Sensitive Detection of Gynecological Cancer Recurrence Using Circulating Tumor DNA and Digital PCR: A Comparative Study with Serum Biochemical Markers
Abstract
1. Introduction
2. Results
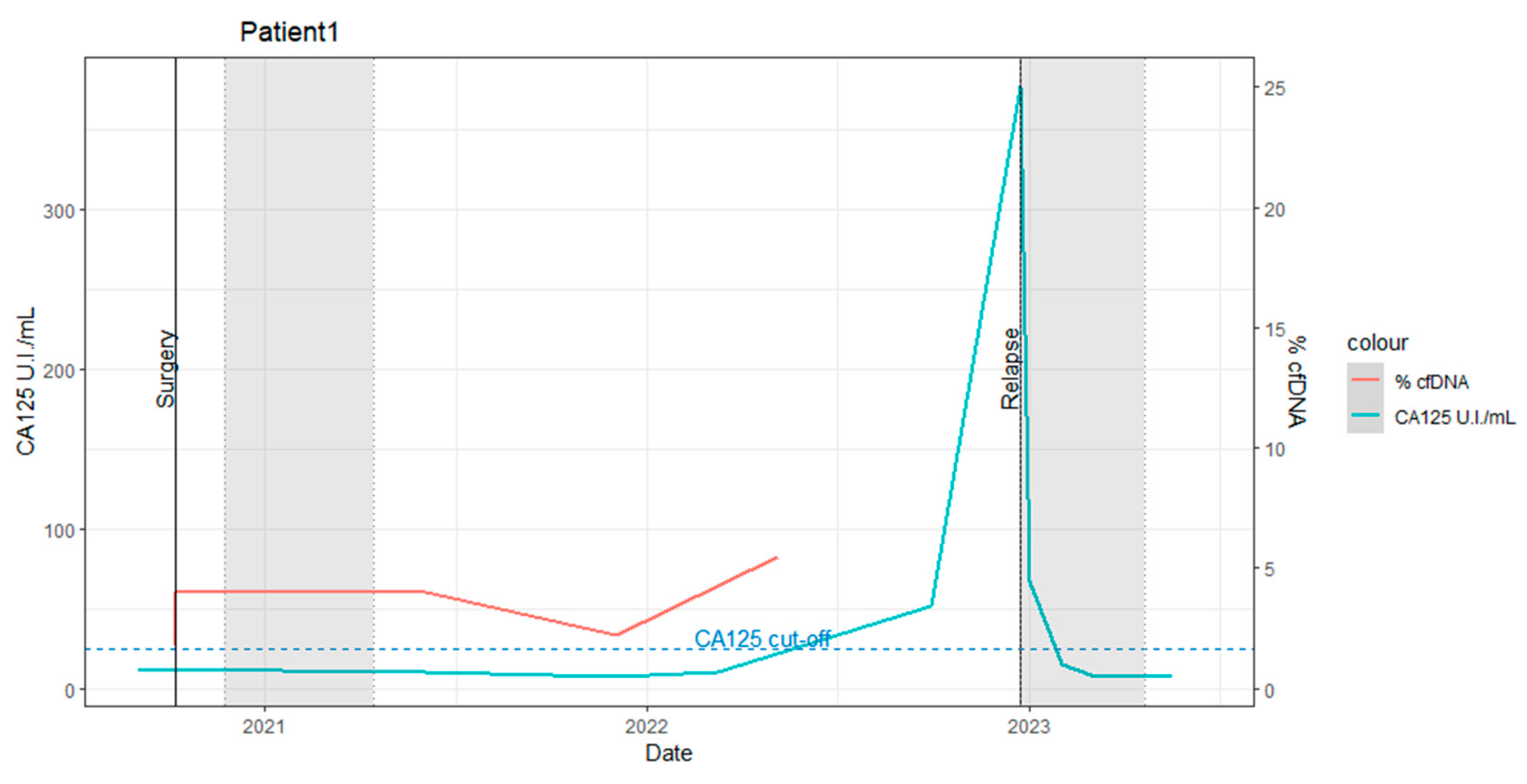
2.1. Patient 1
- Pre-surgery: The baseline levels of biochemical markers were within normal ranges. The percentage of ctDNA carrying the TP53 SNV in the plasma sample was 1.8%.
- Post-surgery: The ctDNA percentage slightly increased to 4%, suggesting incomplete tumoral removal or metastasis. This underscores the utility of dPCR in estimating the efficacy of surgical interventions. The patient subsequently underwent 5 months of adjuvant chemo-radiotherapy (the first gray zone in Figure 1).
- Two months post-adjuvant therapy: The ctDNA percentage remained stable at 4%.
- Eleven months post-adjuvant therapy: The ctDNA percentage increased to 5.5% at 11 months (as indicated in Figure 1, (ctDNA%: red line)).
- Eighteen months post-adjuvant therapy: A slight elevation in CA125 (51.49 UI/mL) was observed without clinical evidence of disease progression.
- Nineteen months post-adjuvant therapy: Clinical recurrence was detected via a positron emission tomography (PET)-computed tomography (CT) scan.
2.2. Patient 2
- Pre-staging laparoscopy (S-LPS): The patient presented with elevated CA125 levels (120 UI/mL).
- Post-chemotherapy: Minimal residual disease was detected in plasma (2.7% ctDNA), with a concomitant decrease in CA125 levels. Laparotomic interval debulking surgery (IDS), performed in April 2020, revealed a 5 mm macroscopic residual lesion, consistent with the ctDNA finding.
- Post-IDS: Despite normalized CA125 levels, the persistence and increase in the PIK3CA mutation (2.7% to 5.6% ctDNA) suggested a potentially incomplete surgical resection. The patient received three additional cycles of adjuvant chemotherapy.
- Eight months post-chemotherapy and IDS: The ctDNA percentage increased to 13%, indicating potential chemotherapy resistance. Thus, in this case, a change in medication is advisable for the subsequent cycle.
- Ten months post-chemotherapy and IDS: Recurrence was detected via a contrast-enhanced chest-abdomen CT scan.
- Thirteen months post-chemotherapy and IDS: The ctDNA percentage further increased to ~45%.
- Eleven months post-recurrence detection: CA125 levels began to rise.
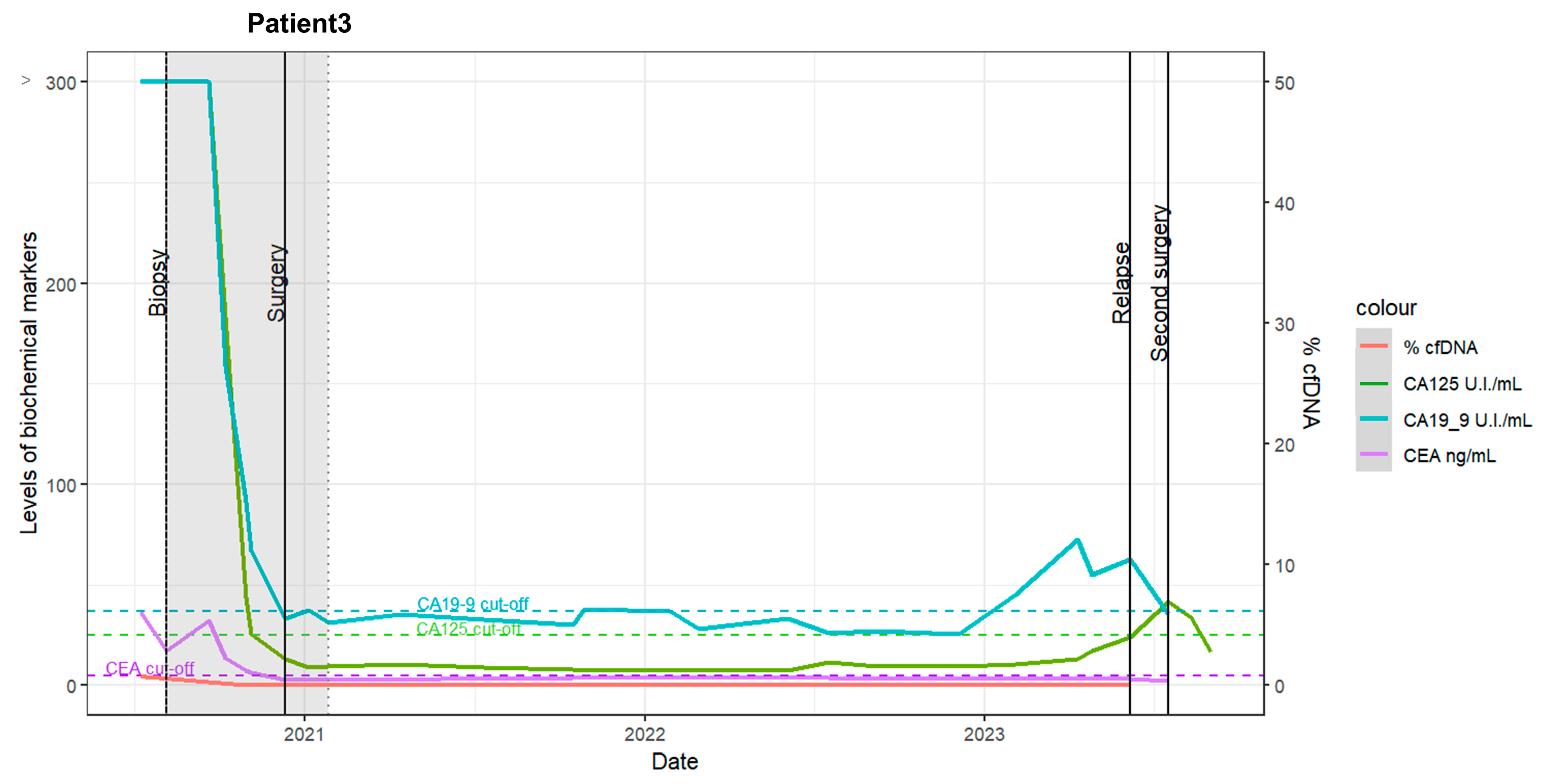
2.3. Patient 3
- Pre- and post-staging laparoscopy (S-LPS): The patient initially underwent S-LPS, followed by four cycles of neoadjuvant chemotherapy. Elevated cancer antigen 125 (CA125) and carbohydrate antigen 19-9 (CA19-9) marker levels were observed pre-chemotherapy, with minimal ctDNA detected for both targets (<1%) before and after S-LPS (Figure 3, red line).
- Post-chemotherapy: A marked decrease in biochemical marker levels was observed following chemotherapy.
- Laparotomic interval debulking surgery (IDS) and adjuvant chemotherapy: The patient underwent laparotomic IDS and four additional chemotherapy cycles.
- Recurrence: Tumor recurrence was detected via a positron emission tomography-computed tomography (PET-CT) scan 27 months after the completion of chemotherapy.
3. Discussion
4. Materials and Methods
4.1. Patient Enrolment
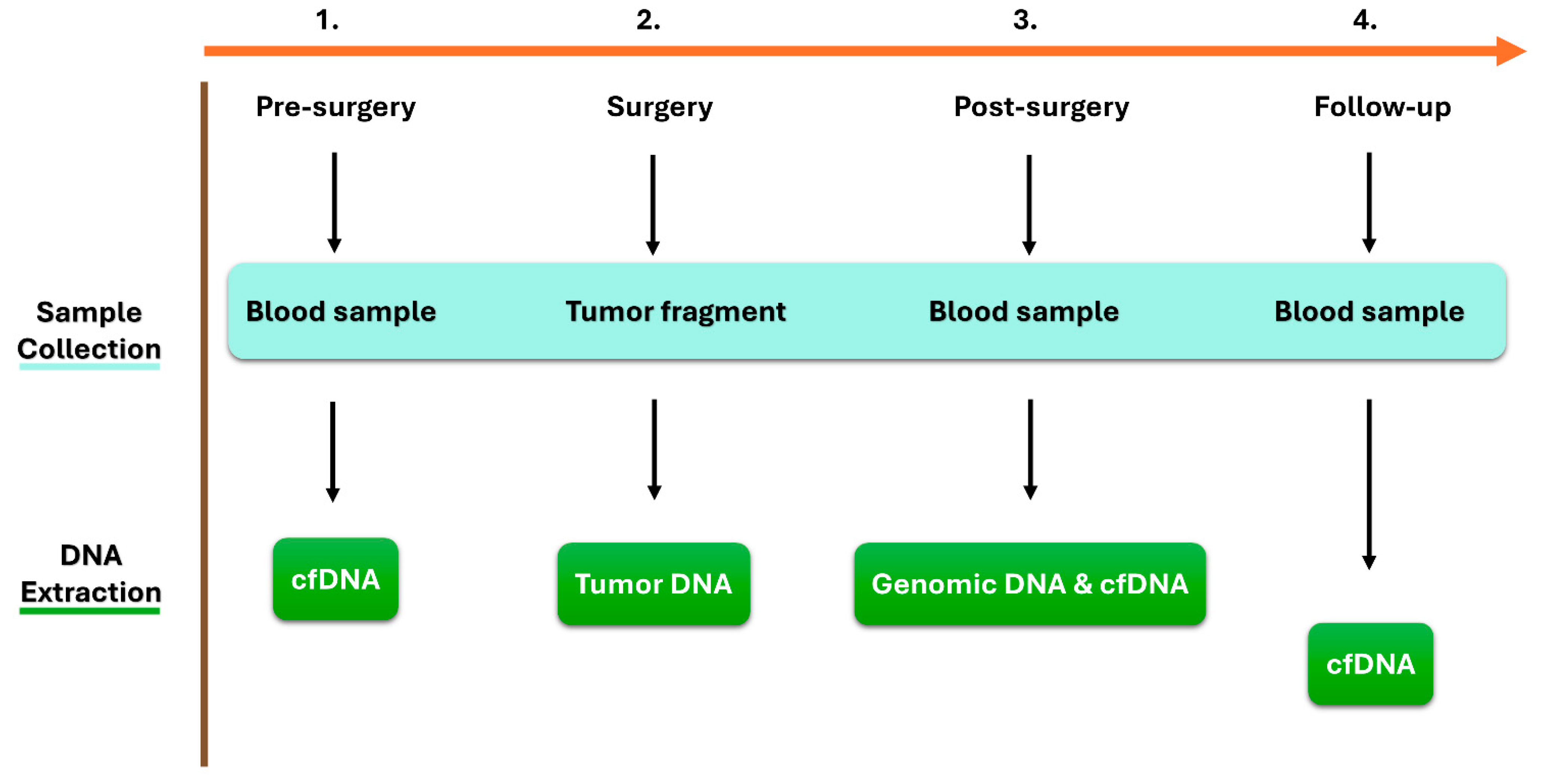
4.2. Sample Collection and DNA Extraction
- Whole blood samples pre-surgery, forty-eight hours post-surgery, and during the follow-up period every three months were collected using QIAGEN PAXgene® Blood ccfDNA Tubes (catalog number: 768165, Becton, NJ, USA). The blood was centrifuged at 1900× g 4 °C for 10 min, and the supernatant plasma was transferred to separate Eppendorf Tubes. The plasma was further centrifuged at 1900× g 4 °C for 10 min to remove the remaining leukocytes. The obtained plasma samples (5 mL) were frozen and stored at −80 °C until the purification of circulating cell-free DNA (ccfDNA). The ccfDNA was extracted from 5 mL of plasma using the QIAGEN QIAamp® DSP Circulating NA Kit (catalog number: 61504, Hilden, Germany), according to the manufacturer’s instructions.
- Genomic DNA (gDNA) was extracted from an aliquot of blood collected post-surgery in an EDTA tube. gDNA was extracted using the QIAGEN QIAamp® DNA Blood Mini Kit (catalog number: 51104, Hilden, Germany), according to the manufacturer’s instructions, and used as healthy DNA.
- Tumor biopsies were collected at the time of initial surgery. Surgically removed cancer tissues were macroscopically selected based on the corresponding hematoxylin-eosin staining. The obtained tumor biopsies were stored at −80 °C until the extraction of tumor DNA. Tumor DNA was then extracted using the salting-out extraction procedure described by Miller et al. [32].
4.3. Whole-Exome Sequencing and Bioinformatic Analysis
4.4. Validation of the Data by Sanger Sequencing
4.5. Digital PCR
of positive partitions from both signals) × 100.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Khanlarkhani, N.; Azizi, E.; Amidi, F.; Khodarahmian, M.; Salehi, E.; Pazhohan, A.; Farhood, B.; Mortezae, K.; Goradel, N.H.; Nashtaei, M.S. Metabolic risk factors of ovarian cancer: A review. JBRA Assist. Reprod. 2022, 26, 335–347. [Google Scholar] [CrossRef] [PubMed]
- González-Martín, A.; Harter, P.; Leary, A.; Lorusso, D.; Miller, R.; Pothuri, B.; Ray-Coquard, I.; Tan, D.; Bellet, E.; Oaknin, A.; et al. Newly diagnosed and relapsed epithelial ovarian cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 833–848. [Google Scholar] [CrossRef]
- Vistad, I.; Bjørge, L.; Solheim, O.; Fiane, B.; Sachse, K.; Tjugum, J.; Skrøppa, S.; Bentzen, A.G.; Stokstad, T.; Iversen, G.A.; et al. A national, prospective observational study of first recurrence after primary treatment for gynecological cancer in Norway. Acta Obstet. et Gynecol. Scand. 2017, 96, 1162–1169. [Google Scholar] [CrossRef]
- Duffy, M.J. Clinical uses of tumor markers: A critical review. Crit. Rev. Clin. Lab. Sci. 2001, 38, 225–262. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J. Use of biomarkers in screening for cancer. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2015; p. 867. [Google Scholar] [CrossRef]
- De Rubis, G.; Krishnan, S.R.; Bebawy, M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef]
- Bratman, S.V.; Yang, S.Y.C.; Iafolla, M.A.J.; Liu, Z.; Hansen, A.R.; Bedard, P.L.; Lheureux, S.; Spreafico, A.; Razak, A.A.; Shchegrova, S.; et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat. Cancer 2020, 1, 873–881. [Google Scholar] [CrossRef]
- Heo, J.; Kim, Y.-N.; Shin, S.; Lee, K.; Lee, J.-H.; Lee, Y.J.; Choi, Z.; Park, J.; Min, S.; Kim, S.W.; et al. Serial Circulating Tumor DNA Analysis with a Tumor-Naïve Next-Generation Sequencing Panel Detects Minimal Residual Disease and Predicts Outcome in Ovarian Cancer. Cancer Res. 2024, 84, 468–478. [Google Scholar] [CrossRef]
- Hou, J.Y.; Chapman, J.S.; Kalashnikova, E.; Pierson, W.; Smith-McCune, K.; Pineda, G.; Vattakalam, R.M.; Ross, A.; Mills, M.; Suarez, C.J.; et al. Circulating tumor DNA monitoring for early recurrence detection in epithelial ovarian cancer. Gynecol. Oncol. 2022, 167, 334–341. [Google Scholar] [CrossRef]
- Lu, Y.; Li, L. The Prognostic Value of Circulating Tumor DNA in Ovarian Cancer: A Meta-Analysis. Technol. Cancer Res. Treat. 2021, 20, 15330338211043784. [Google Scholar] [CrossRef]
- Taliento, C.; Morciano, G.; Nero, C.; Froyman, W.; Vizzielli, G.; Pavone, M.; Salvioli, S.; Tormen, M.; Fiorica, F.; Scutiero, G.; et al. Circulating tumor DNA as a biomarker for predicting progression-free survival and overall survival in patients with epithelial ovarian cancer: A systematic review and meta-analysis. Int. J. Gynecol. Cancer 2024, 34, 906–918. [Google Scholar] [CrossRef] [PubMed]
- Calapre, L.; Giardina, T.; Beasley, A.B.; Reid, A.L.; Stewart, C.; Amanuel, B.; Meniawy, T.M.; Gray, E.S. Identification of TP53 mutations in circulating tumour DNA in high grade serous ovarian carcinoma using next generation sequencing technologies. Sci. Rep. 2023, 13, 278. [Google Scholar] [CrossRef]
- Yousefi, M.; Rajaie, S.; Keyvani, V.; Bolandi, S.; Hasanzadeh, M.; Pasdar, A. Clinical significance of circulating tumor cell related markers in patients with epithelial ovarian cancer before and after adjuvant chemotherapy. Sci. Rep. 2021, 11, 10524. [Google Scholar] [CrossRef]
- Shen, Y.; Shi, R.; Zhao, R.; Wang, H. Clinical application of liquid biopsy in endometrial carcinoma. Med. Oncol. 2023, 40, 92. [Google Scholar] [CrossRef]
- Recio, F.; Scalise, C.B.; Loar, P.; Lumish, M.; Berman, T.; Peddada, A.; Kalashnikova, E.; Rivero-Hinojosa, S.; Beisch, T.; Nicosia, B.; et al. Post-surgical ctDNA-based molecular residual disease detection in patients with stage I uterine malignancies. Gynecol. Oncol. 2024, 182, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.; Camacho-Vanegas, O.; Anand, S.; Sebra, R.; Camacho, S.C.; Garnar-Wortzel, L.; Nair, N.; Moshier, E.; Wooten, M.; Uzilov, A.; et al. Personalized Circulating Tumor DNA Biomarkers Dynamically Predict Treatment Response and Survival In Gynecologic Cancers. PLoS ONE 2015, 10, e0145754. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Mughal, T.I.; Razavi, P.; Dawson, S.-J.; Moss, E.L.; Govindan, R.; Tan, I.B.; Yap, Y.-S.; Robinson, W.A.; Morris, C.D.; et al. Liquid biopsies for residual disease and recurrence. Med 2021, 2, 1292–1313. [Google Scholar] [CrossRef]
- Bauml, J.M.; Li, B.T.; Velcheti, V.; Govindan, R.; Curioni-Fontecedro, A.; Dooms, C.; Takahashi, T.; Duda, A.W.; Odegaard, J.I.; Cruz-Guilloty, F.; et al. Clinical validation of Guardant360 CDx as a blood-based companion diagnostic for sotorasib. Lung Cancer 2022, 166, 270–278. [Google Scholar] [CrossRef]
- Ribeiro, T.B.; Ribeiro, A.; Rodrigues, L.O.; Harada, G.; Nobre, M.R.C. U.S. Food and Drug Administration anticancer drug approval trends from 2016 to 2018 for lung, colorectal, breast, and prostate cancer. Int. J. Technol. Assess. Health Care 2020, 36, 20–28. [Google Scholar] [CrossRef]
- Kawasaki, E.S.; Clark, S.S.; Coyne, M.Y.; Smith, S.D.; Champlin, R.; Witte, O.N.; McCormick, F.P. Diagnosis of chronic myeloid and acute lymphocytic leukemias by detection of leukemia-specific mRNA sequences amplified in vitro. Proc. Natl. Acad. Sci. USA 1988, 85, 5698–5702. [Google Scholar] [CrossRef]
- Crucitta, S.; Ruglioni, M.; Novi, C.; Manganiello, M.; Arici, R.; Petrini, I.; Pardini, E.; Cucchiara, F.; Marmorino, F.; Cremolini, C.; et al. Comparison of digital PCR systems for the analysis of liquid biopsy samples of patients affected by lung and colorectal cancer. Clin. Chim. Acta 2023, 541, 117239. [Google Scholar] [CrossRef] [PubMed]
- Freed-Pastor, W.A.; Prives, C. Mutant p53: One name, many proteins. Genes Dev. 2012, 26, 1268–1286. [Google Scholar] [CrossRef]
- CROOK, T. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell 1994, 79, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Vavalà, T.; Monica, V.; Iacono, M.L.; Mele, T.; Busso, S.; Righi, L.; Papotti, M.; Scagliotti, G.V.; Novello, S. Precision medicine in age-specific non-small-cell-lung-cancer patients: Integrating biomolecular results into clinical practice-A new approach to improve personalized translational research. Lung Cancer 2017, 107, 84–90. [Google Scholar] [CrossRef]
- Tahara, T.; Tahara, S.; Horiguchi, N.; Okubo, M.; Terada, T.; Yamada, H.; Yoshida, D.; Omori, T.; Osaki, H.; Maeda, K.; et al. Molecular subtyping of gastric cancer combining genetic and epigenetic anomalies provides distinct clinicopathological features and prognostic impacts. Hum. Mutat. 2019, 40, 347–354. [Google Scholar] [CrossRef]
- Dogruluk, T.; Tsang, Y.H.; Espitia, M.; Chen, F.; Chen, T.; Chong, Z.; Appadurai, V.; Dogruluk, A.; Eterovic, A.K.; Bonnen, P.E.; et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res. 2015, 75, 5341–5354. [Google Scholar] [CrossRef]
- Ng, P.K.-S.; Li, J.; Jeong, K.J.; Shao, S.; Chen, H.; Tsang, Y.H.; Sengupta, S.; Wang, Z.; Bhavana, V.H.; Tran, R.; et al. Systematic Functional Annotation of Somatic Mutations in Cancer. Cancer Cell 2018, 33, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Minato, T.; Ito, S.; Li, B.; Fujimori, H.; Mochizuki, M.; Yamaguchi, K.; Tamai, K.; Shimada, M.; Tokunaga, H.; Shigeta, S.; et al. Liquid biopsy with droplet digital PCR targeted to specific mutations in plasma cell-free tumor DNA can detect ovarian cancer recurrence earlier than CA125. Gynecol. Oncol. Rep. 2021, 38, 100847. [Google Scholar] [CrossRef]
- Shintani, D.; Hihara, T.; Ogasawara, A.; Sato, S.; Yabuno, A.; Tai, K.; Fujiwara, K.; Watanabe, K.; Hasegawa, K. Tumor-related mutations in cell-free DNA in pre-operative plasma as a prognostic indicator of recurrence in endometrial cancer. Int. J. Gynecol. Cancer 2020, 30, 1340–1346. [Google Scholar] [CrossRef]
- Feng, W.; Jia, N.; Jiao, H.; Chen, J.; Chen, Y.; Zhang, Y.; Zhu, M.; Zhu, C.; Shen, L.; Long, W. Circulating tumor DNA as a prognostic marker in high-risk endometrial cancer. J. Transl. Med. 2021, 19, 51. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Megna, R.; Petretta, M.; Alfano, B.; Cantoni, V.; Green, R.; Daniele, S.; Acampa, W.; Nappi, C.; Gaudieri, V.; Assante, R.; et al. A New Relational Database Including Clinical Data and Myocardial Perfusion Imaging Findings in Coronary Artery Disease. Curr. Med. Imaging Former. Curr. Med. Imaging Rev. 2018, 15, 661–671. [Google Scholar] [CrossRef]
- Fagotti, A.; Ferrandina, G.; Fanfani, F.; Garganese, G.; Vizzielli, G.; Carone, V.; Salerno, M.G.; Scambia, G. Prospective validation of a laparoscopic predictive model for optimal cytoreduction in advanced ovarian carcinoma. Am. J. Obstet. Gynecol. 2008, 199, e1–e642. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balasan, N.; Kharrat, F.; Di Lorenzo, G.; Athanasakis, E.; Bianco, A.M.; Conti, A.; Di Stazio, M.T.; Butera, G.; Cicogna, S.; Mangogna, A.; et al. Sensitive Detection of Gynecological Cancer Recurrence Using Circulating Tumor DNA and Digital PCR: A Comparative Study with Serum Biochemical Markers. Int. J. Mol. Sci. 2024, 25, 11997. https://doi.org/10.3390/ijms252211997
Balasan N, Kharrat F, Di Lorenzo G, Athanasakis E, Bianco AM, Conti A, Di Stazio MT, Butera G, Cicogna S, Mangogna A, et al. Sensitive Detection of Gynecological Cancer Recurrence Using Circulating Tumor DNA and Digital PCR: A Comparative Study with Serum Biochemical Markers. International Journal of Molecular Sciences. 2024; 25(22):11997. https://doi.org/10.3390/ijms252211997
Chicago/Turabian StyleBalasan, Nour, Feras Kharrat, Giovanni Di Lorenzo, Emmanouil Athanasakis, Anna Monica Bianco, Andrea Conti, Maria Teresa Di Stazio, Giulia Butera, Stefania Cicogna, Alessandro Mangogna, and et al. 2024. "Sensitive Detection of Gynecological Cancer Recurrence Using Circulating Tumor DNA and Digital PCR: A Comparative Study with Serum Biochemical Markers" International Journal of Molecular Sciences 25, no. 22: 11997. https://doi.org/10.3390/ijms252211997
APA StyleBalasan, N., Kharrat, F., Di Lorenzo, G., Athanasakis, E., Bianco, A. M., Conti, A., Di Stazio, M. T., Butera, G., Cicogna, S., Mangogna, A., Romano, F., Ricci, G., & d’Adamo, A. P. (2024). Sensitive Detection of Gynecological Cancer Recurrence Using Circulating Tumor DNA and Digital PCR: A Comparative Study with Serum Biochemical Markers. International Journal of Molecular Sciences, 25(22), 11997. https://doi.org/10.3390/ijms252211997





