Potential Cryptic Diversity in the Genus Scoliodon (Carcharhiniformes: Carcharhinidae): Insights from Mitochondrial Genome Sequencing
Abstract
1. Introduction
2. Results
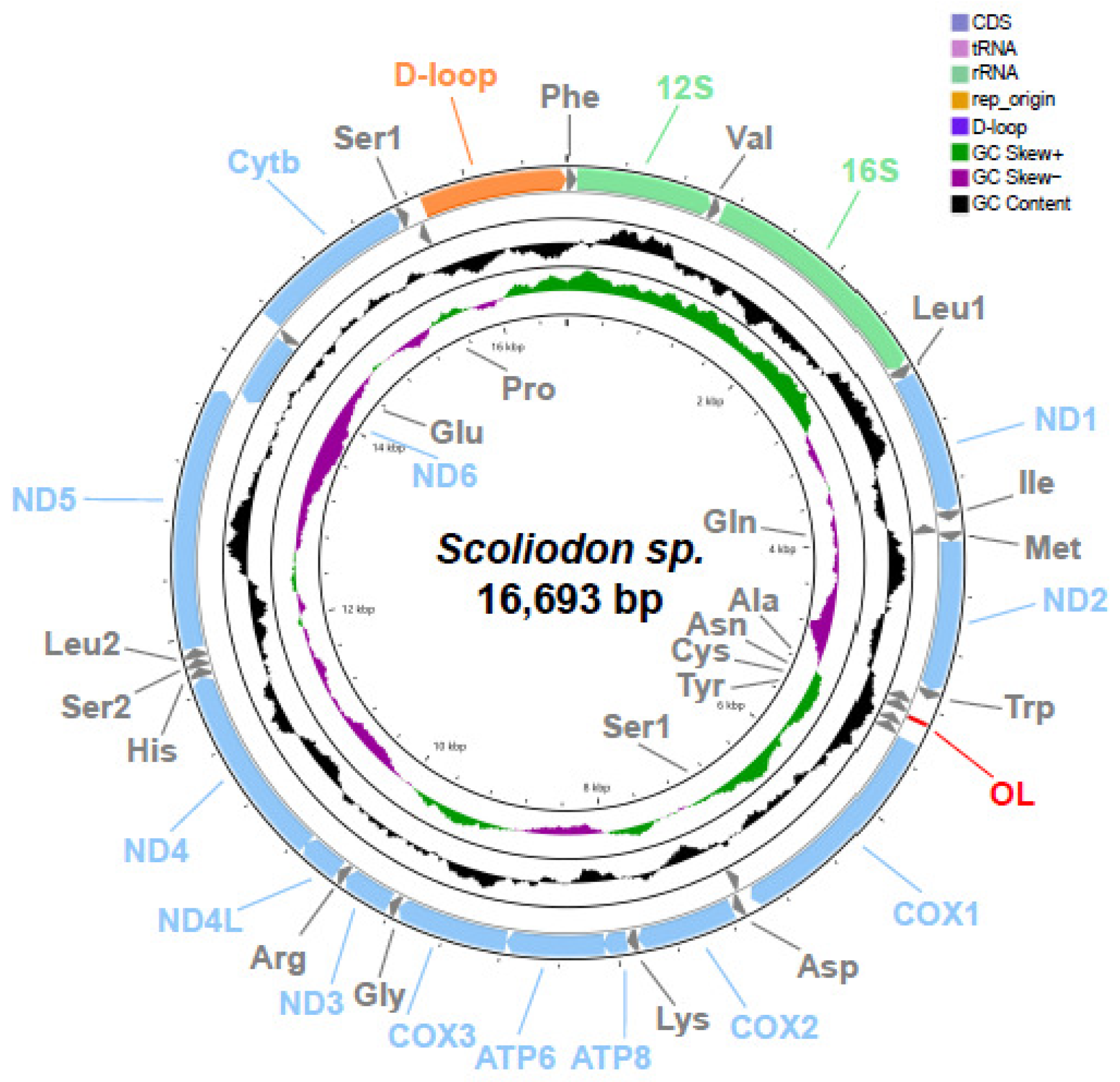
2.1. Genome Organization and Nucleotide Composition
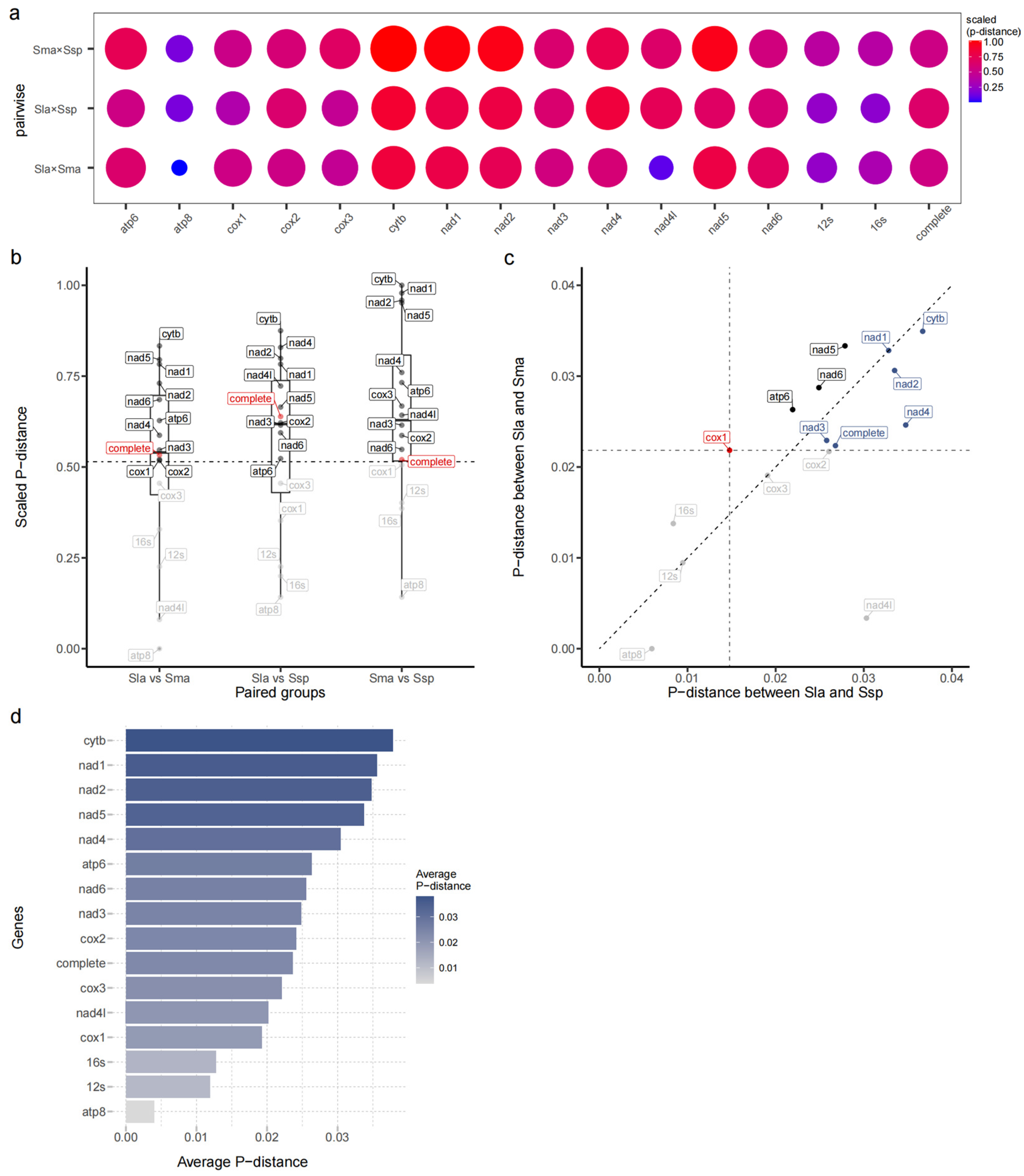
2.2. Multi-Gene Association Genetic Distance Analysis of the Mitochondrial Genome
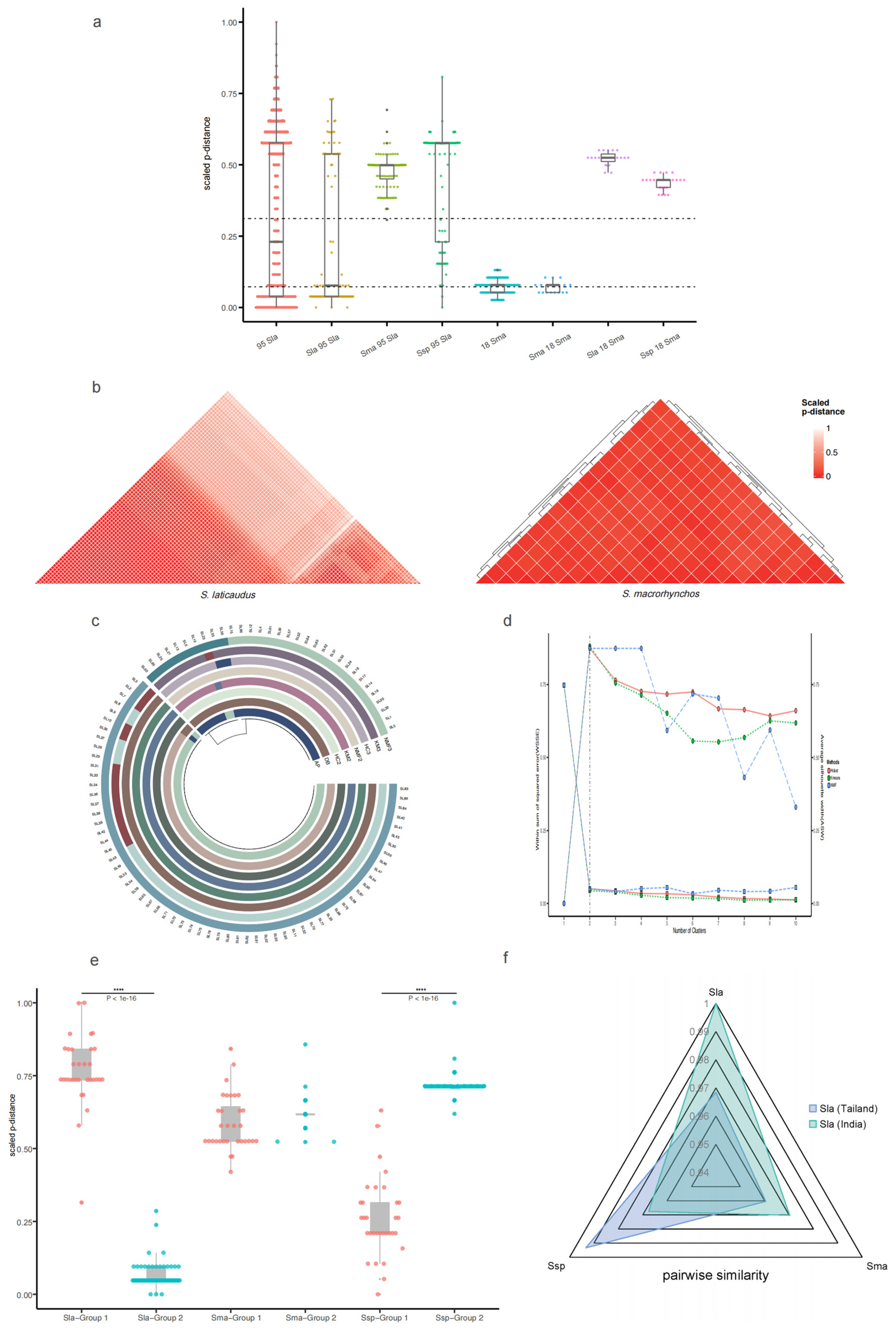
2.3. Population Genetic Distance Analysis Based on the Control Region Sequence
2.4. Verification of the Grouping Results
2.5. Phylogenetic Analysis and Divergence Time Estimates of Genus Scoliodon
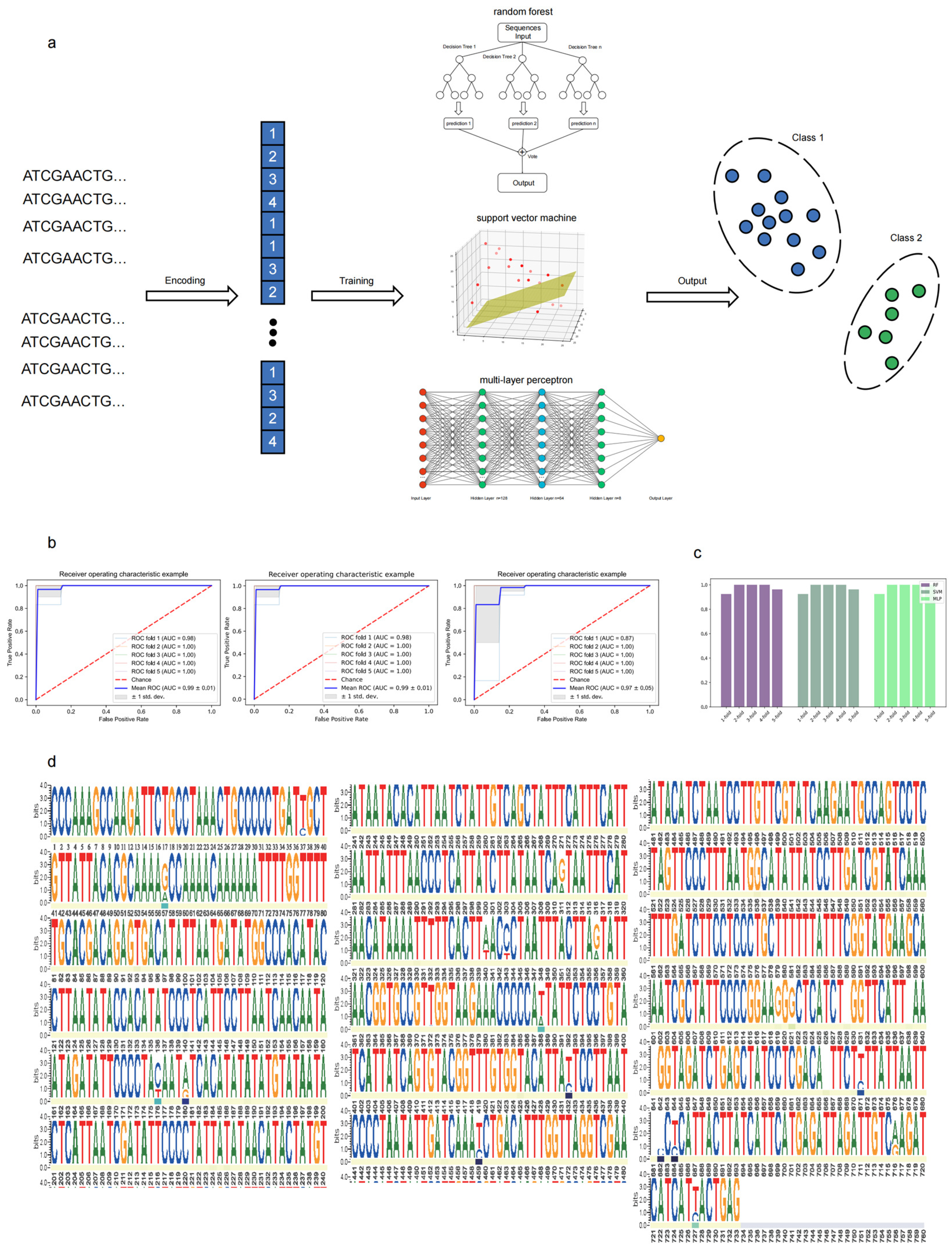
2.6. Cryptic Species Classification Using Machine Learning
3. Discussion
4. Materials and Methods
4.1. Specimen Collection, DNA Extraction, PCR Amplification, and Sequencing
4.2. Sequence Assembly, Annotation, and Basic Analysis
4.3. Genetic Distance Analysis
4.4. Phylogenetic Analyses and Divergence Time Analyses
4.5. Machine Learning
4.5.1. Sequence Dataset Preprocessing
4.5.2. Model Development
4.5.3. Model Evaluation
4.5.4. Feature Importance
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Compagno, L.J.V. Sharks of the world: An annotated and illustrated catalogue of shark species known to date. Part 2. Carcharhiniformes; Food and Agriculture Organization: Rome, Italy, 1984; Volume 4, pp. 533–534. [Google Scholar]
- White, W.; Last, P.; Naylor, G. Scoliodon macrorhynchos (Bleeker, 1852), a second species of spadenose shark from the Western Pacific (Carcharhiniformes: Carcharhinidae). Descr. New Sharks Rays Borneo CSIRO Mar. Atmos. Res. Pap. 2010, 32, 61–76. [Google Scholar]
- Setna, S.; Sarangdhar, P. Description, bionomics and development of Scoliodon sorrakowah (Cuvier). Rec. Zool. Surv. India 1948, 46, 25–53. [Google Scholar] [CrossRef]
- Müller, J. Gattungen der Haifische und Rochen nach einer von ihm mit Hrn. Henle unternommenen gemeinschaftlichen Arbeit über die Naturgeschichte der Knorpelfische. Berichte Der Königlichen Preuss. Akad. Der Wiss. Zu Berl. 1837, 1837, 111–118. [Google Scholar]
- Bleeker, P. Bijdrage tot de Kennis der Plagiostomen van den Indischen Archipel; Lange: New York, NY, USA, 1852; Volume 24. [Google Scholar]
- Springer, V.G. A revision of the carcharhinid shark genera Scoliodon, Loxodon, and Rhizoprionodon. Proc. United States Natl. Mus. 1964, 115, 559–632. [Google Scholar] [CrossRef]
- Müller, J.; Henle, J. Systematische beschreibung der Plagiostomen; Veit und Comp: Berlin, Germany, 1841. [Google Scholar]
- Lim, K.C.; White, W.T.; Then, A.Y.; Naylor, G.J.; Arunrugstichai, S.; Loh, K.-H. Integrated taxonomy revealed genetic differences in morphologically similar and non-sympatric Scoliodon macrorhynchos and S. laticaudus. Animals 2022, 12, 681. [Google Scholar] [CrossRef]
- Austin, J.J.; Melville, J. Incorporating historical museum specimens into molecular systematic and conservation genetics research. Mol. Ecol. Notes 2006, 6, 1089–1092. [Google Scholar] [CrossRef]
- Hebert, P.D.; Gregory, T.R. The promise of DNA barcoding for taxonomy. Syst. Biol. 2005, 54, 852–859. [Google Scholar] [CrossRef]
- Rach, J.; DeSalle, R.; Sarkar, I.N.; Schierwater, B.; Hadrys, H. Character-based DNA barcoding allows discrimination of genera, species and populations in Odonata. Proc. R. Soc. B Biol. Sci. 2008, 275, 237–247. [Google Scholar] [CrossRef]
- Lara, A.; Ponce de León, J.L.; Rodriguez, R.; Casane, D.; Cote, G.; Bernatchez, L.; García-Machado, E. DNA barcoding of Cuban freshwater fishes: Evidence for cryptic species and taxonomic conflicts. Mol. Ecol. Resour. 2010, 10, 421–430. [Google Scholar] [CrossRef]
- Winterbottom, R.; Hanner, R.H.; Burridge, M.; Zur, M. A cornucopia of cryptic species-a DNA barcode analysis of the gobiid fish genus Trimma (Percomorpha, Gobiiformes). ZooKeys 2014, 381, 79–111. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Kartavtsev, Y.P. Divergence at Cyt-b and Co-1 mtDNA genes on different taxonomic levels and genetics of speciation in animals. Mitochondrial DNA 2011, 22, 55–65. [Google Scholar] [CrossRef]
- Zhu, Z.Y.; Yue, G.H. The complete mitochondrial genome of red grouper Plectropomus leopardus and its applications in identification of grouper species. Aquaculture 2008, 276, 44–49. [Google Scholar] [CrossRef]
- Ma, C.; Cheng, Q.; Zhang, Q.; Zhuang, P.; Zhao, Y. Genetic variation of Coilia ectenes (Clupeiformes: Engraulidae) revealed by the complete cytochrome b sequences of mitochondrial DNA. J. Exp. Mar. Biol. Ecol. 2010, 385, 14–19. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. London. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Souza, H.; Marchesin, S.; Itoyama, M. Analysis of the mitochondrial COI gene and its informative potential for evolutionary inferences in the families Coreidae and Pentatomidae (Heteroptera). Genet. Mol. Res. 2016, 15, 1–14. [Google Scholar] [CrossRef]
- Von der Heyden, S.; Lipinski, M.R.; Matthee, C.A. Remarkably low mtDNA control region diversity in an abundant demersal fish. Mol. Phylogenetics Evol. 2010, 55, 1183–1188. [Google Scholar] [CrossRef]
- Wang, C.; Chen, H.; Tian, S.; Yang, C.; Chen, X. Novel gene rearrangement and the complete mitochondrial genome of Cynoglossus monopus: Insights into the envolution of the family Cynoglossidae (Pleuronectiformes). Int. J. Mol. Sci. 2020, 21, 6895. [Google Scholar] [CrossRef]
- Chen, X.; Wang, J.-J.; Ai, W.-M.; Chen, H.; Lin, H.-D. Phylogeography and genetic population structure of the spadenose shark (Scoliodon macrorhynchos) from the Chinese coast. Mitochondrial Dna Part A 2018, 29, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- Boyko, N.; Kmetyk-Podubinska, K.; Andrusiak, I. Application of Ensemble Methods of Strengthening in Search of Legal Information. In Proceedings of the 2021 International Scientific Conference “Intellectual Systems of Decision Making and Problem of Computational Intelligence”, Zalizniy Port, Ukraine, 24–28 May 2021; Springer: Cham, Switzerland, 2021; pp. 188–200. [Google Scholar]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Angermueller, C.; Pärnamaa, T.; Parts, L.; Stegle, O. Deep learning for computational biology. Mol. Syst. Biol. 2016, 12, 878. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- MacQueen, J. Classification and analysis of multivariate observations. In Proceedings of 5th Berkeley Symposium on Mathematical Statistics and Probability; University of California Press: Berkeley, CA, USA, 1967; pp. 281–297. [Google Scholar]
- Aggarwal, C.; Reddy, C. Data Clustering Algorithms and Applications; Taylor & Francis Group LLC.: Abingdon, UK, 2013. [Google Scholar]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Frey, B.J.; Dueck, D. Clustering by passing messages between data points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef]
- Bholowalia, P.; Kumar, A. EBK-means: A clustering technique based on elbow method and k-means in WSN. Int. J. Comput. Appl. 2014, 105. [Google Scholar]
- Hruschka, E.R.; de Castro, L.N.; Campello, R.J. Evolutionary Algorithms for Clustering Gene-Expression Data. In Proceedings of the 4th IEEE International Conference on Data Mining (ICDM’04), Brighton, UK, 1–4 November 2004; IEEE: Piscataway, NJ, USA, 2004; pp. 403–406. [Google Scholar]
- Tibshirani, R.; Walther, G.; Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2001, 63, 411–423. [Google Scholar] [CrossRef]
- Wang, C.; Lai, T.; Ye, P.; Yan, Y.; Feutry, P.; He, B.; Huang, Z.; Zhu, T.; Wang, J.; Chen, X. Novel duplication remnant in the first complete mitogenome of Hemitriakis japanica and the unique phylogenetic position of family Triakidae. Gene 2022, 820, 146232. [Google Scholar] [CrossRef]
- Huang, Y.; Bian, C.; Liu, Z.; Wang, L.; Xue, C.; Huang, H.; Yi, Y.; You, X.; Song, W.; Mao, X. The first genome survey of the Antarctic Krill (Euphausia superba) provides a valuable genetic resource for polar biomedical research. Mar. Drugs 2020, 18, 185. [Google Scholar] [CrossRef] [PubMed]
- Krück, N.C.; Tibbetts, I.R.; Ward, R.D.; Johnson, J.W.; Loh, W.K.; Ovenden, J.R. Multi-gene barcoding to discriminate sibling species within a morphologically difficult fish genus (Sillago). Fish. Res. 2013, 143, 39–46. [Google Scholar] [CrossRef]
- Raje, S.; Sivakami, S.; Mohanraj, G.; Manojkumar, P.; Raju, A.; Joshi, K. Atlas on the Elasmobranch fishery resources of India. CMFRI Spec. Publ. 2007, 95, 122–125. [Google Scholar]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. In Bioinformatics Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2000; pp. 71–91. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Wang, W. The Molecular Detection of Corynespora Cassiicola on Cucumber by PCR Assay Using DNAman Software and NCBI. In Proceedings of the International Conference on Computer and Computing Technologies in Agriculture, Beijing, China, 27–30 September 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 248–258. [Google Scholar]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36 (Suppl. S2), W181–W184. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 12 May 2023).
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis; Taylor & Francis: Abingdon, UK, 2019. [Google Scholar]
- Wickham, H.; Henry, L. Tidyr: Tidy messy data. R Package Version 2020, 1, 397. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Slowikowski, K. ggrepel: Automatically position non-overlapping text labels with “ggplot2.”. R package version 0.8.0. 2018. Available online: https://CRAN.R-project.org/package=ggrepel (accessed on 12 May 2023).
- Clarke, E.; Sherrill-Mix, S. ggbeeswarm: Categorical scatter (violin point) plots. R package version 0.6.0. Retrieved from 2017. Available online: https://CRAN.R-project.org/package=ggbeeswarm (accessed on 12 May 2023).
- Arnold, J.B.; Daroczi, G.; Werth, B.; Weitzner, B.; Kunst, J.; Auguie, B. ggthemes: Extra Themes, Scales and Geoms for ’ggplot2’; R package version 4.2.0. 2019. Available online: https://CRAN.R-project.org/package=ggthemes (accessed on 12 May 2023).
- Gaujoux, R.; Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 2010, 11, 367. [Google Scholar] [CrossRef]
- Bodenhofer, U.; Kothmeier, A.; Hochreiter, S. APCluster: An R package for affinity propagation clustering. Bioinformatics 2011, 27, 2463–2464. [Google Scholar] [CrossRef]
- Hahsler, M.; Piekenbrock, M.; Doran, D. dbscan: Fast density-based clustering with R. J. Stat. Softw. 2019, 91, 1–30. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A.; Mundt, F. Factoextra: Extract and visualize the results of multivariate data analyses. R Package Version 2017, 1, 337–354. [Google Scholar]
- Tiedemann, F. Ggpol: Visualizing Social Science Data with ’ggplot2’; R package version 0.0.7. 2020. Available online: https://CRAN.R-project.org/package=ggpol (accessed on 12 May 2023).
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Cluster: Cluster analysis basics and extensions. R Package Version 2012, 1, 56. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Ranwez, V.; Douzery, E.J.; Cambon, C.; Chantret, N.; Delsuc, F. MACSE v2: Toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 2018, 35, 2582–2584. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nylander, J.A.; Ronquist, F.; Huelsenbeck, J.P.; Nieves-Aldrey, J. Bayesian phylogenetic analysis of combined data. Syst. Biol. 2004, 53, 47–67. [Google Scholar] [CrossRef] [PubMed]
- Sitnikova, T. Bootstrap method of interior-branch test for phylogenetic trees. Mol. Biol. Evol. 1996, 13, 605–611. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Douglas, J.; Zhang, R.; Bouckaert, R. Adaptive dating and fast proposals: Revisiting the phylogenetic relaxed clock model. PLoS Comput. Biol. 2021, 17, e1008322. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Suykens, J.A. Support vector machines: A nonlinear modelling and control perspective. Eur. J. Control. 2001, 7, 311–327. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the Difficulty of Training Deep Feedforward Neural Networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13–15 May 2010; Proceedings of Machine Learning Research. PMLR: Birmingham, UK, 2010; pp. 249–256. [Google Scholar]
- Poole, D.I.; Goebel, R.G.; Mackworth, A.K. Computational Intelligence; Oxford University Press: New York, NY, USA, 1998; Volume 1. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Lee, Y.W.; Choi, J.W.; Shin, E.-H. Machine learning model for predicting malaria using clinical information. Comput. Biol. Med. 2021, 129, 104151. [Google Scholar] [CrossRef]
- Balasubramaniam, S.; Kumar, K.S. Optimal Ensemble Learning Model for COVID-19 detection using chest X-ray images. Biomed. Signal Process. Control. 2022, 81, 104392. [Google Scholar] [CrossRef]
- Ahmed, A.M.; Aly, S.F. Egyptian License Plates Recognition System Using Morphologial Operations and Multi Layered Perceptron. In Proceedings of the International Conference on ICT in Our Lives, Alexandria, Egypt; ResearchGate: Berlin, Germany, 2019; pp. 21–23. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]

| Sequence Name | GenBank ID | Region |
|---|---|---|
| Complete mitochondrial genome of S. laticaudus | NC_042504 | Complete mitochondrial genome |
| Complete mitochondrial genome of S. macrorhynchos | NC_018052 | Complete mitochondrial genome |
| 95 control region sequences of S. laticaudus | MW974731-MW974825 | Control regions |
| 115 COI sequences of S. laticaudus | MW974616-MW974730 | COI genes |
| 18 control region sequences of S. macrorhynchos | KX657688-KX657705 | Control regions |
| ND2 sequence of S. laticaudus (India 1) | JQ518654 | ND2 gene |
| ND2 sequence of S. laticaudus (Thailand 2) | JQ518655 | ND2 gene |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, P.; Miao, Y.; Wang, C.; Sonchaeng, P.; Siriwong, S.; Chen, S.; Wang, J.; Chen, X. Potential Cryptic Diversity in the Genus Scoliodon (Carcharhiniformes: Carcharhinidae): Insights from Mitochondrial Genome Sequencing. Int. J. Mol. Sci. 2024, 25, 11851. https://doi.org/10.3390/ijms252111851
Ye P, Miao Y, Wang C, Sonchaeng P, Siriwong S, Chen S, Wang J, Chen X. Potential Cryptic Diversity in the Genus Scoliodon (Carcharhiniformes: Carcharhinidae): Insights from Mitochondrial Genome Sequencing. International Journal of Molecular Sciences. 2024; 25(21):11851. https://doi.org/10.3390/ijms252111851
Chicago/Turabian StyleYe, Peiyuan, Yuanxiang Miao, Chen Wang, Pichai Sonchaeng, Sarawut Siriwong, Shaobo Chen, Junjie Wang, and Xiao Chen. 2024. "Potential Cryptic Diversity in the Genus Scoliodon (Carcharhiniformes: Carcharhinidae): Insights from Mitochondrial Genome Sequencing" International Journal of Molecular Sciences 25, no. 21: 11851. https://doi.org/10.3390/ijms252111851
APA StyleYe, P., Miao, Y., Wang, C., Sonchaeng, P., Siriwong, S., Chen, S., Wang, J., & Chen, X. (2024). Potential Cryptic Diversity in the Genus Scoliodon (Carcharhiniformes: Carcharhinidae): Insights from Mitochondrial Genome Sequencing. International Journal of Molecular Sciences, 25(21), 11851. https://doi.org/10.3390/ijms252111851






