Iodinated PSMA Ligands as XFI Tracers for Targeted Cell Imaging and Characterization of Nanoparticles
Abstract
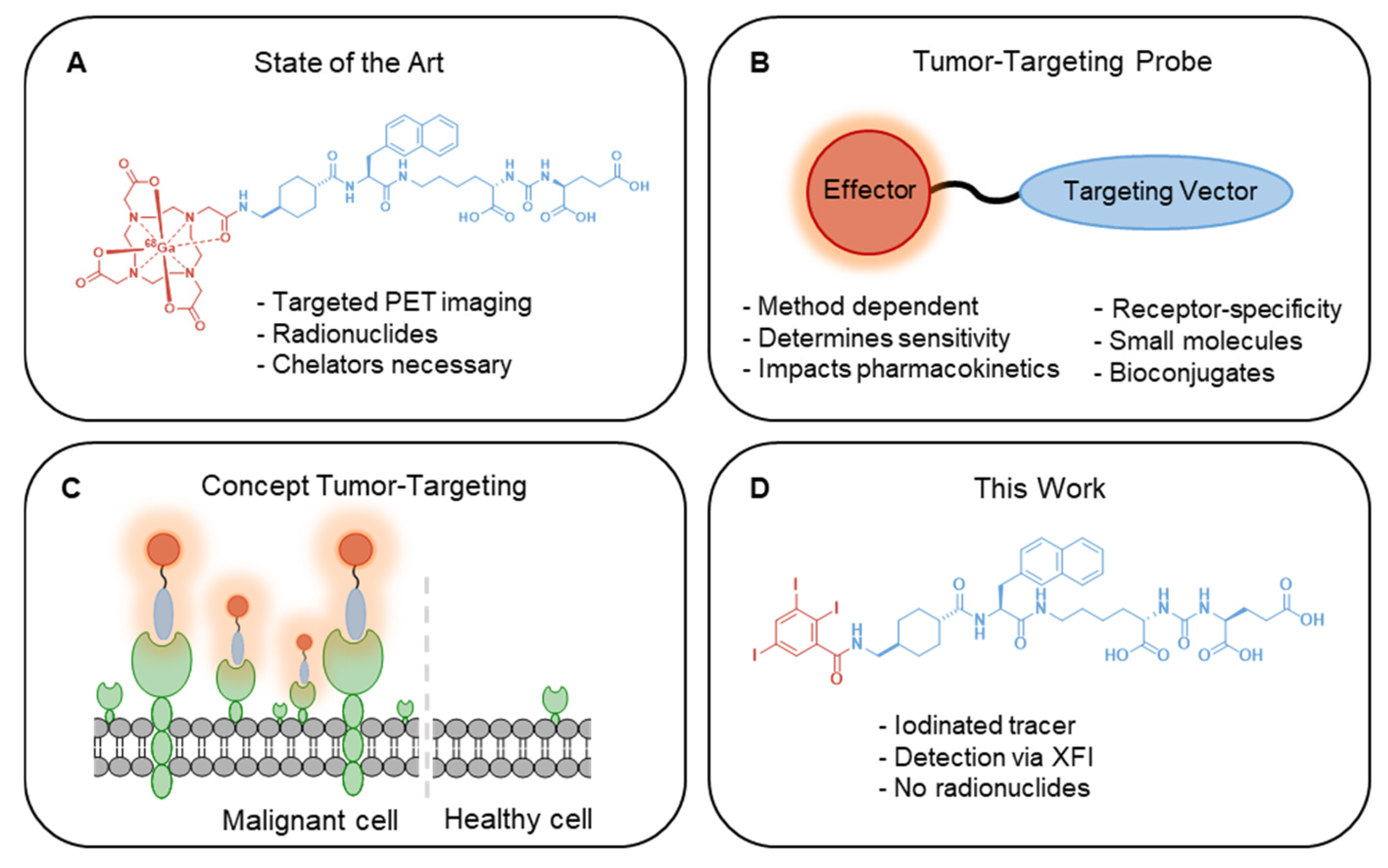
1. Introduction
2. Results
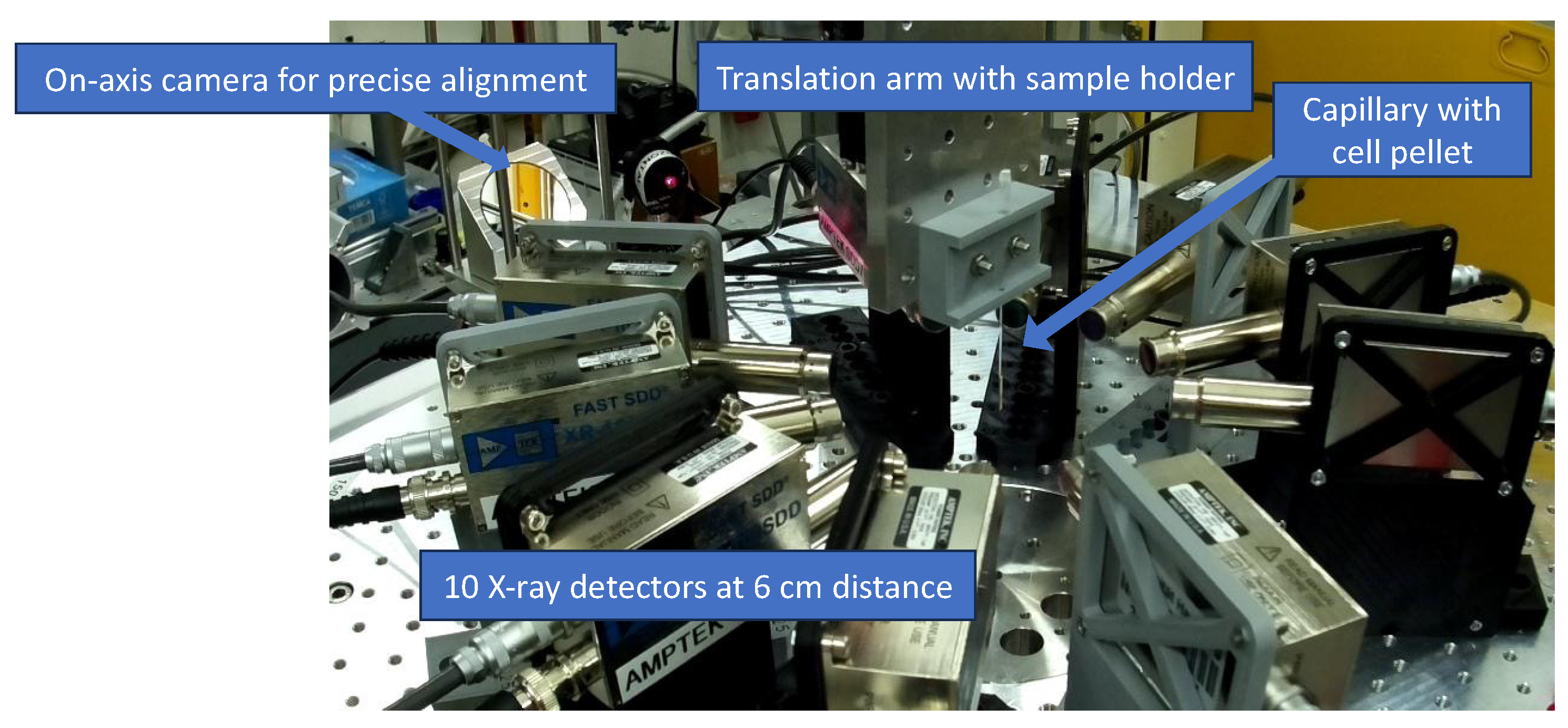
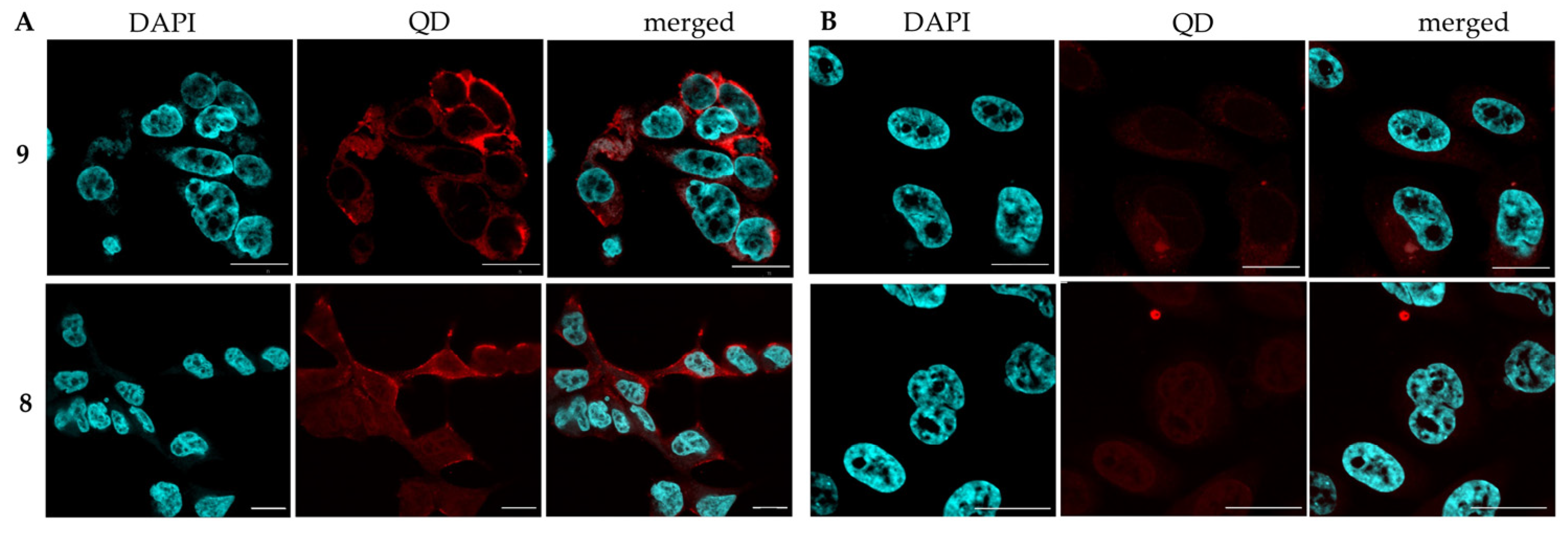
2.1. XFI Measurements with Prostate Cancer Cell Lines
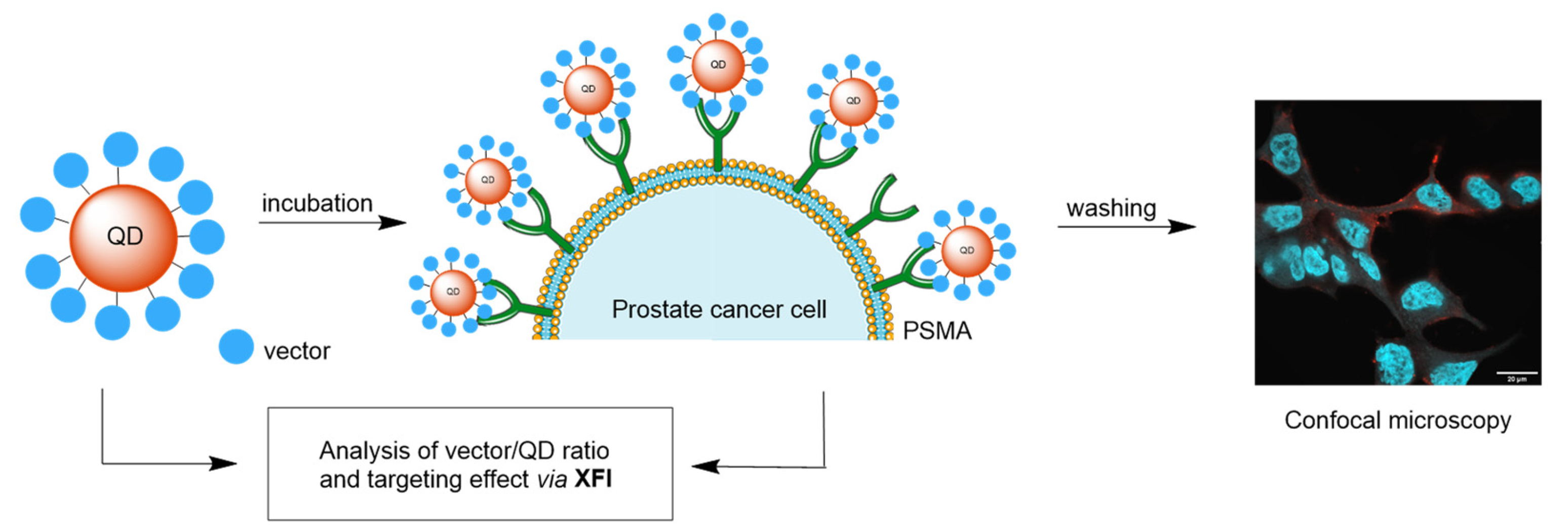
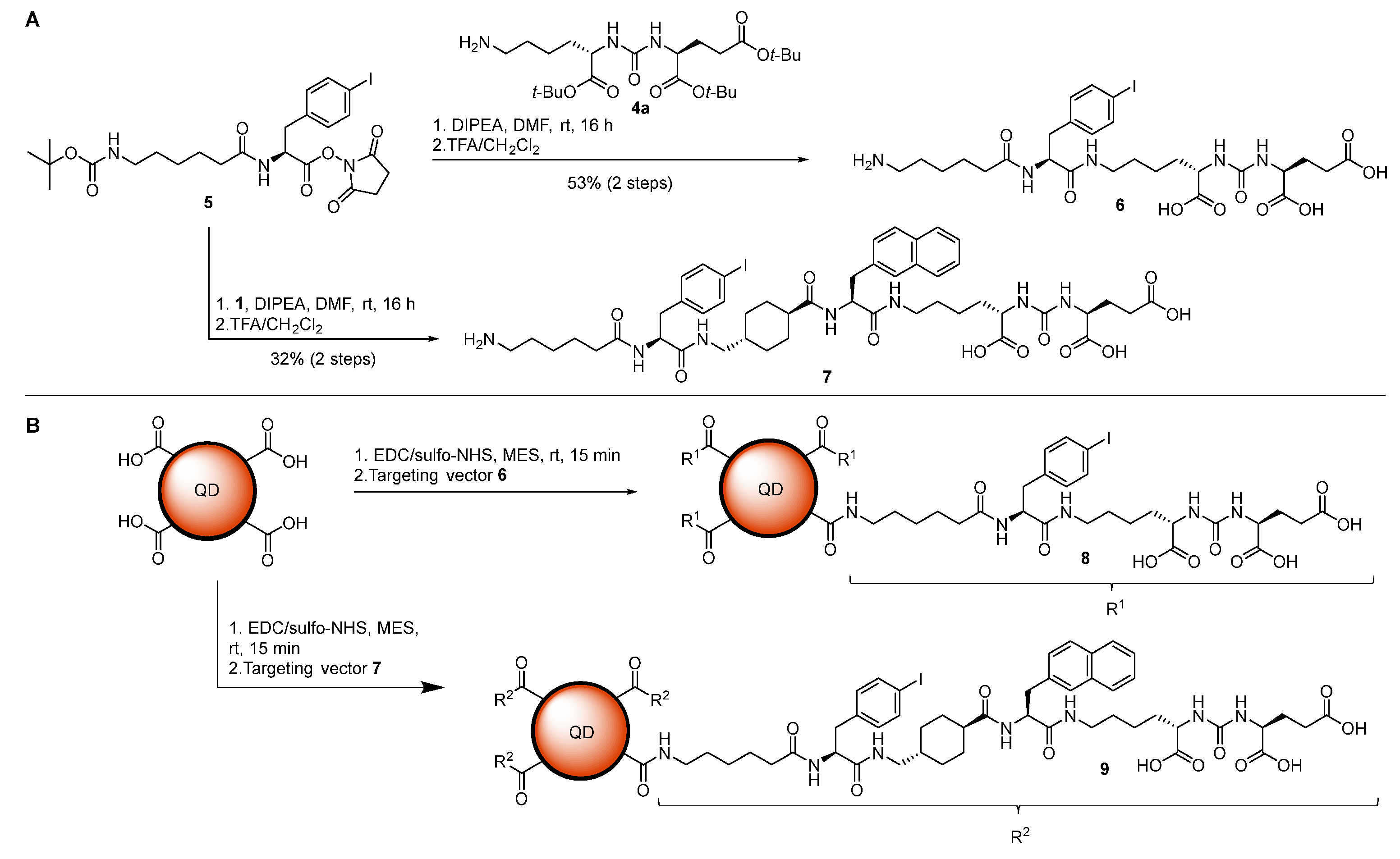
2.2. Iodinated Tracers for Characterization of Nanoparticle Conjugates via XFI and ICP-MS
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- James, M.L.; Gambhir, S.S. A molecular imaging primer: Modalities, imaging agents, and applications. Physiol. Rev. 2012, 92, 897–965. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.D.; Paudel, R.; Liu, J.; Ma, C.; Zhang, Z.S.; Zhou, S.K. MRI contrast agents: Classification and application (Review). Int. J. Mol. Med. 2016, 38, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Wahsner, J.; Gale, E.M.; Rodriguez-Rodriguez, A.; Caravan, P. Chemistry of MRI Contrast Agents: Current Challenges and New Frontiers. Chem. Rev. 2019, 119, 957–1057. [Google Scholar] [CrossRef]
- Derlin, T.; Grunwald, V.; Steinbach, J.; Wester, H.J.; Ross, T.L. Molecular Imaging in Oncology Using Positron Emission Tomography. Dtsch. Arztebl. Int. 2018, 115, 175–181. [Google Scholar] [CrossRef]
- Ceci, F.; Castellucci, P.; Fanti, S. Current application and future perspectives of prostate specific membrane antigen PET imaging in prostate cancer. Q. J. Nucl. Med. Mol. Imaging 2019, 63, 7–18. [Google Scholar] [CrossRef]
- Rahmim, A.; Zaidi, H. PET versus SPECT: Strengths, limitations and challenges. Nucl. Med. Commun. 2008, 29, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Gawne, P.J.; Man, F.; Blower, P.J.; TM de Rosales, R. Direct Cell Radiolabeling for in Vivo Cell Tracking with PET and SPECT Imaging. Chem. Rev. 2022, 122, 10266–10318. [Google Scholar] [CrossRef]
- Frangioni, J.V. New technologies for human cancer imaging. J. Clin. Oncol. 2008, 26, 4012–4021. [Google Scholar] [CrossRef]
- Joshi, B.P.; Wang, T.D. Targeted Optical Imaging Agents in Cancer: Focus on Clinical Applications. Contrast Media Mol. Imaging 2018, 2018, 2015237. [Google Scholar] [CrossRef]
- Sim, N.; Parker, D. Critical design issues in the targeted molecular imaging of cell surface receptors. Chem. Soc. Rev. 2015, 44, 2122–2134. [Google Scholar] [CrossRef]
- Roberts, M.J.; Maurer, T.; Perera, M.; Eiber, M.; Hope, T.A.; Ost, P.; Siva, S.; Hofman, M.S.; Murphy, D.G.; Emmett, L.; et al. Using PSMA imaging for prognostication in localized and advanced prostate cancer. Nat. Rev. Urol. 2023, 20, 23–47. [Google Scholar] [CrossRef] [PubMed]
- El Fakiri, M.; Geis, N.M.; Ayada, N.; Eder, M.; Eder, A.C. PSMA-Targeting Radiopharmaceuticals for Prostate Cancer Therapy: Recent Developments and Future Perspectives. Cancers 2021, 13, 3967. [Google Scholar] [CrossRef] [PubMed]
- Baranski, A.C.; Schafer, M.; Bauder-Wust, U.; Roscher, M.; Schmidt, J.; Stenau, E.; Simpfendorfer, T.; Teber, D.; Maier-Hein, L.; Hadaschik, B.; et al. PSMA-11-Derived Dual-Labeled PSMA Inhibitors for Preoperative PET Imaging and Precise Fluorescence-Guided Surgery of Prostate Cancer. J. Nucl. Med. 2018, 59, 639–645. [Google Scholar] [CrossRef]
- Murphy, G.P.; Elgamal, A.A.; Su, S.L.; Bostwick, D.G.; Holmes, E.H. Current evaluation of the tissue localization and diagnostic utility of prostate specific membrane antigen. Cancer 1998, 83, 2259–2269. [Google Scholar] [CrossRef]
- Widjaja, L.; Werner, R.A.; Ross, T.L.; Bengel, F.M.; Derlin, T. PSMA Expression Predicts Early Biochemical Response in Patients with Metastatic Castration-Resistant Prostate Cancer under (177)Lu-PSMA-617 Radioligand Therapy. Cancers 2021, 13, 2938. [Google Scholar] [CrossRef]
- Gourni, E.; Henriksen, G. Metal-Based PSMA Radioligands. Molecules 2017, 22, 523. [Google Scholar] [CrossRef] [PubMed]
- Umbricht, C.A.; Benesova, M.; Schmid, R.M.; Turler, A.; Schibli, R.; van der Meulen, N.P.; Muller, C. (44)Sc-PSMA-617 for radiotheragnostics in tandem with (177)Lu-PSMA-617-preclinical investigations in comparison with (68)Ga-PSMA-11 and (68)Ga-PSMA-617. EJNMMI Res. 2017, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Benešová, M.; Schäfer, M.; Bauder-Wüst, U.; Afshar-Oromieh, A.; Kratochwil, C.; Mier, W.; Haberkorn, U.; Kopka, K.; Eder, M. Preclinical Evaluation of a Tailor-Made DOTA-Conjugated PSMA Inhibitor with Optimized Linker Moiety for Imaging and Endoradiotherapy of Prostate Cancer. J. Nucl. Med. 2015, 56, 914–920. [Google Scholar] [CrossRef]
- Schwarzenboeck, S.M.; Rauscher, I.; Bluemel, C.; Fendler, W.P.; Rowe, S.P.; Pomper, M.G.; Afshar-Oromieh, A.; Herrmann, K.; Eiber, M. PSMA Ligands for PET Imaging of Prostate Cancer. J. Nucl. Med. 2017, 58, 1545–1552. [Google Scholar] [CrossRef]
- Horoszewicz, J.S.; Kawinski, E.; Murphy, G.P. Monoclonal-Antibodies to a New Antigenic Marker in Epithelial Prostatic Cells and Serum of Prostatic-Cancer Patients. Anticancer Res. 1987, 7, 927–936. [Google Scholar]
- Israeli, R.S.; Powell, C.T.; Fair, W.R.; Heston, W.D. Molecular cloning of a complementary DNA encoding a prostate-specific membrane antigen. Cancer Res. 1993, 53, 227–230. [Google Scholar] [PubMed]
- Chang, S.S.; Heston, W.D. The clinical role of prostate-specific membrane antigen (PSMA). Urol. Oncol. 2002, 7, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Henzlova, M.J. SPECT: Workhorse of state of the art nuclear cardiology. J. Nucl. Cardiol. 2018, 25, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Moses, W.W. Fundamental Limits of Spatial Resolution in PET. Nucl. Instrum. Methods Phys. Res. A 2011, 648 (Suppl. 1), S236–S240. [Google Scholar] [CrossRef]
- Hosono, M.; Takenaka, M.; Monzen, H.; Tamura, M.; Kudo, M.; Nishimura, Y. Cumulative radiation doses from recurrent PET-CT examinations. Br. J. Radiol. 2021, 94, 20210388. [Google Scholar] [CrossRef]
- Keereweer, S.; Van Driel, P.B.; Snoeks, T.J.; Kerrebijn, J.D.; Baatenburg de Jong, R.J.; Vahrmeijer, A.L.; Sterenborg, H.J.; Lowik, C.W. Optical image-guided cancer surgery: Challenges and limitations. Clin. Cancer Res. 2013, 19, 3745–3754. [Google Scholar] [CrossRef]
- Notni, J. RGD Forever!-Past, Present, and Future of a 3-Letter-Code in Radiopharmacy and Life Sciences. Pharmaceuticals 2022, 16, 56. [Google Scholar] [CrossRef]
- Wang, F.; Li, Z.; Feng, X.; Yang, D.; Lin, M. Advances in PSMA-targeted therapy for prostate cancer. Prostate Cancer Prostatic Dis. 2022, 25, 11–26. [Google Scholar] [CrossRef]
- Pang, X.; He, X.; Qiu, Z.; Zhang, H.; Xie, R.; Liu, Z.; Gu, Y.; Zhao, N.; Xiang, Q.; Cui, Y. Targeting integrin pathways: Mechanisms and advances in therapy. Signal Transduct. Target. Ther. 2023, 8, 1. [Google Scholar] [CrossRef]
- Sindhu, R.K.; Yadav, S.K.; Kaur, A.; Kumar, M.; Kumar, P. X-Ray Fluorescence. In X-Ray Fluorescence in Biological Sciences; John Wiley & Sons: Hoboken, NJ, USA, 2022; pp. 623–646. [Google Scholar]
- Sanchez-Cano, C.; Alvarez-Puebla, R.A.; Abendroth, J.M.; Beck, T.; Blick, R.; Cao, Y.; Caruso, F.; Chakraborty, I.; Chapman, H.N.; Chen, C.; et al. X-ray-Based Techniques to Study the Nano-Bio Interface. ACS Nano 2021, 15, 3754–3807. [Google Scholar] [CrossRef]
- George, G.N.; Pickering, I.J. X-ray Fluorescence Imaging: Elemental and Chemical Speciation Mapping of Biological Systems. In Encyclopedia of Biophysics; Roberts, G., Watts, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–6. [Google Scholar]
- Strotton, M.; Hosogane, T.; di Michiel, M.; Moch, H.; Varga, Z.; Bodenmiller, B. Multielement Z-tag imaging by X-ray fluorescence microscopy for next-generation multiplex imaging. Nat. Methods 2023, 20, 1310–1322. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Foss, C.A.; Byun, Y.; Nimmagadda, S.; Pullambhatla, M.; Fox, J.J.; Castanares, M.; Lupold, S.E.; Babich, J.W.; Mease, R.C.; et al. Radiohalogenated prostate-specific membrane antigen (PSMA)-based ureas as imaging agents for prostate cancer. J. Med. Chem. 2008, 51, 7933–7943. [Google Scholar] [CrossRef] [PubMed]
- Gruner, F.; Blumendorf, F.; Schmutzler, O.; Staufer, T.; Bradbury, M.; Wiesner, U.; Rosentreter, T.; Loers, G.; Lutz, D.; Richter, B.; et al. Localising functionalised gold-nanoparticles in murine spinal cords by X-ray fluorescence imaging and background-reduction through spatial filtering for human-sized objects. Sci. Rep. 2018, 8, 16561. [Google Scholar] [CrossRef] [PubMed]
- Schmutzler, O.; Graf, S.; Behm, N.; Mansour, W.Y.; Blumendorf, F.; Staufer, T.; Körnig, C.; Salah, D.; Kang, Y.; Peters, J.N.; et al. X-ray Fluorescence Uptake Measurement of Functionalized Gold Nanoparticles in Tumor Cell Microsamples. Int. J. Mol. Sci. 2021, 22, 3691. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Wang, Y.; Wang, L.; Zhang, K.; Guan, Y.; Guo, Y.; Chen, C. In situ label-free X-ray imaging for visualizing the localization of nanomedicines and subcellular architecture in intact single cells. Nat. Protoc. 2024, 19, 30–59. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, X.; Nandi, P.; Lin, Y.; Wang, L.; Chu, Y.S.; Paape, T.; Yang, Y.; Xiao, X.; Liu, Q. Correlative single-cell hard X-ray computed tomography and X-ray fluorescence imaging. Commun. Biol. 2024, 7, 280. [Google Scholar] [CrossRef]
- Staufer, T.; Körnig, C.; Liu, B.; Liu, Y.; Lanzloth, C.; Schmutzler, O.; Bedke, T.; Machicote, A.; Parak, W.J.; Feliu, N.; et al. Enabling X-ray fluorescence imaging for in vivo immune cell tracking. Sci. Rep. 2023, 13, 11505. [Google Scholar] [CrossRef]
- Staufer, T.; Grüner, F. Review of Development and Recent Advances in Biomedical X-ray Fluorescence Imaging. Int. J. Mol. Sci. 2023, 24, 10990. [Google Scholar] [CrossRef]
- Manohar, N.; Reynoso, F.J.; Diagaradjane, P.; Krishnan, S.; Cho, S.H. Quantitative imaging of gold nanoparticle distribution in a tumor-bearing mouse using benchtop x-ray fluorescence computed tomography. Sci. Rep. 2016, 6, 22079. [Google Scholar] [CrossRef]
- Pushie, M.J.; Pickering, I.J.; Korbas, M.; Hackett, M.J.; George, G.N. Elemental and chemically specific X-ray fluorescence imaging of biological systems. Chem. Rev. 2014, 114, 8499–8541. [Google Scholar] [CrossRef]
- Petrov, S.A.; Yusubov, M.S.; Beloglazkina, E.K.; Nenajdenko, V.G. Synthesis of Radioiodinated Compounds. Classical Approaches and Achievements of Recent Years. Int. J. Mol. Sci. 2022, 23, 13789. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, M.; Mutas, M.; Chandralingam, S.; von Salisch, C.; Peric, N.; Segelke, T.; Fischer, M.; Chakraborty, I.; Parak, W.J.; Frangioni, J.V.; et al. Nonradioactive Cell Assay for the Evaluation of Modular Prostate-Specific Membrane Antigen Targeting Ligands via Inductively Coupled Plasma Mass Spectrometry. J. Med. Chem. 2019, 62, 10912–10918. [Google Scholar] [CrossRef] [PubMed]
- Todorov, T.I.; Gray, P.J. Analysis of iodine in food samples by inductively coupled plasma-mass spectrometry. Food Addit. Contam. 2016, 33, 282–290. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Izmer, A.V.; Boulyga, S.F.; Becker, J.S. Determination of 129I/127I isotope ratios in liquid solutions and environmental soil samples by ICP-MS with hexapole collision cell. J. Anal. At. Spectrom. 2003, 18, 1339–1345. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef]
- Liz-Marzan, L.M.; Artzi, N.; Bals, S.; Buriak, J.M.; Chan, W.C.W.; Chen, X.; Hersam, M.C.; Kim, I.D.; Millstone, J.E.; Mulvaney, P.; et al. Celebrating a Nobel Prize to the “Discovery of Quantum Dots, an Essential Milestone in Nanoscience”. ACS Nano 2023, 17, 19474–19475. [Google Scholar] [CrossRef] [PubMed]
- Höeg, F.; Schulz, J.; Graf, S.; Salah, D.; Chandralingam, S.; Maison, W.; Parak, W.J.; Schulz, F. Defined Coadsorption of Prostate Cancer Targeting Ligands and PEG on Gold Nanoparticles for Significantly Reduced Protein Adsorption in Cell Media. J. Phys. Chem. C 2022, 126, 20594–20604. [Google Scholar] [CrossRef]
- Alkilany, A.M.; Zhu, L.; Weller, H.; Mews, A.; Parak, W.J.; Barz, M.; Feliu, N. Ligand density on nanoparticles: A parameter with critical impact on nanomedicine. Adv. Drug Deliv. Rev. 2019, 143, 22–36. [Google Scholar] [CrossRef]
- Liu, Z.; Escudero, A.; Carrillo-Carrion, C.; Chakraborty, I.; Zhu, D.; Gallego, M.; Parak, W.J.; Feliu, N. Biodegradation of Bi-Labeled Polymer-Coated Rare-Earth Nanoparticles in Adherent Cell Cultures. Chem. Mater. 2020, 32, 245–254. [Google Scholar] [CrossRef]
- Eder, M.; Schäfer, M.; Bauder-Wüst, U.; Hull, W.-E.; Wängler, C.; Mier, W.; Haberkorn, U.; Eisenhut, M. 68Ga-Complex Lipophilicity and the Targeting Property of a Urea-Based PSMA Inhibitor for PET Imaging. Bioconjt. Chem. 2012, 23, 688–697. [Google Scholar] [CrossRef]
- Horoszewicz, J.S.; Leong, S.S.; Kawinski, E.; Karr, J.P.; Rosenthal, H.; Chu, T.M.; Mirand, E.A.; Murphy, G.P. Lncap Model of Human Prostatic-Carcinoma. Cancer Res. 1983, 43, 1809–1818. [Google Scholar] [PubMed]
- Humblet, V.; Misra, P.; Bhushan, K.R.; Nasr, K.; Ko, Y.S.; Tsukamoto, T.; Pannier, N.; Frangioni, J.V.; Maison, W. Multivalent scaffolds for affinity maturation of small molecule cell surface binders and their application to prostate tumor targeting. J. Med. Chem. 2009, 52, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Criscione, J.M.; Dobrucki, L.W.; Zhuang, Z.W.; Papademetris, X.; Simons, M.; Sinusas, A.J.; Fahmy, T.M. Development and Application of a Multimodal Contrast Agent for SPECT/CT Hybrid Imaging. Bioconjt. Chem. 2011, 22, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Wang, X.; Klein, E.; Heston, W.D.W. Novel Role of Prostate-Specific Membrane Antigen in Suppressing Prostate Cancer Invasiveness. Cancer Res. 2005, 65, 727–731. [Google Scholar] [CrossRef]
- Gong, M.C.; Latouche, J.B.; Krause, A.; Heston, W.D.; Bander, N.H.; Sadelain, M. Cancer patient T cells genetically targeted to prostate-specific membrane antigen specifically lyse prostate cancer cells and release cytokines in response to prostate-specific membrane antigen. Neoplasia 1999, 1, 123–127. [Google Scholar] [CrossRef]
- Hühn, J.; Carrillo-Carrion, C.; Soliman, M.G.; Pfeiffer, C.; Valdeperez, D.; Masood, A.; Chakraborty, I.; Zhu, L.; Gallego, M.; Yue, Z.; et al. Selected Standard Protocols for the Synthesis, Phase Transfer, and Characterization of Inorganic Colloidal Nanoparticles. Chem. Mater. 2016, 29, 399–461. [Google Scholar] [CrossRef]
- Kerpa, S.; Schulze, V.R.; Holzapfel, M.; Cvancar, L.; Fischer, M.; Maison, W. Decoration of 1, 4, 7, 10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) with N-oxides increases the T(1) relaxivity of Gd-complexes. ChemistryOpen 2024, 13, e202300298. [Google Scholar] [CrossRef]
- Mixdorf, J.C.; Hoffman, S.L.V.; Aluicio-Sarduy, E.; Barnhart, T.E.; Engle, J.W.; Ellison, P.A. Copper-Mediated Radiobromination of (Hetero)Aryl Boronic Pinacol Esters. J. Org. Chem. 2023, 88, 2089–2094. [Google Scholar] [CrossRef]
- Kim, J.H.; Ali, K.H.; Oh, Y.J.; Seo, Y.H. Design, synthesis, and biological evaluation of histone deacetylase inhibitor with novel salicylamide zinc binding group. Medicine 2022, 101, e29049. [Google Scholar] [CrossRef]


| Sample | Mass Cd by XFI (µg) | Mass I (ng) | Mass Cd by ICP-MS (µg) | Ratio (I/QD) |
|---|---|---|---|---|
| 8 | 2.1 | 68.8 | 1.7 | 26 |
| 9 | 2.4 | 28.6 | 2.4 | 11 |
| QDs | 1.8 | 0 | 1.8 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerpa, S.; Holzapfel, M.; Staufer, T.; Kuhrwahl, R.; Mutas, M.; Werner, S.; Schulze, V.R.; Nakielski, P.; Feliu, N.; Oetjen, E.; et al. Iodinated PSMA Ligands as XFI Tracers for Targeted Cell Imaging and Characterization of Nanoparticles. Int. J. Mol. Sci. 2024, 25, 11880. https://doi.org/10.3390/ijms252211880
Kerpa S, Holzapfel M, Staufer T, Kuhrwahl R, Mutas M, Werner S, Schulze VR, Nakielski P, Feliu N, Oetjen E, et al. Iodinated PSMA Ligands as XFI Tracers for Targeted Cell Imaging and Characterization of Nanoparticles. International Journal of Molecular Sciences. 2024; 25(22):11880. https://doi.org/10.3390/ijms252211880
Chicago/Turabian StyleKerpa, Svenja, Malte Holzapfel, Theresa Staufer, Robert Kuhrwahl, Marina Mutas, Stefan Werner, Verena R. Schulze, Pascal Nakielski, Neus Feliu, Elke Oetjen, and et al. 2024. "Iodinated PSMA Ligands as XFI Tracers for Targeted Cell Imaging and Characterization of Nanoparticles" International Journal of Molecular Sciences 25, no. 22: 11880. https://doi.org/10.3390/ijms252211880
APA StyleKerpa, S., Holzapfel, M., Staufer, T., Kuhrwahl, R., Mutas, M., Werner, S., Schulze, V. R., Nakielski, P., Feliu, N., Oetjen, E., Haak, J., Ziegler, F., Buchin, R., Han, J., Parak, W. J., Grüner, F., & Maison, W. (2024). Iodinated PSMA Ligands as XFI Tracers for Targeted Cell Imaging and Characterization of Nanoparticles. International Journal of Molecular Sciences, 25(22), 11880. https://doi.org/10.3390/ijms252211880







