Mitochondrial Genome Assembly and Structural Characteristics Analysis of Gentiana rigescens
Abstract
1. Introduction
2. Results
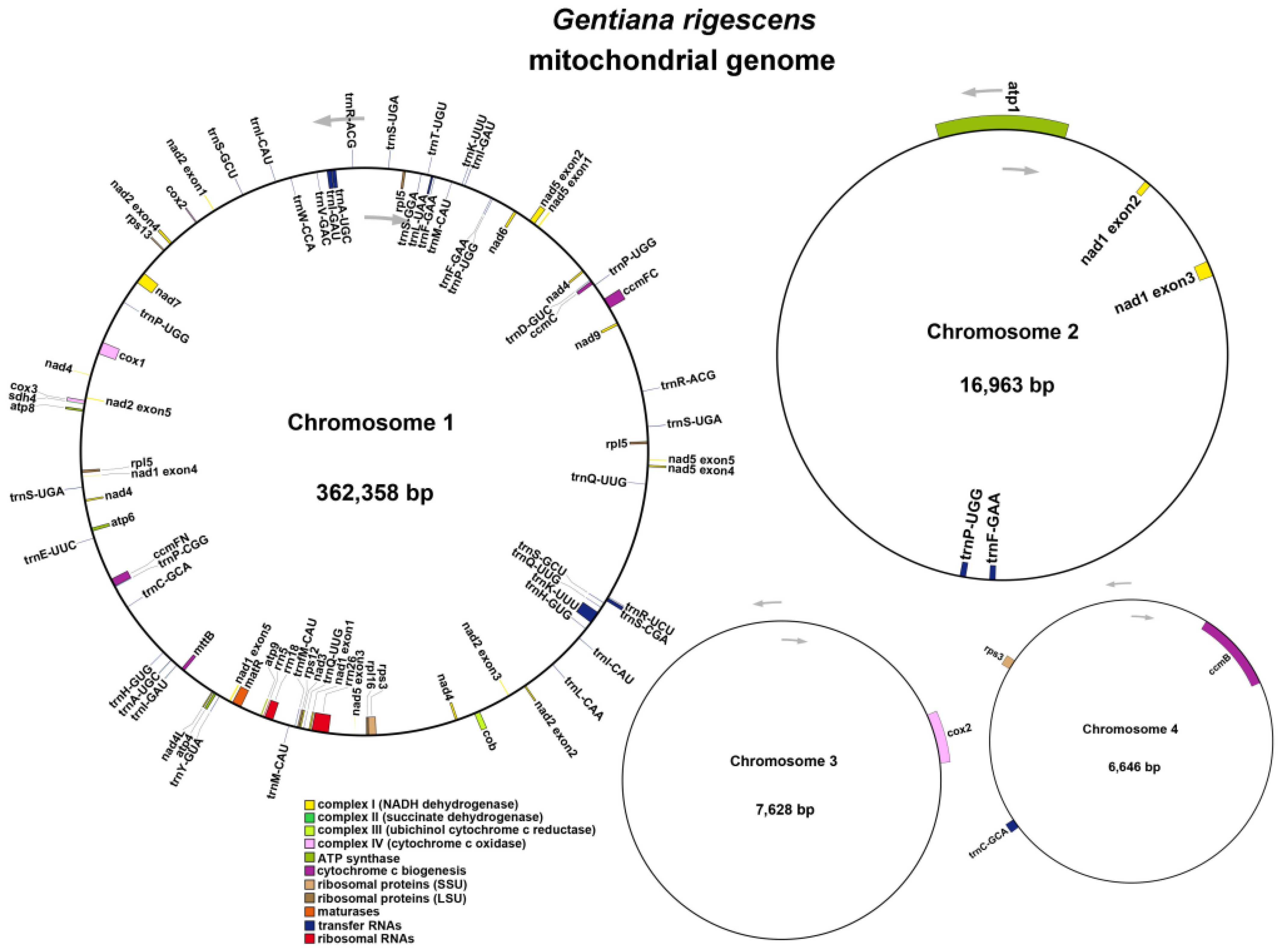
2.1. Mitochondrial Structure and Gene Content of G. rigescens
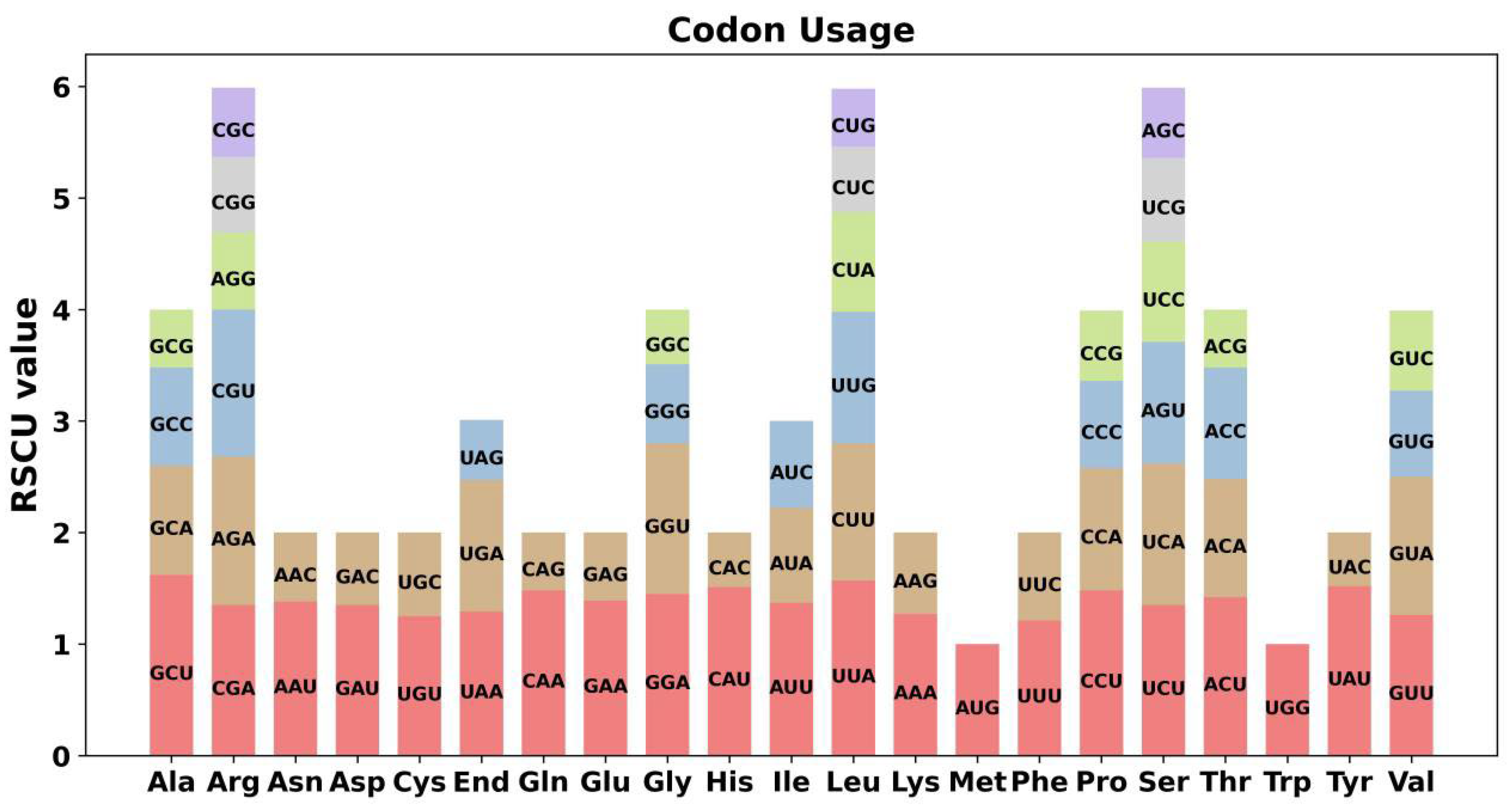
2.2. Codon Usage Analysis of PCGs
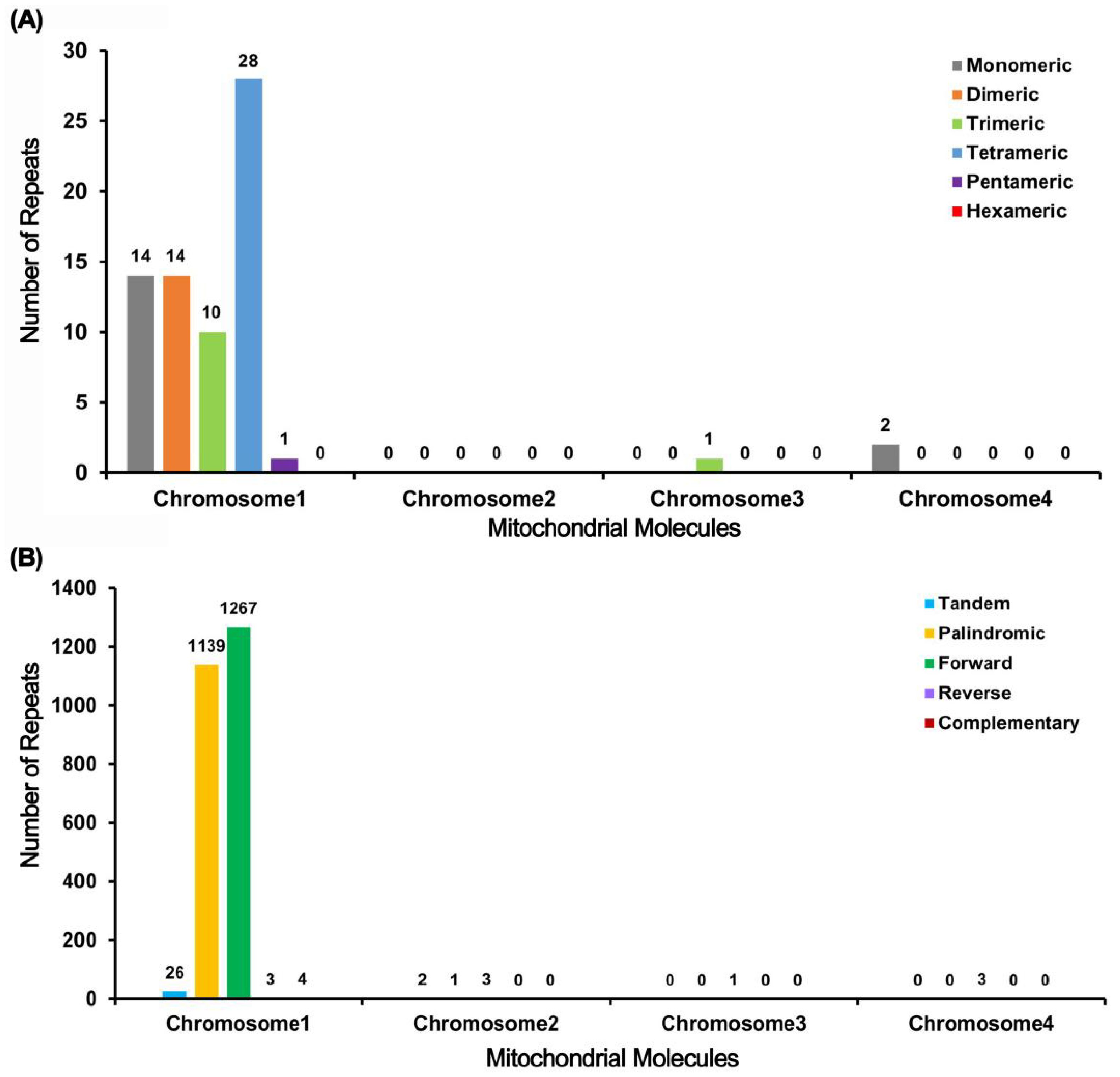
2.3. Repeat Elements
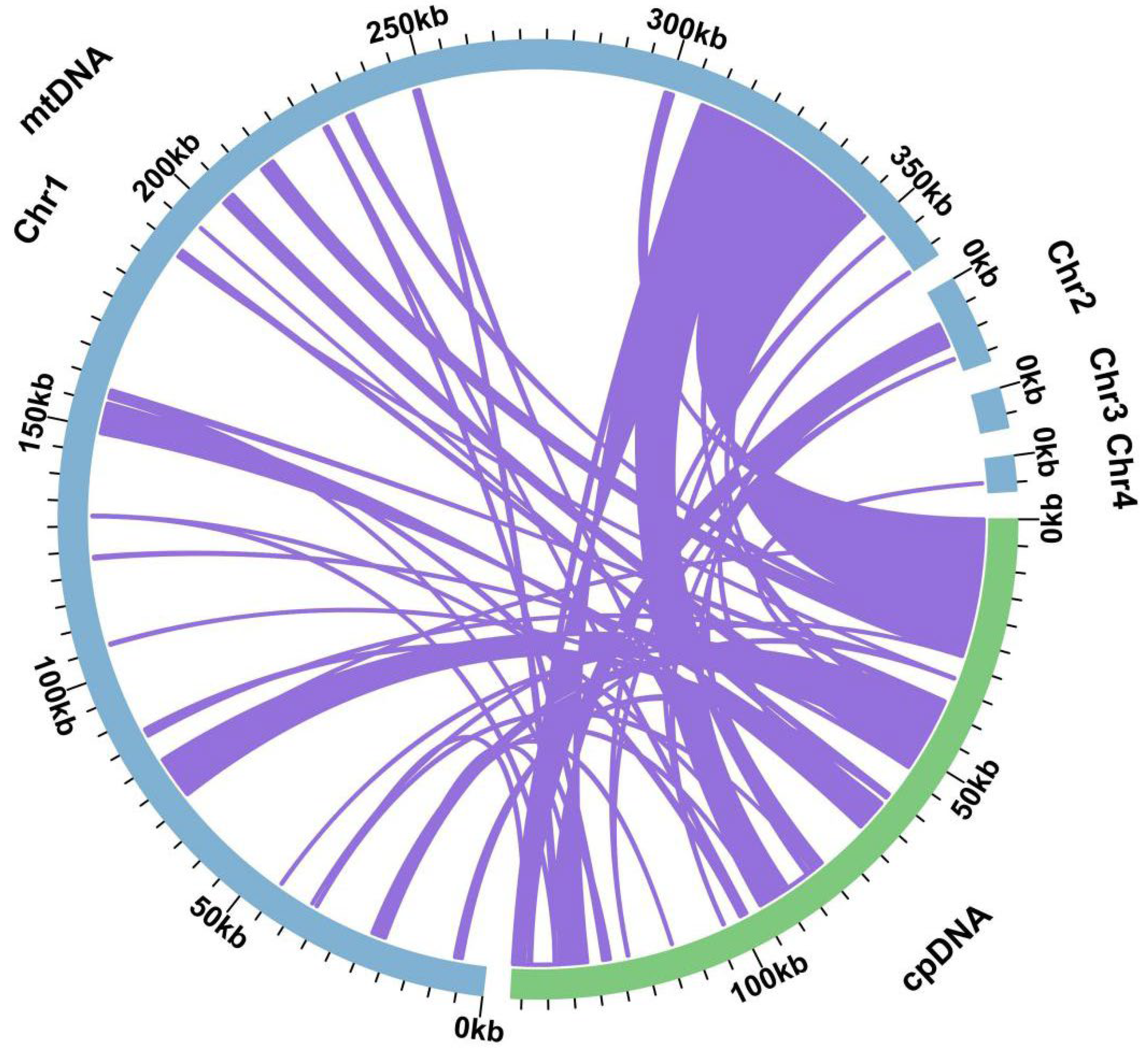
2.4. Plastid DNA Insertion in Mitogenome
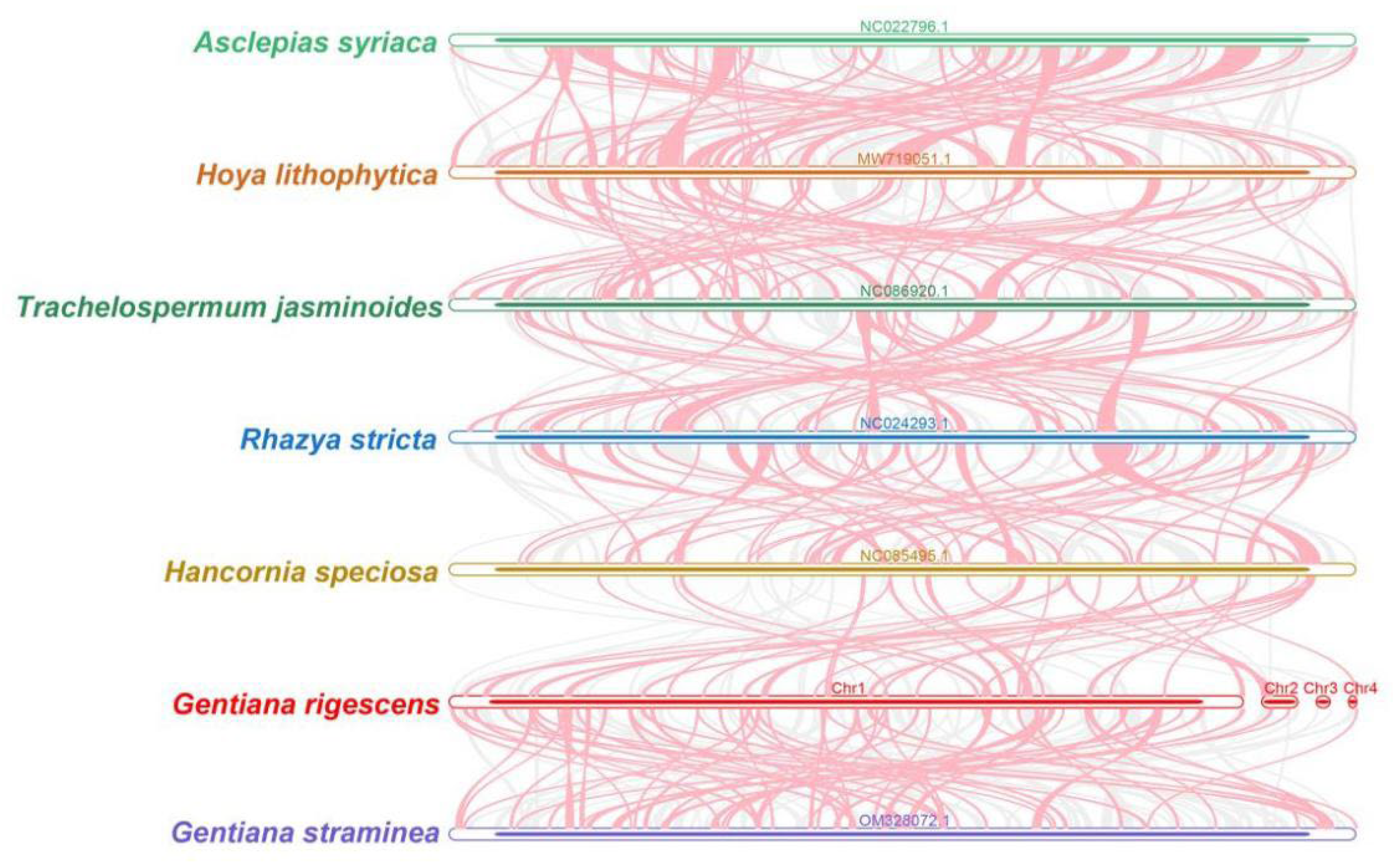
2.5. Synteny and Phylogenetic Analysis
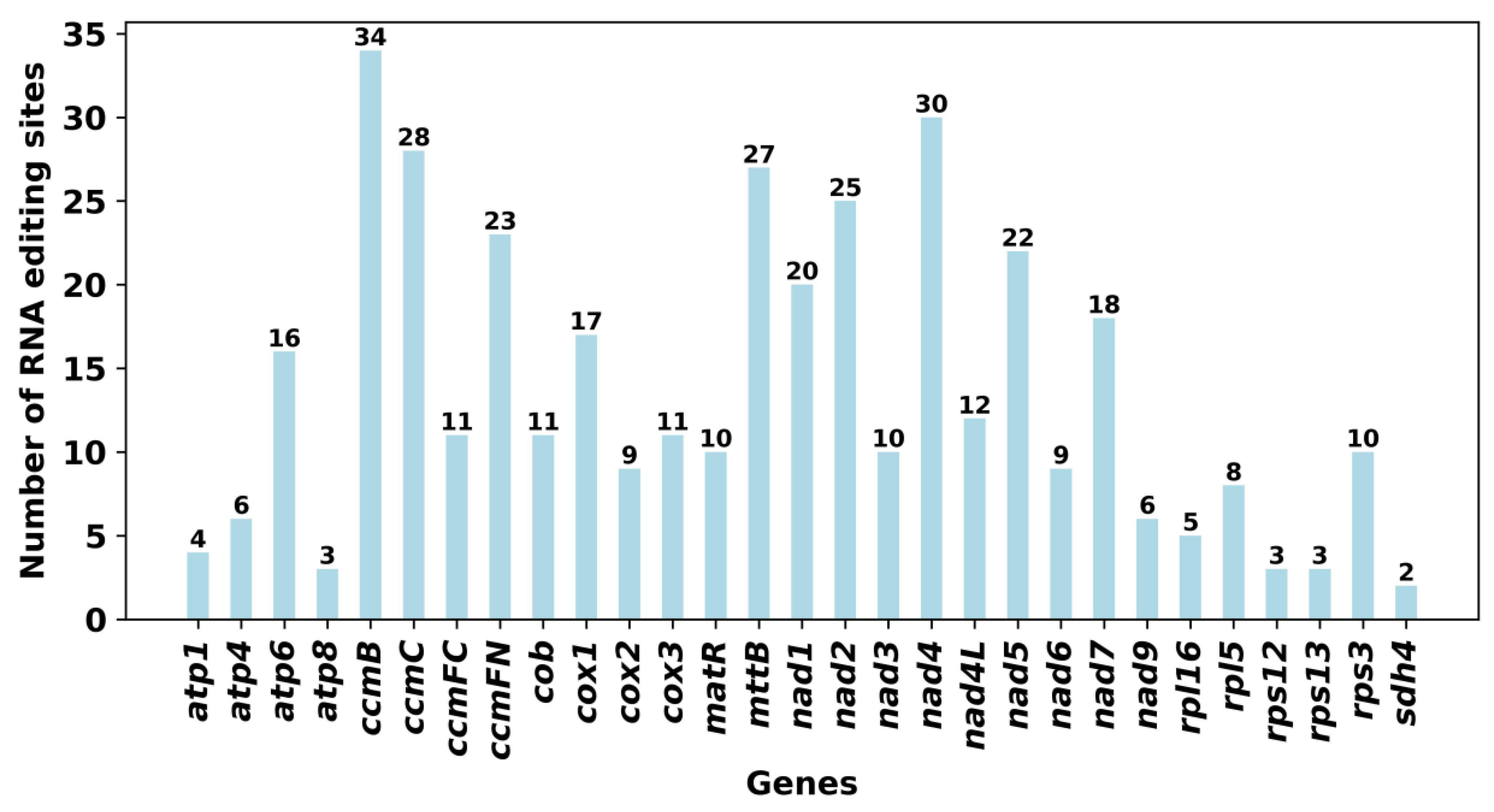
2.6. RNA Editing Events
3. Discussion
4. Materials and Methods
4.1. Plant Sampling, DNA Extraction, and Sequencing
4.2. Mitochondrial Genome Assembly and Annotation
4.3. Analysis of Codon Usage
4.4. Analysis of Repeat Elements
4.5. Analysis of Chloroplast DNA Insertion in Mitogenome
4.6. Synteny and Phylogenetic Analysis
4.7. Analysis of RNA Editing Events
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China: 2020 Edition; China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- Picault, N.; Hodges, M.; Palmieri, L.; Palmieri, F. The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci. 2004, 9, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Dyall, S.D.; Brown, M.T.; Johnson, P.J. Ancient invasions: From endosymbionts to organelles. Science 2004, 304, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Takemura, M.; Oda, K.; Yamato, K.; Ohta, E.; Nakamura, Y.; Nozato, N.; Akashi, K.; Ohyama, K. Gene clusters for ribosomal proteins in the mitochondrial genome of a liverwort, Marchantia polymorpha. Nucleic Acids Res. 1992, 20, 3199–3205. [Google Scholar] [CrossRef][Green Version]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef]
- Wynn, E.L.; Christensen, A.C. Repeats of Unusual Size in Plant Mitochondrial Genomes: Identification, Incidence and Evolution. G3 Genes Genomes Genet. 2018, 9, 549–559. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef]
- Varré, J.S.; D’Agostino, N.; Touzet, P.; Gallina, S.; Tamburino, R.; Cantarella, C.; Ubrig, E.; Cardi, T.; Drouard, L.; Gualberto, J.M.; et al. Complete Sequence, Multichromosomal Architecture and Transcriptome Analysis of the Solanum tuberosum Mitochondrial Genome. Int. J. Mol. Sci. 2019, 20, 4788. [Google Scholar] [CrossRef]
- Mower, J.P. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef]
- Lukeš, J.; Kaur, B.; Speijer, D.; Dave, S. RNA Editing in Mitochondria and Plastids: Weird and Widespread. Trends Genet. 2021, 37, 99–102. [Google Scholar] [CrossRef]
- Qiu, Y.-L.; Li, L.; Wang, B.; Xue, J.-Y.; Hendry, T.A.; Li, R.-Q.; Brown, J.W.; Liu, Y.; Hudson, G.T.; Chen, Z.-D. Angiosperm phylogeny inferred from sequences of four mitochondrial genes. J. Syst. Evol. 2010, 8, 391–425. [Google Scholar] [CrossRef]
- Ala, K.; Zhao, Z.; Ni, L.; Wang, Z. Comparative analysis of mitochondrial genomes of two alpine medicinal plants of Gentiana (Gentianaceae). PLoS ONE 2023, 18, e0281134. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.N.; Zhou, Z.L.; Ni, L.H.; Gaawe, D.; Mi, M. The identification of Sect. Cruciata (Gentiana) species using mtDNA nad1/b-c and nad5/d-e fragments. Acta Pharm. Sin. 2019, 54, 166–172. [Google Scholar]
- Skippington, E.; Barkman, T.J.; Rice, D.W.; Palmer, J.D. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. USA 2015, 112, E3515–E3524. [Google Scholar] [CrossRef] [PubMed]
- Putintseva, Y.A.; Bondar, E.I.; Simonov, E.P.; Sharov, V.V.; Oreshkova, N.V.; Kuzmin, D.A.; Konstantinov, Y.M.; Shmakov, V.N.; Belkov, V.I.; Sadovsky, M.G.; et al. Siberian larch (Larix sibirica Ledeb.) mitochondrial genome assembled using both short and long nucleotide sequence reads is currently the largest known mitogenome. BMC Genom. 2020, 21, 654. [Google Scholar] [CrossRef] [PubMed]
- Straub, S.C.; Cronn, R.C.; Edwards, C.; Fishbein, M.; Liston, A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (apocynaceae). Genome Biol. Evol. 2013, 5, 1872–1885. [Google Scholar] [CrossRef]
- Park, S.; Ruhlman, T.A.; Sabir, J.S.; Mutwakil, M.H.; Baeshen, M.N.; Sabir, M.J.; Baeshen, N.A.; Jansen, R.K. Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genom. 2014, 15, 405. [Google Scholar] [CrossRef]
- Yang, J.X.; Nicolas, D.; Bai, M.Z.; Guo, Y.Y. Multichromosomal Mitochondrial Genome of Paphiopedilum micranthum: Compact and Fragmented Genome, and Rampant Intracellular Gene Transfer. Int. J. Mol. Sci. 2023, 24, 3976. [Google Scholar] [CrossRef]
- Liu, D.; Guo, H.L.; Zhu, J.L.; Qu, K.; Chen, Y.; Guo, Y.T.; Ding, P.; Yang, H.P.; Xu, T.; Jing, Q.; et al. Complex Physical Structure of Complete Mitochondrial Genome of Quercus acutissima (Fagaceae): A Significant Energy Plant. Genes 2022, 13, 1321. [Google Scholar] [CrossRef]
- Zhang, X.F.; Jacob, B.L.; Wang, H.X.; Zhi-Xin Zhu, Z.X.; Wang, H.F. Comparative analysis of chloroplast genome structure and molecular dating in Myrtales. BMC Plant Biol. 2021, 21, 219. [Google Scholar] [CrossRef]
- Mahajan, S.; Agashe, D. Evolutionary jumps in bacterial GC content. G3 Genes Genomes Genet. 2022, 12, jkac108. [Google Scholar] [CrossRef]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2021, 49, 539–565. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Q.; Liu, J.F.; Jiang, S.C.; Guo, B.Q.; Guo, J.; Cao, L.L.; Zhang, W. Mitochondrial genome characteristics and phylogenetic analysis of 21 species of Poaceae. Genom. Appl. Biol. 2024, 1–16. Available online: http://kns.cnki.net/kcms/detail/45.1369.q.20240705.0927.002.html (accessed on 1 August 2024).
- Yu, R.; Sun, C.; Zhong, Y.; Liu, Y.; Sanchez-Puerta, M.V.; Mower, J.P.; Zhou, R. The minicircular and extremely heteroplasmic mitogenome of the holoparasitic plant Rhopalocnemis phalloides. Curr. Biol. 2021, 32, 470–479.e5. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.S.; Zhao, C.X.; Chen, F.; Liu, Y.H.; Zhang, S.Z.; Wu, H.; Zhang, L.S.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef] [PubMed]
- Li, J.L.; Xu, Y.C.; Shan, Y.Y.; Pei, X.Y.; Yong, S.Y.; Liu, C.; Yu, J. Assembly of the complete mitochondrial genome of an endemic plant, Scutellaria tsinyunensis, revealed the existence of two conformations generated by a repeat-mediated recombination. Planta 2021, 254, 36. [Google Scholar] [CrossRef]
- Zhang, X.; Shan, Y.Y.; Li, J.L.; Qin, Q.L.; Yu, J.; Deng, H.P. Assembly of the Complete Mitochondrial Genome of Pereskia aculeata Revealed That Two Pairs of Repetitive Elements Mediated the Recombination of the Genome. Int. J. Mol. Sci. 2023, 24, 8366. [Google Scholar] [CrossRef]
- Chun, Y.H.; Nicole, G.; Nahal, A.; Jeremy, N.T.; William, M. Mutational decay and age of chloroplast and mitochondrial genomes transferred recently to angiosperm nuclear chromosomes. Plant Physiol. 2005, 138, 1723–1733. [Google Scholar]
- Turmel, M.; Otis, C.; Lemieux, C. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: Insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc. Natl. Acad. Sci. USA 2002, 99, 11275–11280. [Google Scholar] [CrossRef]
- Li, Y.Y.; Liu, Y.Y.; Zeng, X.; Wu, P.; Li, Q.M.; Guo, S.X.; Hao, Z.G. Complete mitochondrial genome of Angelica dahurica and its implications on evolutionary analysis of complex mitochondrial genome architecture in Apiaceae. Front. Plant Sci. 2024, 15, 1367299. [Google Scholar] [CrossRef]
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2019, 101, 1040–1056. [Google Scholar] [CrossRef]
- Bi, C.W.; Lu, N.; Xu, Y.Q.; He, C.P.; Lu, Z.H. Characterization and Analysis of the Mitochondrial Genome of Common Bean (Phaseolus vulgaris) by Comparative Genomic Approaches. Int. J. Mol. Sci. 2020, 21, 3778. [Google Scholar] [CrossRef]
- Yang, H.X.; Li, W.H.; Yu, X.L.; Zhang, X.Y.; Zhang, Z.Y.; Liu, Y.X.; Wang, W.X.; Tian, X.X. Insights into molecular structure, genome evolution and phylogenetic implication through mitochondrial genome sequence of Gleditsia sinensis. Sci. Rep. 2021, 11, 14850. [Google Scholar] [CrossRef] [PubMed]
- Grewe, F.; Edger, P.P.; Keren, I.; Sultan, L.; Pires, J.C.; Ostersetzer-Biran, O.; Mower, J.P. Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea. Mitochondrion 2014, 19 Pt B, 135–143. [Google Scholar] [CrossRef]
- Aboul-Maaty, N.A.-F.; Oraby, H.A.-S. Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull. Natl. Res. Cent. 2019, 43, 25. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Heng, L.; Richard, D. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.C.; Zhang, Y.D.; Xu, Y.S. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Lewis, S.E.; Searle, S.M.; Harris, N.; Gibson, M.; Lyer, V.; Richter, J.; Wiel, C.; Bayraktaroglu, L.; Birney, E.; Crosby, M.A.; et al. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, RESEARCH0082. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Zhang, H.G.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Shi, L.C.; Chen, H.M.; Jiang, M.; Wang, L.Q.; Wu, X.; Huang, L.F.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Liu, S.Y.; Ni, Y.; Li, J.L.; Zhang, X.Y.; Yang, H.Y.; Chen, H.M.; Liu, C. CPGView: A package for visualizing detailed chloroplast genome structures. Mol. Ecol. Resour. 2023, 23, 694–704. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Edera, A.A.; Small, I.; Milone, D.H.; Sanchez-Puerta, M.V. Deepred-Mt: Deep representation learning for predicting C-to-U RNA editing in plant mitochondria. Comput. Biol. Med. 2021, 136, 104682. [Google Scholar] [CrossRef]
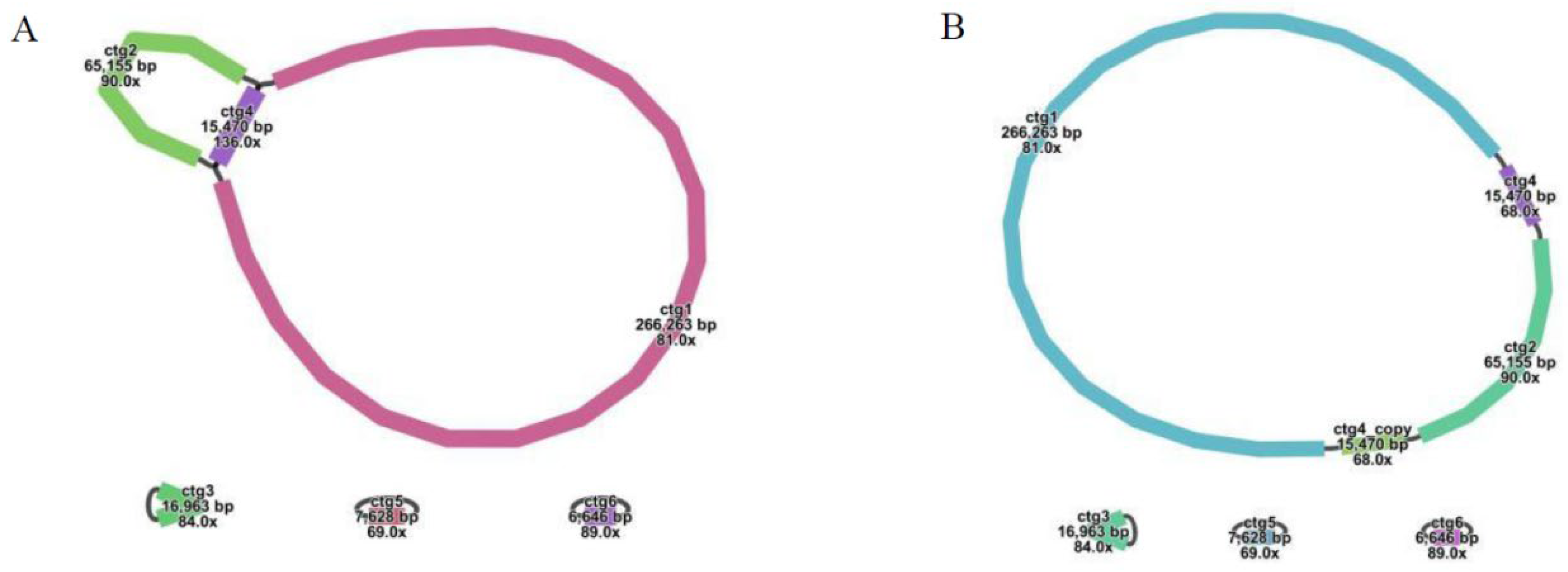

| Node | Length (bp) | Depth (×) |
|---|---|---|
| ctg1 | 266,263 | 81 |
| ctg2 | 65,155 | 90 |
| ctg3 | 16,963 | 84 |
| ctg4 | 15,470 | 136 |
| ctg5 | 7628 | 69 |
| ctg6 | 6646 | 89 |
| Group of Genes | Name of Genes |
|---|---|
| ATP synthase | atp1, atp4, atp6, atp8, atp9 |
| NADH dehydrogenase | nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9 |
| Cytochrome b | cob |
| Cytochrome c biogenesis | ccmB, ccmC, ccmFC, ccmFN |
| Cytochrome c oxidase | cox1, cox2, cox3 |
| Maturases | matR |
| Protein transport subunit | mttB |
| Ribosomal protein large subunit | rpl5(×3), rpl16 |
| Ribosomal protein small subunit | rps3, rps12, rps13 |
| Succinate dehydrogenase | sdh4 |
| Ribosome RNA | rrn5, rrn18, rrn26 |
| Transfer RNA | trnA-UGC(×2), trnC-GCA(×2), trnD-GUC, trnE-UUC, trnF-GAA(×3), trnfM-CAU, trnH-GUG(×2), trnI-CAU(×2), trnI-GAU(×3), trnKUUU(×2), trnL-CAA, trnL-UAA, trnM-CAU(×2), trnP-CGG, trnP-UGG(×4), trnQ-UUG(×3), trnR-ACG(×2), trnR-UCU, trnS-CGA, trnS-GCU(×2), trnS-GGA, trnS-UGA(×3), trnT-UGU, trnV-GAC, trnW-CCA, trnY-GUA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Z.; Zhang, Y.; Wu, L.; Li, G. Mitochondrial Genome Assembly and Structural Characteristics Analysis of Gentiana rigescens. Int. J. Mol. Sci. 2024, 25, 11428. https://doi.org/10.3390/ijms252111428
Xie Z, Zhang Y, Wu L, Li G. Mitochondrial Genome Assembly and Structural Characteristics Analysis of Gentiana rigescens. International Journal of Molecular Sciences. 2024; 25(21):11428. https://doi.org/10.3390/ijms252111428
Chicago/Turabian StyleXie, Zongyi, Yingmin Zhang, Lixin Wu, and Guodong Li. 2024. "Mitochondrial Genome Assembly and Structural Characteristics Analysis of Gentiana rigescens" International Journal of Molecular Sciences 25, no. 21: 11428. https://doi.org/10.3390/ijms252111428
APA StyleXie, Z., Zhang, Y., Wu, L., & Li, G. (2024). Mitochondrial Genome Assembly and Structural Characteristics Analysis of Gentiana rigescens. International Journal of Molecular Sciences, 25(21), 11428. https://doi.org/10.3390/ijms252111428






