Genetic Diversity of the Species of the Genus Deschampsia P.Beauv. (Poaceae) Based on the Analysis of the ITS Region: Polymorphism Proves Distant Hybridization
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. DNA Sampling for Analysis
4.2. DNA Isolation and Sequencing by the Sanger Method
4.3. Molecular Phylogenetic Analysis of the Sequences Obtained by the Sanger Method
4.4. Next-Generation Sequencing
4.5. Molecular Phylogenetic Analysis of NGS Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tzvelev, N.N.; Probatova, N.S. Grasses of Russia; KMK Scientific Press: Moscow, Russia, 2019; 646p. (In Russian) [Google Scholar]
- Rozhevitz, R.Y. Deschampsia, P.B. In Flora of the USSR; Komarov, V.L., Ed.; Izdatel’stvo Akademii Nauk SSSR: Leningrad, Russia, 1934; Volume II, pp. 243–252. (In Russian) [Google Scholar]
- Tzvelev, N.N. Grasses of the USSR; Nauka: Moscow, Russia, 1976; 788p. (In Russian) [Google Scholar]
- Soreng, R.J.; Peterson, P.M.; Romaschenko, K.; Davidse, G.; Zuloaga, F.O.; Judziewic, E.J.; Filgueiras, T.S.; Morrone, O. A worldwide phylogenetic classification of the Poaceae (Gramineae). J. Syst. Evol. 2015, 53, 117–137. [Google Scholar] [CrossRef]
- Soreng, R.J.; Peterson, P.M.; Zuloaga, F.O.; Romaschenko, K.; Clark, L.G.; Teisher, J.K.; Gillespie, L.J.; Barberá, P.; Welker, C.A.D.; Kellogg, E.A.; et al. A worldwide phylogenetic classification of the Poaceae (Gramineae) III: An update. J. Syst. Evol. 2022, 60, 476–521. [Google Scholar] [CrossRef]
- Tkach, N.; Schneider, J.; Döring, E.; Wölk, A.; Hochbach, A.; Nissen, J.; Winterfeld, G.; Meyer, S.; Gabriel, J.; Hoffmann, M.H.; et al. Phylogenetic lineages and the role of hybridization as driving force of evolution in grass supertribe Poodae. Taxon 2020, 69, 234–277. [Google Scholar] [CrossRef]
- Quintanar, A.; Castroviejo, S.; Catalán, P. Phylogeny of tribe Aveneae (Pooideae, Poaceae) inferred from plastid trnT-F and nuclear ITS sequences. Am. J. Bot. 2007, 94, 1554–1569. [Google Scholar] [CrossRef] [PubMed]
- Tzvelev, N.N. Deschampsia Beauv. In Arctic flora of the USSR; Tolmachev, A.I., Ed.; Nauka: Moscow, Russia, 1964; Volume II, pp. 77–92. (In Russian) [Google Scholar]
- Rothera, S.L.; Davy, A.J. Polyploidy and habitat differentiation in Deschampsia cespitosa. New Phytol. 1986, 102, 449–467. [Google Scholar] [CrossRef]
- Ghukasyan, A. Extent of karyological study of Armenian grasses (Poaceae). Flora Veg. Plant Resour. Armen. 2004, 15, 85–89. (In Russian) [Google Scholar]
- Gnutikov, A.A.; Myakoshina, J.A.; Nosov, N.N.; Punina, E.O.; Rodionov, A.V. IAPT chromosome data 32/5 (K. Marhold & J. Kučera (eds.), & al.). Taxon 2020, 69, 1128–1129. [Google Scholar] [CrossRef]
- Chiapella, J.; Probatova, N.S. The Deschampsia cespitosa complex (Poaceae: Aveneae) with special reference to Russia. Bot. J. Lin. Soc. 2003, 142, 213–228. [Google Scholar] [CrossRef]
- Tzvelev, N.N. Problems of Theoretical Morphology and the Evolution of Higher Plants; KMK: St. Petersburg, Russia; Moscow, Russia, 2005; 407p. (In Russian) [Google Scholar]
- Xue, Z.; Chiapella, J.O.; Paun, O.; Volkova, P.; Peintinger, M.; Wasowicz, P.; Tikhomirov, N.; Grigoryan, M.; Barfuss, M.H.J.; Greimler, J. Phylogeographic patterns of Deschampsia cespitosa (Poaceae) in Europe inferred from genomic data. Bot. J. Lin. Soc. 2023, 201, 341–360. [Google Scholar] [CrossRef]
- Fernández Souto, D.P.; Catalano, S.A.; Tosto, D.; Bernasconi, P.; Sala, A.; Wagner, M.; Corach, D. Phylogenetic relationships of Deschampsia antarctica (Poaceae): Insights from nuclear ribosomal ITS. Pl. Syst. Evol. 2006, 261, 1–9. [Google Scholar] [CrossRef]
- González, M.L.; Urdampilleta, J.D.; Fasanella, M.; Premoli, A.C.; Chiapella, J.O. Distribution of rDNA and polyploidy in Deschampsia antarctica E. Desv. in Antarctic and Patagonic populations. Polar Biol. 2016, 39, 1663–1677. [Google Scholar] [CrossRef]
- González, M.L.; Chiapella, J.O.; Urdampilleta, J.D. The Antarctic and South American species of Deschampsia: Phylogenetic relationships and cytogenetic differentiation. Syst. Biodivers. 2021, 19, 453–470. [Google Scholar] [CrossRef]
- González, M.L.; Chiapella, J.O.; Urdampilleta, J.D. Chromosomal Differentiation of Deschampsia (Poaceae) Based on Four Satellite DNA Families. Front. Genet. 2021, 12, 728664. [Google Scholar] [CrossRef]
- Brassac, J.; Blattner, F.R. Species-Level Phylogeny and Polyploid Relationships in Hordeum (Poaceae) Inferred by Next-Generation Sequencing and In Silico Cloning of Multiple Nuclear Loci. Syst. Biol. 2015, 64, 792–808. [Google Scholar] [CrossRef]
- Kawano, S. Cytogeography and evolution of the Deschampsia caespitosa complex. Can. J. Bot. 1963, 41, 719–742. [Google Scholar] [CrossRef]
- Albers, F. Vergleichende Karyologie der Gräser-Subtriben Aristaveninae und Airinae (Poaceae–Aveneae). Pl. Syst. Evol. 1980, 136, 137–167. [Google Scholar] [CrossRef]
- Garcia-Suarez, R.; Alonso-Blanco, C.; Fernandez-Carvajal, M.C.; Fernandez-Prieto, J.A.; Roca, A.; Giraldez, R. Diversity and systematics of Deschampsia sensu lato (Poaceae), inferred from karyotypes, protein electrophoresis, total genomic DNA hybridization and chloroplast DNA analysis. Pl. Syst. Evol. 1997, 205, 99–110. [Google Scholar] [CrossRef]
- Petrovsky, V.V.; Zhukova, P.G. Chromosome numbers and taxonomy of some plant species of Wrangel Island. Bot. Zhurn. 1981, 66, 380–387. (In Russian) [Google Scholar]
- Volkov, R.A.; Komarova, N.Y.; Hemleben, V. Ribosomal DNA in plant hybrids: Inheritance, rearrangement, expression. Syst. Biodiv. 2007, 5, 261–276. [Google Scholar] [CrossRef]
- Runemark, A.; Vallejo-Marin, M.; Meier, J.I. Eukaryote hybrid genomes. PLoS Genet. 2019, 15, e1008404. [Google Scholar] [CrossRef]
- Soreng, R.J. Chloroplast-DNA phylogenetics and biogeography in a reticulating group: Study in Poa (Poaceae). Am. J. Bot. 1990, 77, 1383–1400. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Nosov, N.N.; Kim, E.S.; Machs, E.M.; Punina, E.O.; Probatova, N.S. The origin of polyploid genomes of bluegrasses Poa L. and gene flow between northern Pacific and Sub-Antarctic islands. Rus. J. Gen. 2010, 46, 1407–1416. [Google Scholar] [CrossRef]
- Lövkvist, B.; Hultgård, U.M. Chromosome numbers in south Swedish vascular plants. Opera Botanica 1999, 137, 1–42. [Google Scholar]
- Kamelin, R.V. The peculiarities of angiosperm speciation. Proc. Zool. Inst. RAS 2009, 313 (Suppl. 1), 141–149. [Google Scholar] [CrossRef]
- Krahulcová, A. Chromosome numbers in selected monocotyledons (Czech Republic, Hungary, and Slovakia). Preslia 2003, 75, 97–113. [Google Scholar]
- Amosova, A.V.; Bolsheva, N.L.; Zoshchuk, S.A.; Twardovska, M.O.; Yurkevich, O.Y.; Andreev, I.O.; Samatadze, T.E.; Badaeva, E.D.; Kunakh, V.A.; Muravenko, O.V. Comparative molecular cytogenetic characterization of seven Deschampsia (Poaceae) species. PLoS ONE 2017, 12, e0175760. [Google Scholar] [CrossRef] [PubMed]
- Chiapella, J. A molecular phylogenetic study of Deschampsia (Poaceae: Aveneae) inferred from nuclear ITS and plastid trnL sequence data: Support for the recognition of Avenella and Vahlodea. Taxon 2007, 56, 55–64. [Google Scholar] [CrossRef]
- Saarela, J.M.; Bull, R.D.; Paradis, M.J.; Ebata, S.N.; Peterson, P.M.; Soreng, R.J.; Paszko, B. Molecular phylogenetics of cool-season grasses in the subtribes Agrostidinae, Anthoxanthinae, Aveninae, Brizinae, Calothecinae, Koeleriinae and Phalaridinae (Poaceae, Pooideae, Poeae, Poeae chloroplast group 1). PhytoKeys 2017, 87, 1–139. [Google Scholar] [CrossRef]
- Gnutikov, A.A.; Nosov, N.N.; Punina, E.O.; Probatova, N.S.; Rodionov, A.V. On the placement of Coleanthus subtilis and the subtribe Coleanthinae within Poaceae by new molecular phylogenetic data. Phytotaxa 2020, 468, 243–274. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Gnutikov, A.A.; Nosov, N.N.; Machs, E.M.; Mikhaylova, Y.V.; Shneyer, V.S.; Punina, E.O. Intragenomic Polymorphism of the ITS 1 Region of 35S rRNA Gene in the Group of Grasses with Two-Chromosome Species: Different Genome Composition in Closely Related Zingeria Species. Plants 2020, 9, 1647. [Google Scholar] [CrossRef]
- Hackel, E. Catalogue Raisonne des Graminees du Portugal; Imprimerie de l’Université: Coimbre, Spain, 1880; 33p. [Google Scholar]
- Rivas-Martínez, S.; Díaz, T.E.; Fernández-González, F.; Izco, J.; Loidi, J.; Lousã, M.; Penas, A. Vascular plant communities of Spain and Portugal. Addenda to the syntaxonomical checklist of 2001. Part II. Itinera Geobot. 2002, 15, 698. [Google Scholar]
- Rodionov, A.V.; Shneyer, V.S.; Gnutikov, A.A.; Nosov, N.N.; Punina, E.O.; Zhurbenko, P.M.; Loskutov, I.G.; Muravenko, O.V. Species dialectics: From initial uniformity, through the greatest possible diversity to ultimate uniformity. Bot. Zhurn. 2020, 105, 835–853. (In Russian) [Google Scholar] [CrossRef]
- Peterson, P.M.; Soreng, R.J.; Romaschenko, K.; Barberá, P.; Quintanar, A.; Aedo, C.; Saarela, J.M. Phylogeny and biogeography of Calamagrostis (Poaceae: Pooideae: Poeae: Agrostidinae), description of a new genus, Condilorachia (Calothecinae), and expansion of Greeneochloa and Pentapogon (Echinopogoninae). J. Syst. Evol. 2022, 60, 570–590. [Google Scholar] [CrossRef]
- Ridgway, K.P.; Duck, J.M.; Young, J.P.W. Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron. BMC Ecol. 2003, 3, 8. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Müller, K. SeqState—Primer design and sequence statistics for phylogenetic DNA data sets. App. Bioinf. 2005, 4, 65–69. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef] [PubMed]
- Múrias dos Santos, A.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. TCS BU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Gen. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Amosova, A.V.; Yurkevich, O.Y.; Bolsheva, N.L.; Samatadze, T.E.; Zoshchuk, S.A.; Muravenko, O.V. Repeatome Analyses and Satellite DNA Chromosome Patterns in Deschampsia sukatschewii, D. cespitosa, and D. antarctica (Poaceae). Genes 2022, 13, 762. [Google Scholar] [CrossRef]
- Bateson, W. The progress of genetic research. In Royal Horticultural Society, Report of the Third International Conference 1906 on Genetics; Wilks, W., Ed.; Royal Horticultural Society: London, UK, 1907; pp. 90–97. [Google Scholar]



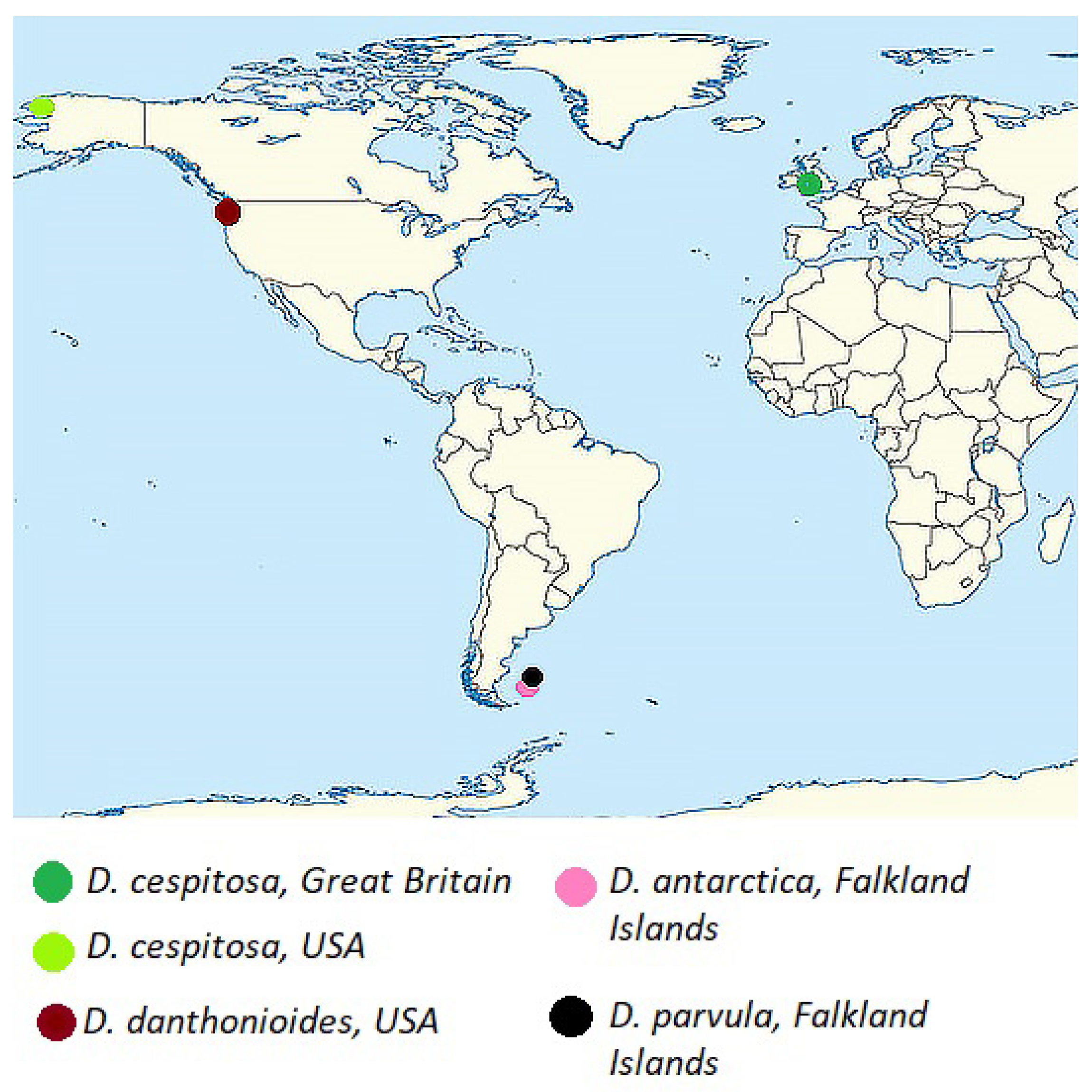














| Species | GenBank Number | Country of Origin | |
|---|---|---|---|
| ITS1–5.8S rRNA Gene–ITS2, Sanger Method | 18S rRNA Gene–ITS1–5.8S rRNA Gene, NGS | ||
| Deschampsia antarctica E.Desv. | OR900972 | Great Britain, Falkland Islands | |
| Deschampsia antarctica E.Desv. | OR900973 | Great Britain, Falkland Islands | |
| Deschampsia antarctica E.Desv. | OR908158–OR908252 | Great Britain, Falkland Islands, Weddel Island | |
| Deschampsia baicalensis Tzvelev | OR903200 | Russia, Altai Republic | |
| Deschampsia brevifolia R.Br. | OR900968 | OR908382–OR908435 | Russia, Krasnoyarsk Krai, Bolshevik Island |
| Deschampsia cespitosa (L.) P.Beauv. | OR900970 | OR907857–OR907948 | Great Britain, Wales |
| Deschampsia cespitosa (L.) P.Beauv. | OR901684 | Russia, Altai Republic | |
| Deschampsia cespitosa (L.) P.Beauv. | OR907949–OR908004 | USA, Alaska | |
| Deschampsia danthonioides Munro | OR900974 | USA, Washington state | |
| Deschampsia glauca Hartm. | OR903202 | Russia, Tyva Republic | |
| Deschampsia koelerioides Regel | OR900969 | Russia, Altai Republic | |
| Deschampsia pamirica Roshev | OR903201 | OR908299–OR908381 | Russia, Altai Republic |
| Deschampsia parvula E.Desv. | OR900975 | OR908005–OR908075 | Great Britain, Falkland Islands |
| Deschampsia submutica (Trautv.) O.D.Nikif. | OR903199 | Russia, Altai Republic | |
| Deschampsia sukatschewii (Popl.) Roshev. | OR900971 | OR908076–OR908157 | Russia, Altai Republic |
| Deschampsia sukatschewii (Popl.) Roshev. | OR900967 | Russia, Yakutia | |
| Deschampsia sp. Alt 15-434 | OR908253–OR908298 | Russia, Altai Republic | |
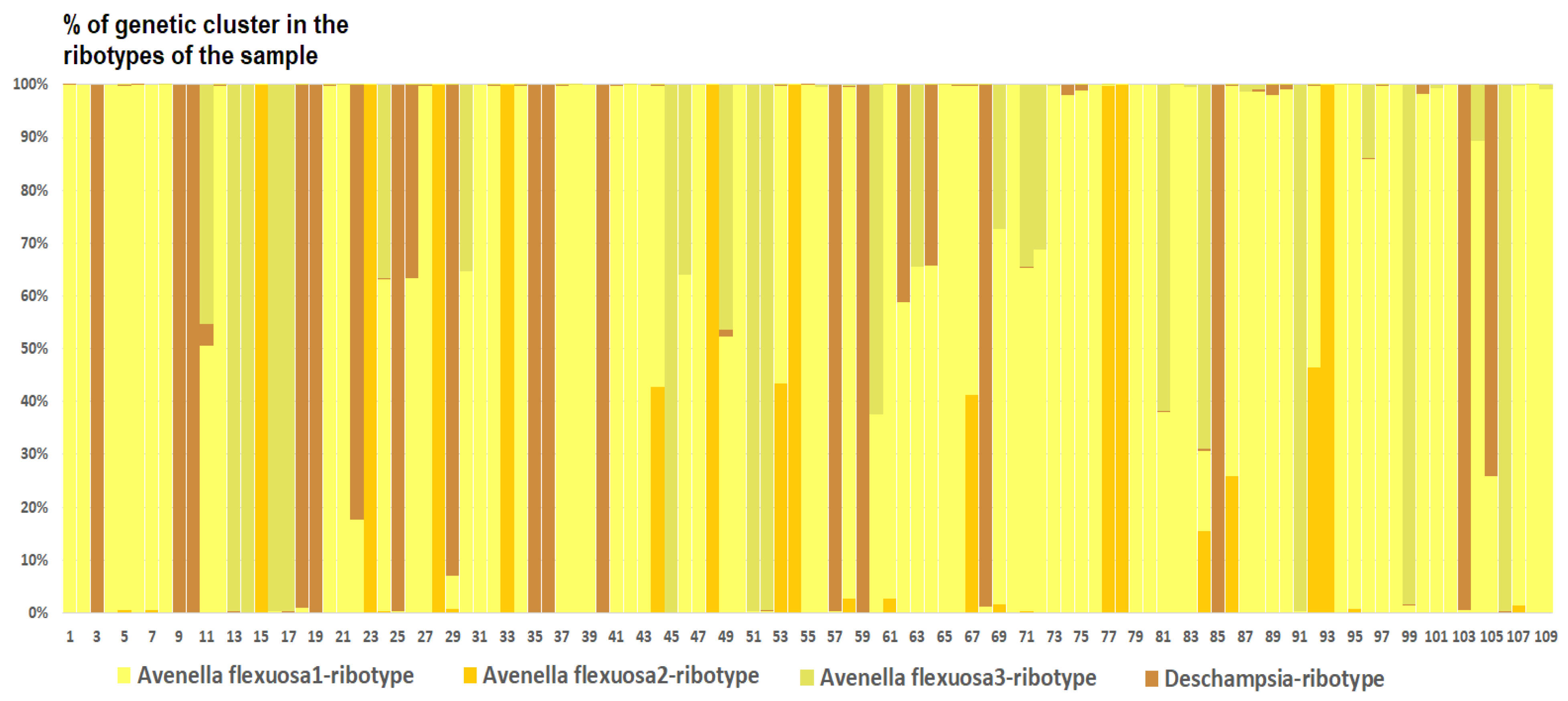
| Avenella flexuosa (L.) Drejer | OR901685 | PQ269223–PQ269266 | Russia, Karachay-Cherkessia Republic |
| Avenella flexuosa (L.) Drejer | OR901686 | Russia, Arkhangelsk Oblast | |
| Avenella flexuosa (L.) Drejer | OR907748–OR907856 | Great Britain | |
| Avenella flexuosa (L.) Drejer | PQ283993–PQ284022 | Russia, Leningrad Oblast | |
| Species | Total Number of Reads | Ribotype Symbol | Number of Reads | % from the Total Number of the Reads |
|---|---|---|---|---|
| Deschampsia antarctica | 23,239 | An1 | 7063 | 30 |
| An2 | 5277 | 23 | ||
| Deschampsia brevifolia | 13,928 | C1 | 1804 | 13 |
| P2/B2 | 1326 | 10 | ||
| S2 | 1162 | 7 | ||
| B4 | 1016 | 7 | ||
| Deschampsia cespitosa, USA | 18,111 | CA1 | 4988 | 28 |
| CA2 | 3259 | 18 | ||
| C1 | 1309 | 13 | ||
| Deschampsia cespitosa, Great Britain | 22,596 | C1 | 5451 | 24 |
| C2 | 3032 | 13 | ||
| C3 | 2926 | 13 | ||
| Deschampsia pamirica | 16,025 | S3 | 2821 | 18 |
| P2/B2 | 1175 | 7 | ||
| P | 1154 | 7 | ||
| Deschampsia parvula | 15,241 | Pa1 | 5379 | 35 |
| Pa2 | 1677 | 11 | ||
| Pa3 | 1034 | 7 | ||
| Deschampsia sukatschewii | 20,619 | S1 | 3511 | 17 |
| S2 | 1594 | 8 | ||
| S3 | 1582 | 8 | ||
| Deschampsia sp. Alt 15-434 | 10,044 | S1 | 2237 | 22 |
| S3 | 1731 | 17 | ||
| Avenella flexuosa | 20,474 | Fl | 8745 | 43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gnutikov, A.A.; Nosov, N.N.; Muravenko, O.V.; Amosova, A.V.; Shneyer, V.S.; Loskutov, I.G.; Punina, E.O.; Rodionov, A.V. Genetic Diversity of the Species of the Genus Deschampsia P.Beauv. (Poaceae) Based on the Analysis of the ITS Region: Polymorphism Proves Distant Hybridization. Int. J. Mol. Sci. 2024, 25, 11348. https://doi.org/10.3390/ijms252111348
Gnutikov AA, Nosov NN, Muravenko OV, Amosova AV, Shneyer VS, Loskutov IG, Punina EO, Rodionov AV. Genetic Diversity of the Species of the Genus Deschampsia P.Beauv. (Poaceae) Based on the Analysis of the ITS Region: Polymorphism Proves Distant Hybridization. International Journal of Molecular Sciences. 2024; 25(21):11348. https://doi.org/10.3390/ijms252111348
Chicago/Turabian StyleGnutikov, Alexander A., Nikolai N. Nosov, Olga V. Muravenko, Alexandra V. Amosova, Victoria S. Shneyer, Igor G. Loskutov, Elizaveta O. Punina, and Alexander V. Rodionov. 2024. "Genetic Diversity of the Species of the Genus Deschampsia P.Beauv. (Poaceae) Based on the Analysis of the ITS Region: Polymorphism Proves Distant Hybridization" International Journal of Molecular Sciences 25, no. 21: 11348. https://doi.org/10.3390/ijms252111348
APA StyleGnutikov, A. A., Nosov, N. N., Muravenko, O. V., Amosova, A. V., Shneyer, V. S., Loskutov, I. G., Punina, E. O., & Rodionov, A. V. (2024). Genetic Diversity of the Species of the Genus Deschampsia P.Beauv. (Poaceae) Based on the Analysis of the ITS Region: Polymorphism Proves Distant Hybridization. International Journal of Molecular Sciences, 25(21), 11348. https://doi.org/10.3390/ijms252111348









