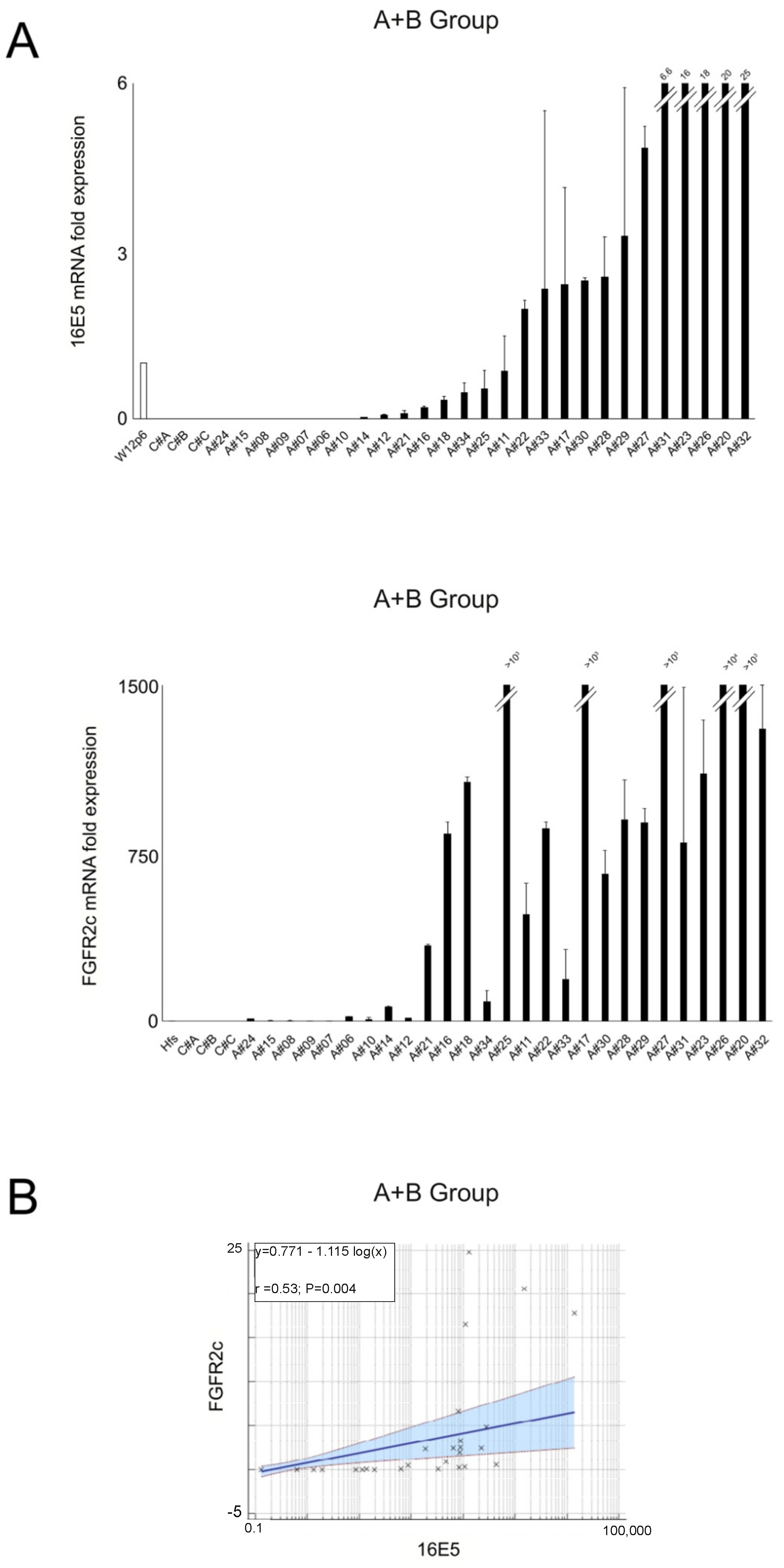
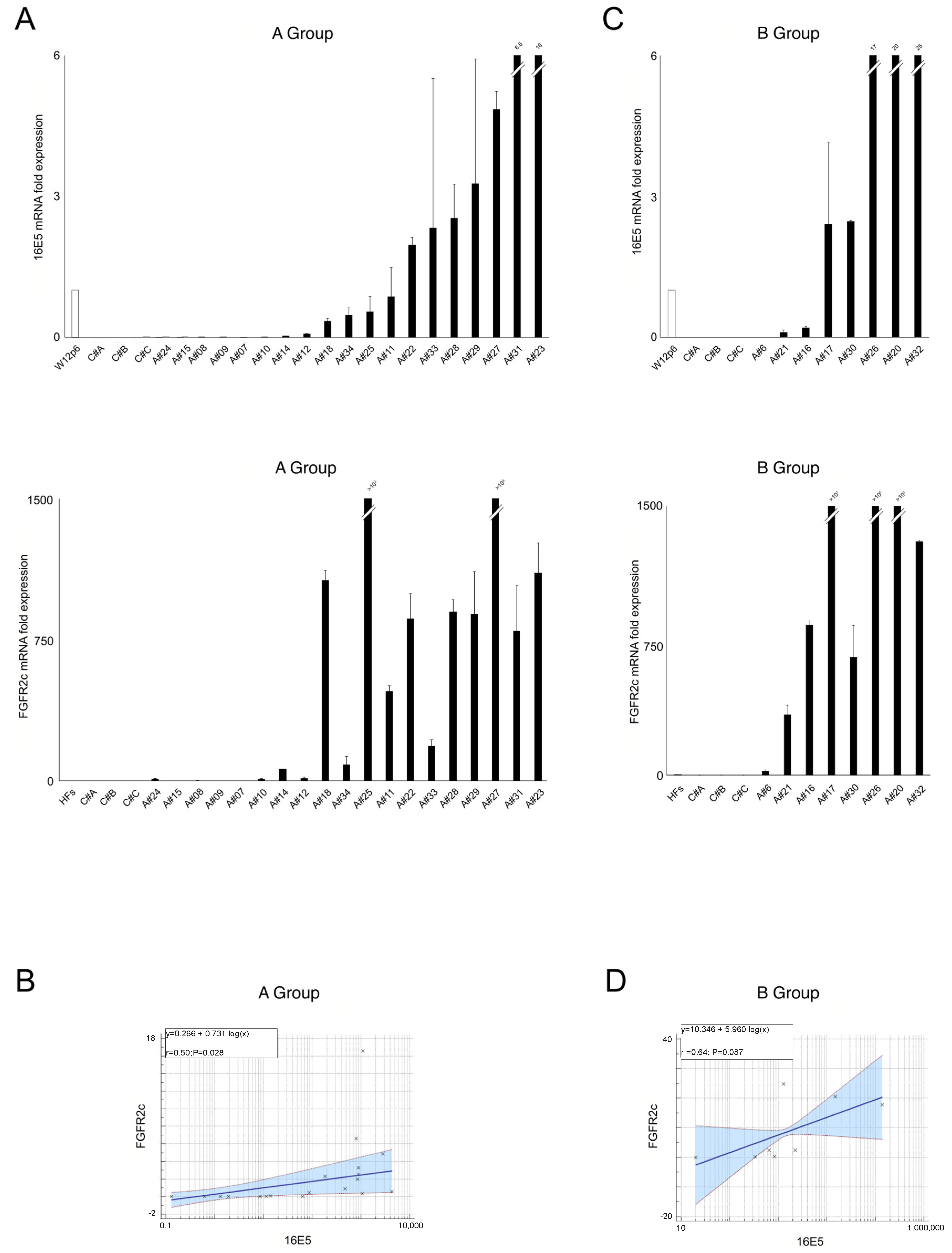
2.1. 16E5 and FGFR2c Expression in Anal Intraepithelial Lesions
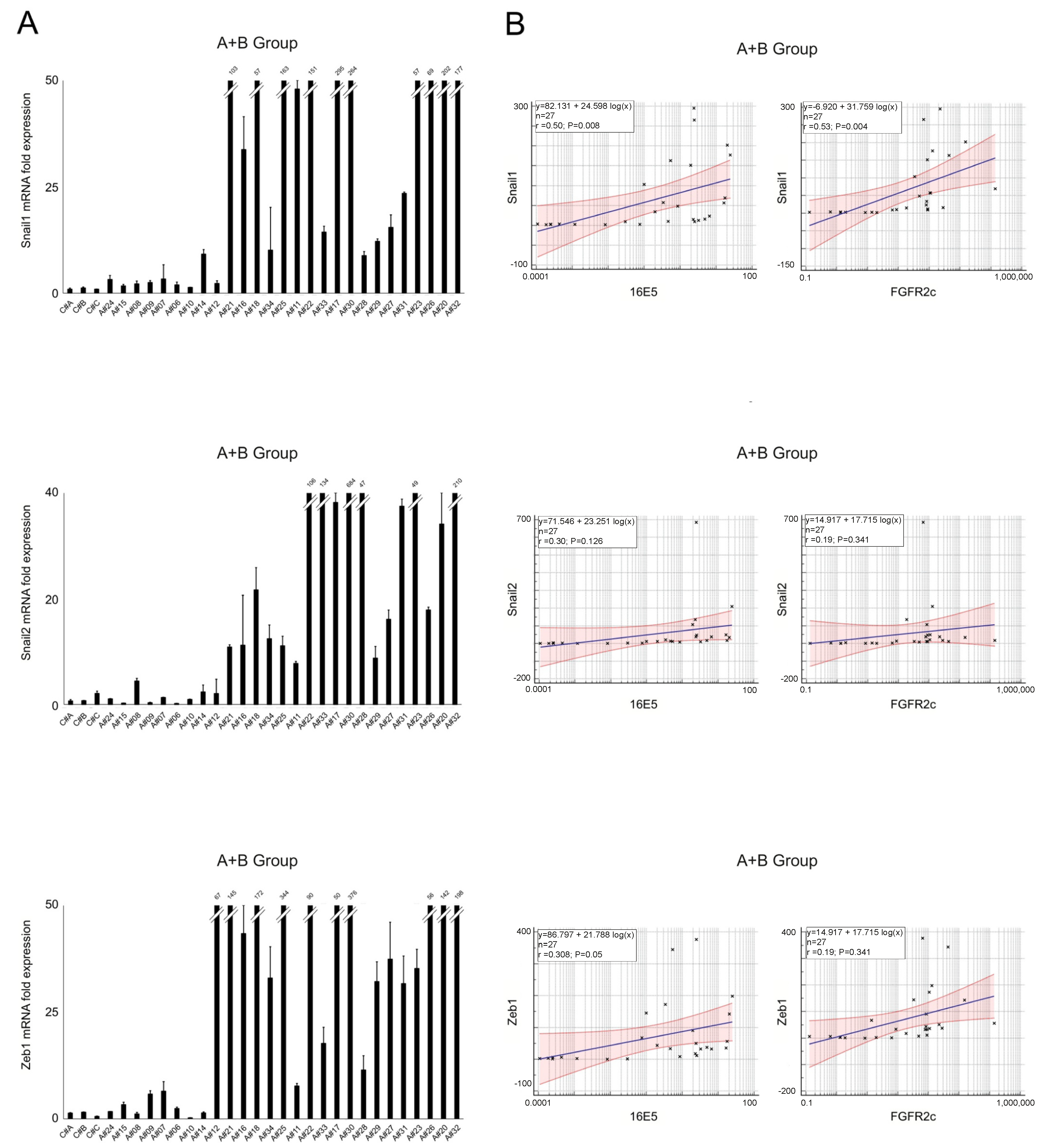
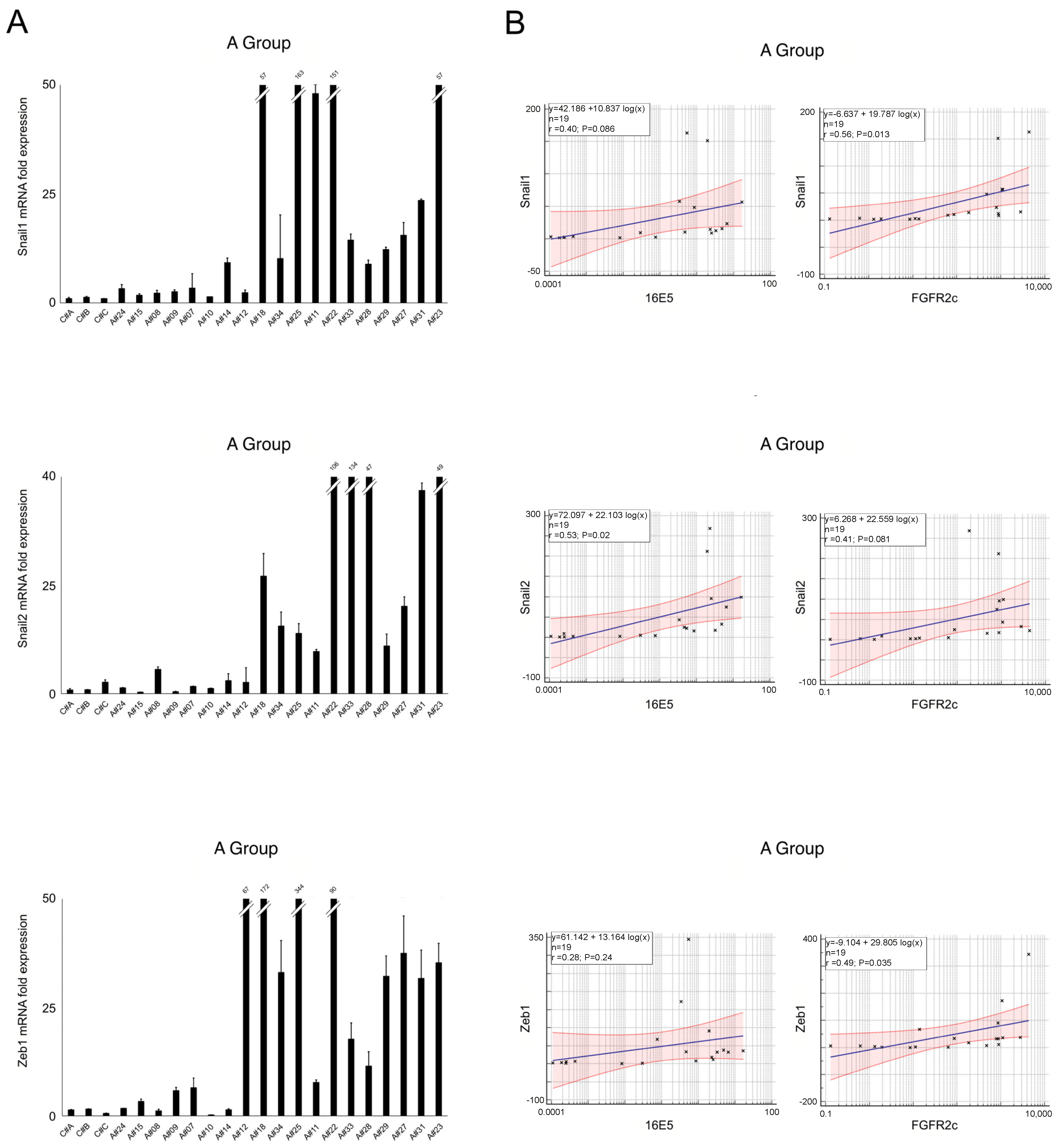
2.2. EMT-Related Transcription Factors and Their Association with 16E5 and FGFR2c Expression
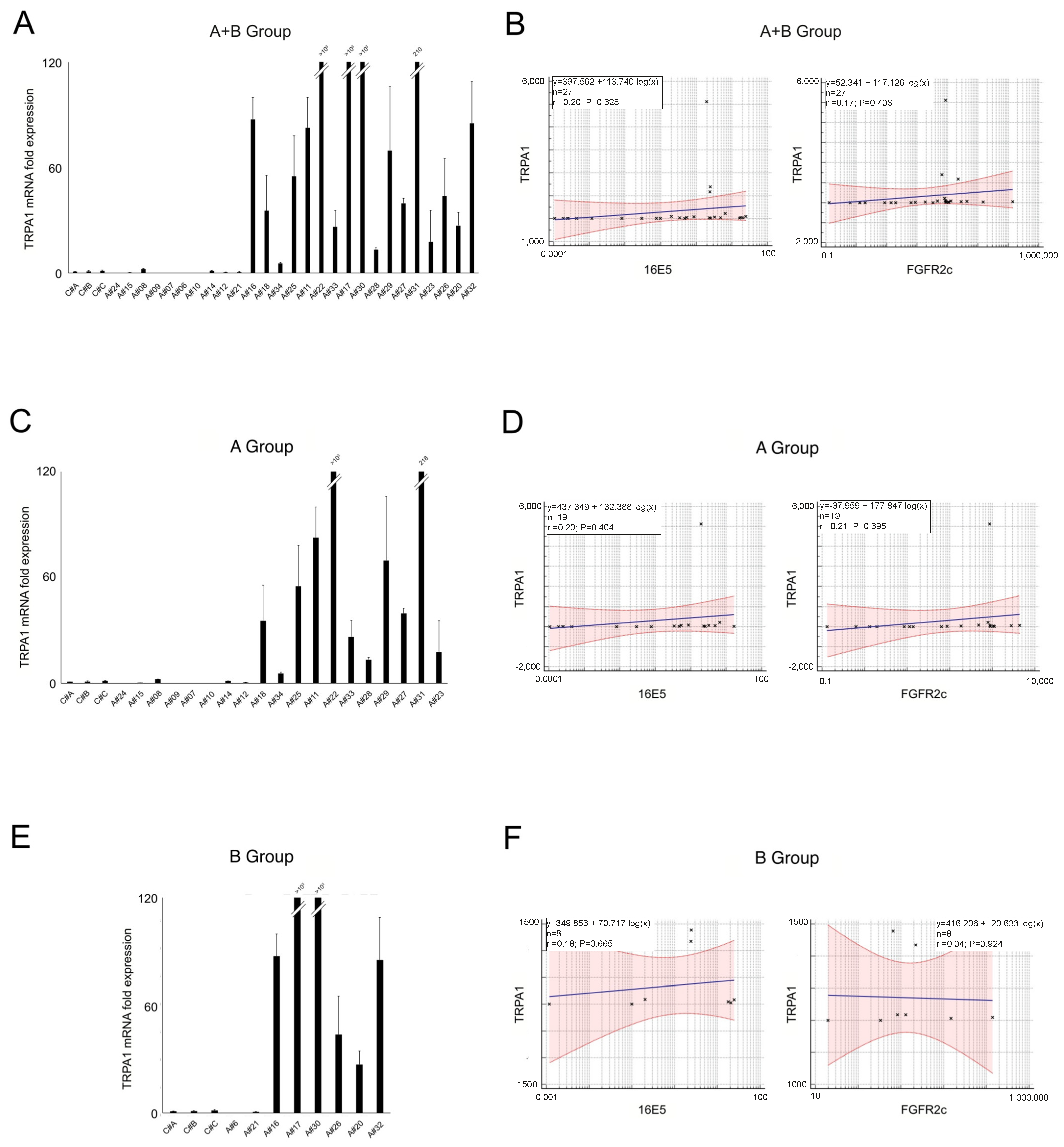
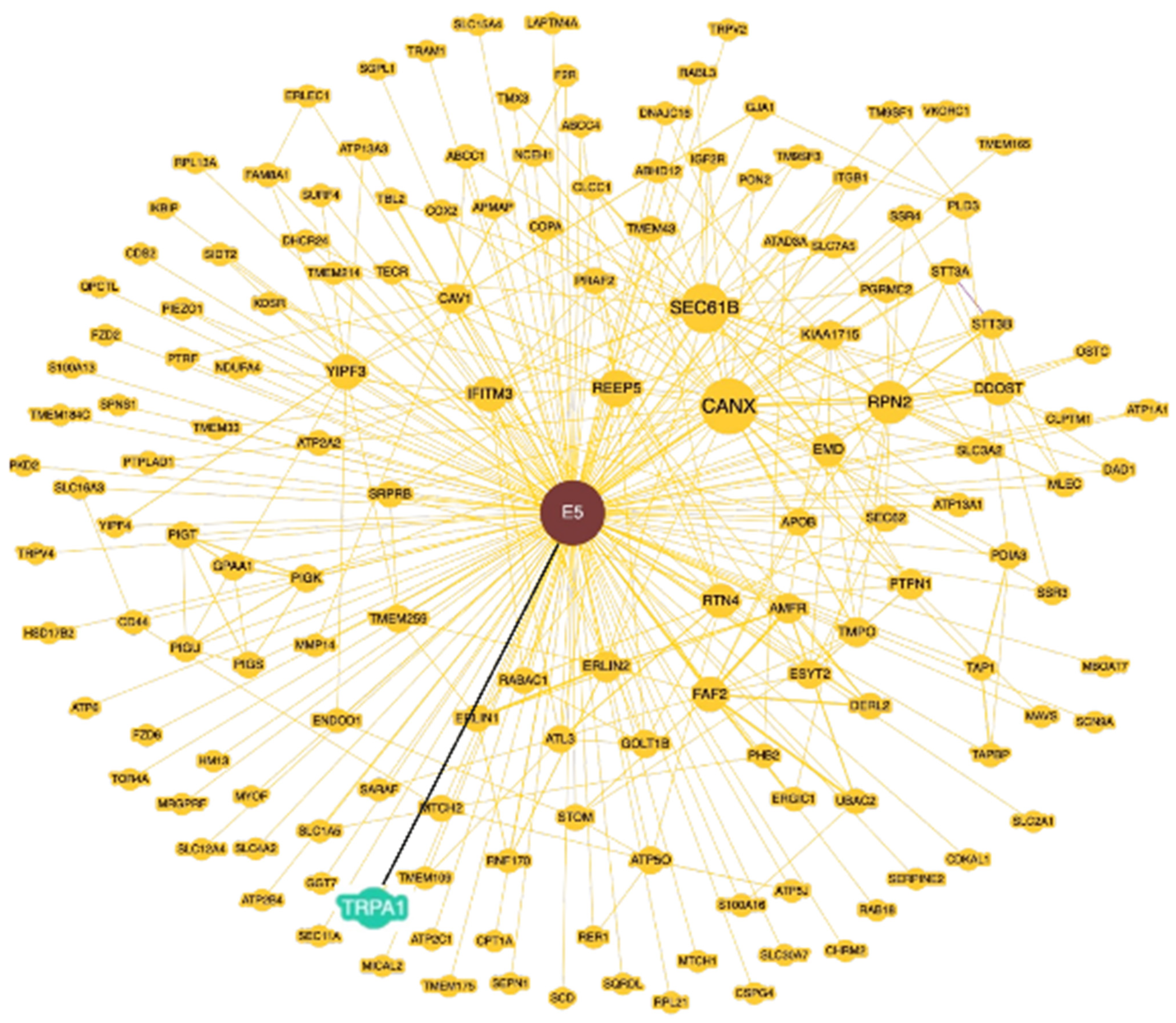
2.3. TRPA1 Expression and Its Relationship with 16E5 and FGFR2c in Anal Lesions
3. Discussion
4. Materials and Methods
4.1. Etichs Statement
4.2. Cytological Samples
4.3. Primers
4.4. RNA Extraction and cDNA Synthesis
4.5. PCR Amplification and Real-Time Quantitation
4.6. Statistical Analysis
4.7. Bioinformatical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Xu, C. Human Papillomavirus-Related Cancers. Adv. Exp. Med. Biol. 2017, 1018, 23–34. [Google Scholar] [PubMed]
- Mukherjee, A.G.; Ramesh Wanjari, U.; Valsala Gopalakrishnan, A.; Jayaraj, R.; Katturajan, R.; Kannampuzha, S.; Murali, R.; Namachivayam, A.; Prince, S.E.; Vellingiriet, B.; et al. HPV-associated cancers: Insights into the mechanistic scenario and latest updates. Med. Oncol. 2023, 40, 212. [Google Scholar] [CrossRef]
- Brant, A.C.; Menezes, A.N.; Felix, S.P.; De Almeida, L.M.; Sammeth, M.; Moreira, M.A. Characterization of HPV integration, viral gene expression and E6E7 alternative transcripts by RNA-Seq: A descriptive study in invasive cervical cancer. Genomics 2019, 111, 1853–1861. [Google Scholar] [CrossRef]
- Matsukura, T.; Koi, S.; Sugase, M. Both episomal and integrated forms of human papillomavirus type 16 are involved in invasive cervical cancers. Virology 1989, 172, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Cullen, A.P.; Reid, R.; Campion, M.; Lörincz, A.T. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J. Virol. 1991, 65, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Hwang, E.S.; Park, S.N.; Ahn, H.K.; Um, S.J.; Kim, C.J.; Kim, S.J.; Namkoong, S.E. Physical status and expression of HPV genes in cervical cancers. Gynecol. Oncol. 1997, 65, 121–129. [Google Scholar] [CrossRef]
- Cavuslu, S.; Starkey, W.G.; Kell, B.; Best, J.M.; Cason, J. Detection of human papillomavirus type 16 in microtitre plate based immuno-enzymatic assays: Use to determine E5 gene expression in cervical carcinomas. Clin. Diagn. Virol. 1996, 5, 215–218. [Google Scholar] [CrossRef]
- Chang, J.L.; Tsao, Y.P.; Liu, D.W.; Huang, S.J.; Lee, W.H.; Chen, S.L. The expression of HPV-16 E5 protein in squamous neoplastic changes in the uterine cervix. J. Biomed. Sci. 2001, 8, 206–213. [Google Scholar] [CrossRef]
- Um, S.H.; Mundi, N.; Yoo, J.; Palma, D.A.; Fung, K.; MacNeil, D.; Wehrli, B.; Mymryk, J.S.; Barret, J.W.; Nichols, A.C. Variable expression of the forgotten oncogene E5 in HPV-positive oropharyngeal cancer. J. Clin. Virol. 2014, 61, 94–100. [Google Scholar] [CrossRef]
- French, D.; Belleudi, F.; Mauro, M.V.; Mazzetta, F.; Raffa, S.; Fabiano, V.; Frega, A.; Torrisi, M.R. Expression of HPV16 E5 down-modulates the TGFbeta signaling pathway. Mol. Cancer 2013, 12, 38. [Google Scholar] [CrossRef]
- Purpura, V.; Belleudi, F.; Caputo, S.; Torrisi, M.R. HPV16 E5 and KGFR/FGFR2b interplay in differentiating epithelial cells. Oncotarget 2013, 4, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, D.; Belleudi, F.; Magenta, A.; Torrisi, M.R. HPV16 E5 expression induces switching from FGFR2b to FGFR2c and epithelial-mesenchymal transition. Int. J. Cancer 2015, 137, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, D.; Rosato, B.; Nanni, M.; Magenta, A.; Belleudi, F.; Torrisi, M.R. Expression of the FGFR2 mesenchymal splicing variant in epithelial cells drives epithelial-mesenchymal transition. Oncotarget 2016, 7, 5440–5460. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, D.; French, D.; Raffa, S.; Guttieri, L.; Torrisi, M.R.; Belleudi, F. Expression of the E5 oncoprotein of HPV16 impacts on the molecular profiles of EMT-related and differentiation genes in ectocervical low-grade lesions. Int. J. Mol. Sci. 2021, 22, 6534. [Google Scholar] [CrossRef]
- Hoots, B.E.; Palefsky, J.M.; Pimenta, J.M.; Smith, J.S. Human papillomavirus type distribution in anal cancer and anal intraepithelial lesions. Int. J. Cancer 2009, 124, 2375–2383. [Google Scholar] [CrossRef]
- Upadhyay, L.; Hartzell, M.; Parikh, A.R.; Strickland, M.R.; Klempner, S.; Malla, M. Recent Advances in the Management of Anal Cancer. Healthcare 2023, 11, 3010. [Google Scholar] [CrossRef] [PubMed]
- Elnaggar, J.H.; Huynh, V.O.; Lin, D.; Hillman, R.T.; Abana, C.O.; El Alam, M.B.; Tomasic, K.C.; Karpinets, T.V.; Kouzy, R.; Phan, J.L.; et al. HPV-related anal cancer is associated with changes in the anorectal microbiome during cancer development. Front. Immunol. 2023, 14, 1051431. [Google Scholar] [CrossRef]
- Steinau, M.; Unger, E.R.; Hernandez, B.Y.; Goodman, M.T.; Copeland, G.; Hopenhayn, C.; Cozen, W.; Saber, M.S.; Huang, Y.; Peters, E.; et al. Human papillomavirus prevalence in invasive anal cancers in the United States before vaccine introduction. J. Low. Genit. Tract Dis. 2013, 17, 397–403. [Google Scholar] [CrossRef]
- Péré, H.; Vernet, R.; Pernot, S.; Pavie, J.; Robillard, N.; Puech, J.; Lameiras, S.; Lucas, M.L.; Nicolas, A.; Badoual, C.; et al. Episomal HPV16 responsible for aggressive and deadly metastatic anal squamous cell carcinoma evidenced in peripheral blood. Sci. Rep. 2021, 11, 4633. [Google Scholar] [CrossRef]
- Wechsler, E.I.; Tugizov, S.; Herrera, R.; Da Costa, M.; Palefsky, J.M. E5 can be expressed in anal cancer and leads to epidermal growth factor receptor-induced invasion in a human papillomavirus 16-transformed anal epithelial cell line. J. Gen. Virol. 2018, 99, 631–644. [Google Scholar] [CrossRef]
- Stanley, M.A.; Browne, H.M.; Appleby, M.; Minson, A.C. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int. J. Cancer 1989, 43, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Mancini, V.; Raffa, S.; Fiorio Pla, A.; French, D.; Torrisi, M.R.; Ranieri, D.; Belleudi, F. TRPA1 Contributes to FGFR2c Signaling and to Its Oncogenic Outcomes in Pancreatic Ductal Adenocarcinoma-Derived Cell Lines. Cancers 2024, 16, 609. [Google Scholar] [CrossRef]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34 (Suppl. S1), D535–D539. [Google Scholar] [CrossRef]
- Rozenblatt-Rosen, O.; Deo, R.C.; Padi, M.; Adelmant, G.; Calderwood, M.A.; Rolland, T.; Grace, M.; Dricot, A.; Askenazi, M.; Tavares, M.; et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 2012, 487, 491–495. [Google Scholar] [CrossRef]
- De Vuyst, H.; Clifford, G.M.; Nascimento, M.C.; Madeleine, M.M.; Franceschi, S. Prevalence and Type Distribution of Human Papillomavirus in Carcinoma and Intraepithelial Neoplasia of the Vulva, Vagina and Anus: A Meta-Analysis. Int. J. Cancer 2009, 124, 1626–1636. [Google Scholar] [CrossRef]
- Parwaiz, I.; MacCabe, T.A.; Thomas, M.G.; Messenger, D.E. A Systematic Review and Meta-Analysis of Prognostic Biomarkers in Anal Squamous Cell Carcinoma Treated with Primary Chemoradiotherapy. Clin. Oncol. 2019, 31, e1–e13. [Google Scholar] [CrossRef] [PubMed]
- Urbute, A.; Rasmussen, C.L.; Belmonte, F.; Obermueller, T.; Prigge, E.S.; Arbyn, M.; Verdoodt, F.; Kjaer, S.K. Prognostic Significance of HPV DNA and p16INK4a in Anal Cancer: A Systematic Review and Meta-Analysis. Cancer Epidemiol. Biomark. Prev. 2020, 29, 703–710. [Google Scholar] [CrossRef]
- Ren, S.; Gaykalova, D.A.; Guo, T.; Favorov, A.V.; Fertig, E.J.; Tamayo, P.; Callejas-Valera, J.L.; Allevato, M.; Gilardi, M.; Santos, J.; et al. HPV E2, E4, E5 drive alternative carcinogenic pathways in HPV positive cancers. Oncogene 2020, 39, 6327–6339. [Google Scholar] [CrossRef]
- Donà, M.G.; Benevolo, M.; Latini, A.; Rollo, F.; Colafigli, M.; Frasca, M.; Zaccarelli, M.; Giglio, A.; Moretto, D.; Pescarmona, E.; et al. Anal cytological lesions and HPV infection in individuals at increased risk for anal cancer. Cancer Cytopathol. 2018, 126, 461–470. [Google Scholar] [CrossRef]
- Darragh, T.M.; Donà, M.G.; Benevolo, M.; Latini, A.; Rollo, F.; Colafigli, M.; Frasca, M.; Zaccarelli, M.; Giglio, A.; Moretto, D.; et al. Anal-rectal cytology. In The Bethesda System for Reporting Cervical Cytology: Definitions, Criteria, and Explanatory Notes, 2nd ed.; Solomon, D., Nayar, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 169–175. [Google Scholar]
- Darragh, T.M.; Palefsky, J.M. Anal cytology. In The Bethesda System for Reporting Cervical Cytology: Definitions, Criteria, and Explanatory Notes, 3rd ed.; Nayar, R., Wilbur, D.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 263–285. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raffa, S.; Mancini, V.; French, D.; Rollo, F.; Benevolo, M.; Giuliani, E.; Donà, M.G.; Ranieri, D.; Belleudi, F. The Expression of HPV-16 E5 Oncoprotein Impacts the Transcript Profiles of FGFR2 and EMT-Related Genes in Preneoplastic Anal Epithelium Lesions. Int. J. Mol. Sci. 2024, 25, 12085. https://doi.org/10.3390/ijms252212085
Raffa S, Mancini V, French D, Rollo F, Benevolo M, Giuliani E, Donà MG, Ranieri D, Belleudi F. The Expression of HPV-16 E5 Oncoprotein Impacts the Transcript Profiles of FGFR2 and EMT-Related Genes in Preneoplastic Anal Epithelium Lesions. International Journal of Molecular Sciences. 2024; 25(22):12085. https://doi.org/10.3390/ijms252212085
Chicago/Turabian StyleRaffa, Salvatore, Vanessa Mancini, Deborah French, Francesca Rollo, Maria Benevolo, Eugenia Giuliani, Maria Gabriella Donà, Danilo Ranieri, and Francesca Belleudi. 2024. "The Expression of HPV-16 E5 Oncoprotein Impacts the Transcript Profiles of FGFR2 and EMT-Related Genes in Preneoplastic Anal Epithelium Lesions" International Journal of Molecular Sciences 25, no. 22: 12085. https://doi.org/10.3390/ijms252212085
APA StyleRaffa, S., Mancini, V., French, D., Rollo, F., Benevolo, M., Giuliani, E., Donà, M. G., Ranieri, D., & Belleudi, F. (2024). The Expression of HPV-16 E5 Oncoprotein Impacts the Transcript Profiles of FGFR2 and EMT-Related Genes in Preneoplastic Anal Epithelium Lesions. International Journal of Molecular Sciences, 25(22), 12085. https://doi.org/10.3390/ijms252212085






