An Analysis of the Mechanism About CO2 Enrichment Promoting Carbohydrate Metabolism in Cucumber (Cucumis sativus L.) Leaves
Abstract
1. Introduction
2. Results
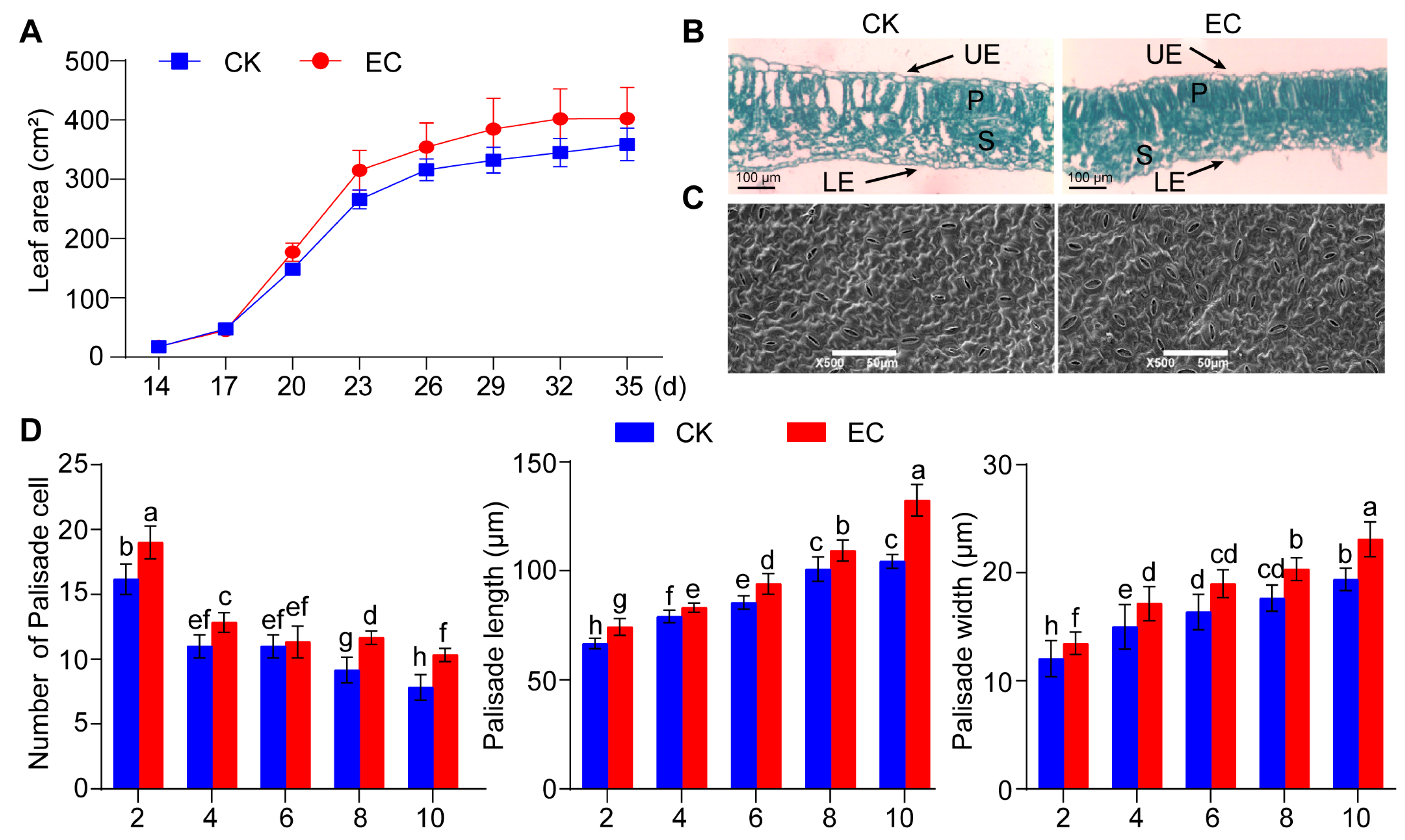
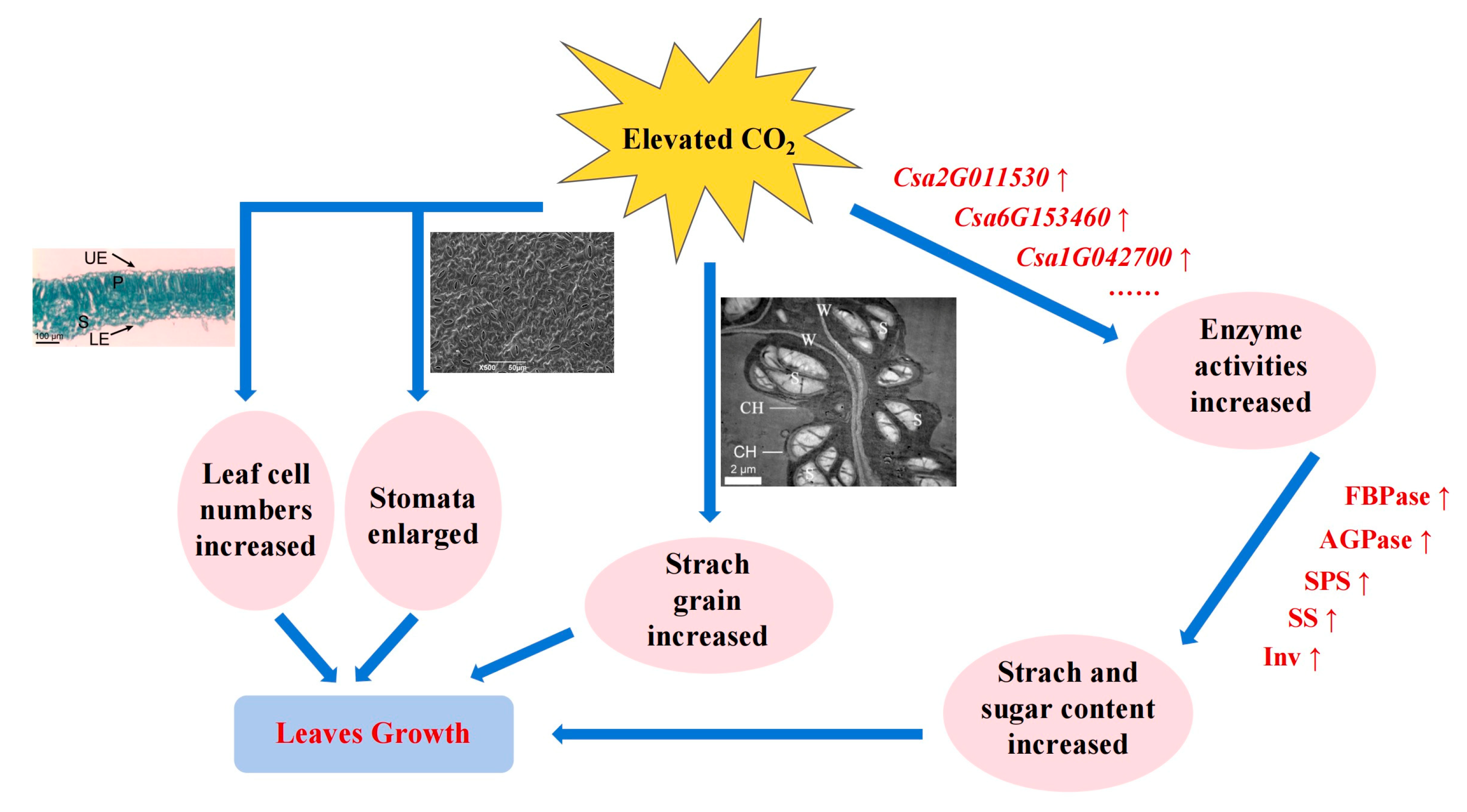
2.1. Effect of CO2 Enrichment on the Growth and Development of Leaves
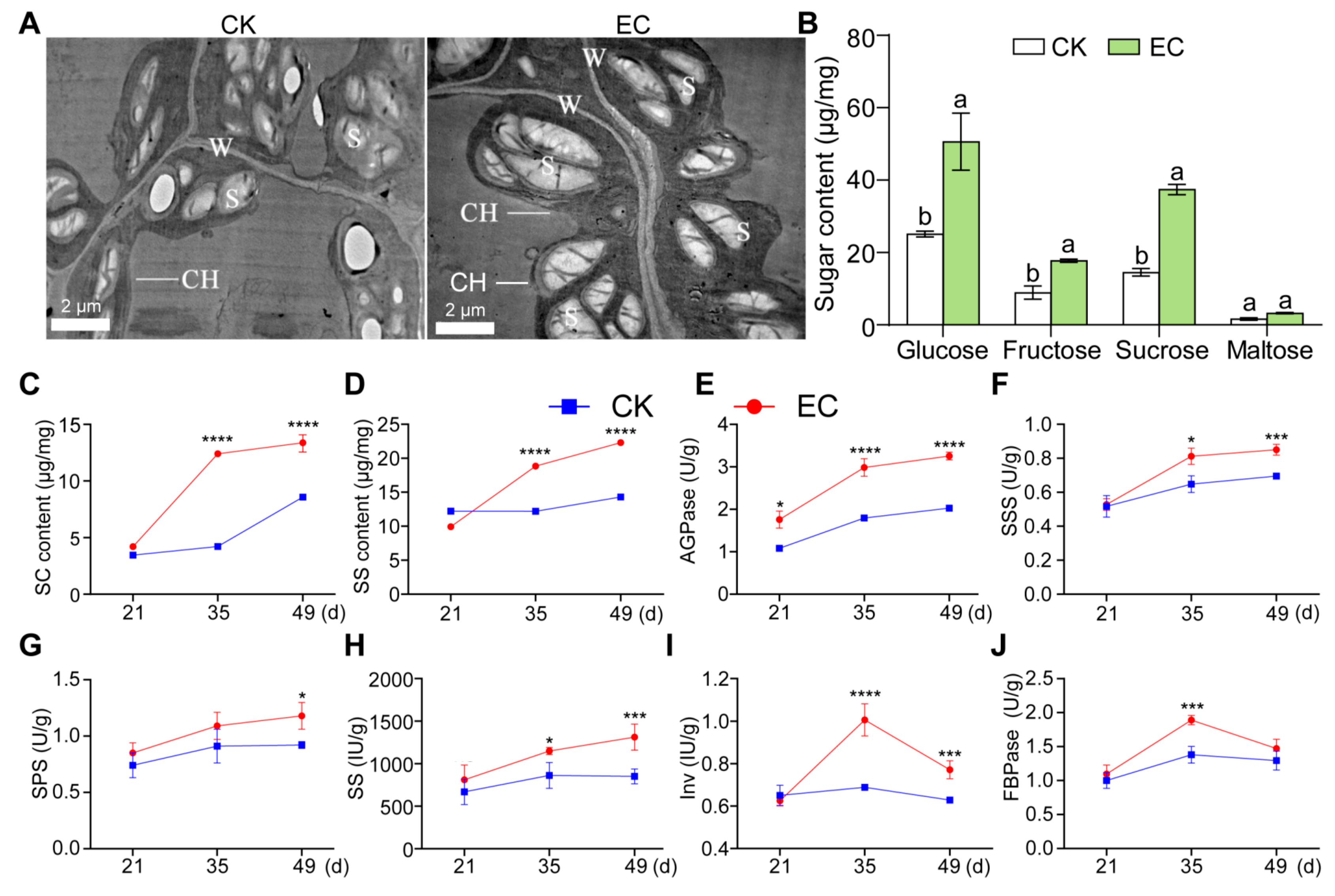
2.2. Effect of CO2 Enrichment on Carbohydrate Accumulation and Enzyme Activity of Leaves
2.3. Transcriptome Analysis of Leaves under Different CO2 Conditions
2.4. Analysis of Genes-Encoding Carbohydrate Metabolism Enzymes in Leaves under CO2 Enrichment
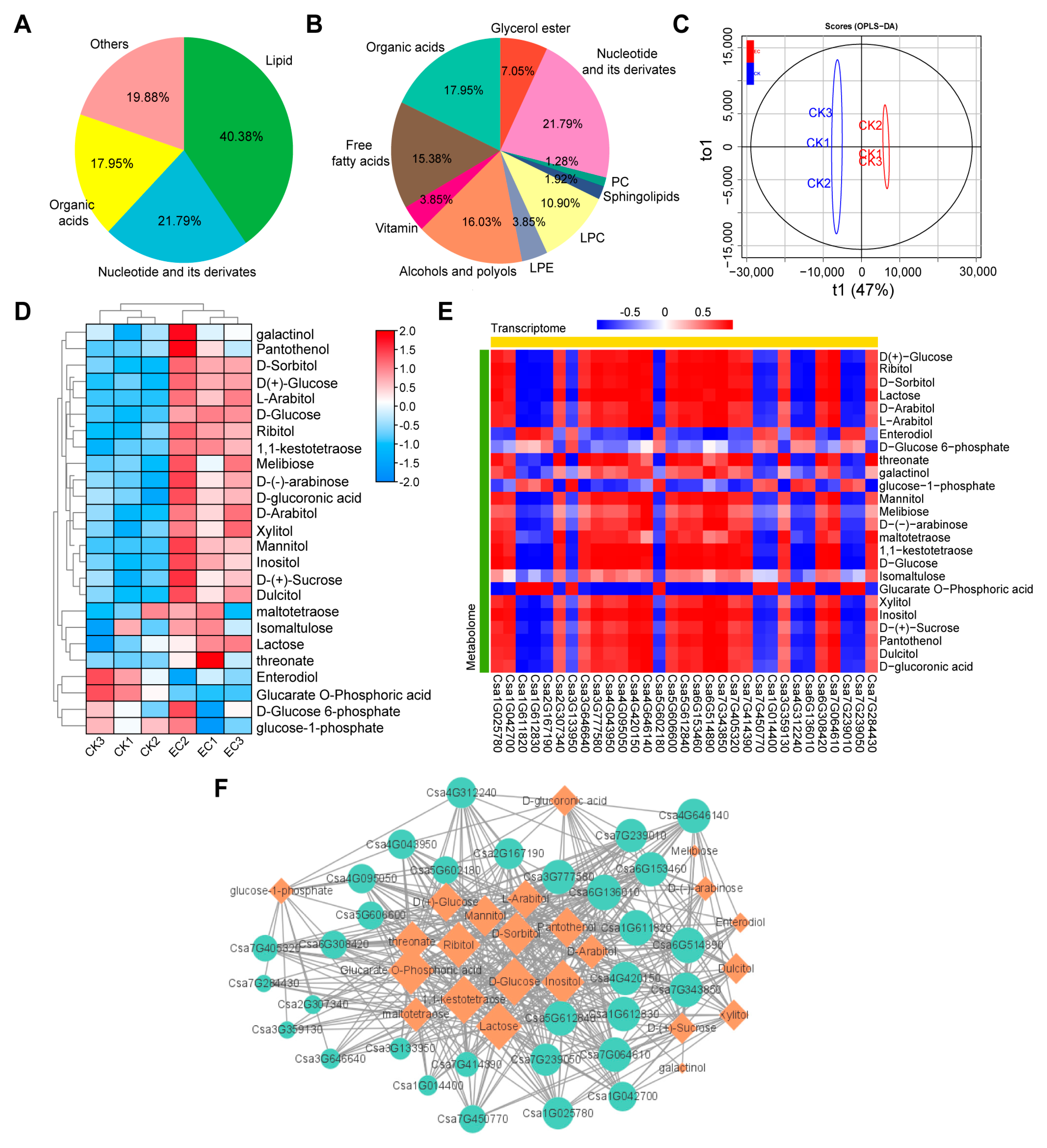
2.5. Correlation Analysis of Transcriptome and Metabolome
2.6. RT-qPCR Verification
3. Discussion
4. Materials and Methods
4.1. Plants and Growth Conditions
4.2. Material Selection and Sampling
4.3. Observation of Leaf Growth Dynamics
4.4. Microscopic and Ultramicroscopic Observations
4.5. Determination of Soluble Sugar and Starch Content
4.6. Determination of Sucrose, Glucose, and Fructose Cotent
4.7. Determination of Enzyme Activities
4.8. RNA Extraction and Transcriptome Analysis
4.9. Determination and Analysis of Metabolome
4.10. Quantitative Real-Time PCR
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seneweera, S.; Aben, S.K.; Basra, A.S.; Jones, B.; Conroy, J.P. Involvement of ethylene in the morphological and developmental response of rice to elevated atmospheric CO2 concentrations. Plant Growth Regul. 2003, 39, 143–153. [Google Scholar] [CrossRef]
- Wu, Y.; Li, Q.M.; Zou, Z.R.; Deng, Y.L. Effects of CO2 concentration doubling and drought stress on leaf translocase activity and sugar metabolism in cucumber seedlings. J. Northwest A F Univ. (Nat. Sci. Ed.) 2008, 36, 121–126. (In Chinese) [Google Scholar] [CrossRef]
- Aranjuelo, I.; Erice, G.; Sanz-Sáez, A.; Abadie, C.; Gilard, F.; Gil-Quintana, E.; Avice, J.C.; Staudinger, C.; Wienkoop, S.; Araus, J.L.; et al. Differential effect on primary carbon metabolism of flag leaves in durum wheat (Triticum durum Desf.). Plant Cell Environ. 2015, 38, 2780–2794. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Foyer, C.H. Sink regulation of photosynthesis. J. Exp. Bot. 2001, 52, 1383–1400. [Google Scholar] [CrossRef]
- Watanabe, C.K.; Sato, S.; Yanagisawa, S.; Uesono, Y.; Terashima, I.; Noguchi, K. Effects of elevated CO2 on levels of primary metabolites and transcripts of genes encoding respiratory enzymes and their diurnal patterns in Arabidopsis thaliana: Possible relationships with respiratory rates. Plant Cell Physiol. 2014, 55, 341–357. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, L.; Zhang, M.; Zhang, Y.; Wang, Q. Effect of carbon dioxide enrichment on health-promoting compounds and organoleptic properties of tomato fruits grown in greenhouse. Food Chem. 2014, 153, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, G.; Sun, B.; Zhang, S.; Zhang, Y.; Liao, Y.; Zhou, Y.; Xia, X.; Shi, K.; Yu, J. Stimulated leaf dark respiration in tomato in an elevated carbon dioxide atmosphere. Sci. Rep. 2013, 3, 3433. [Google Scholar] [CrossRef]
- Elansary, H.O. Impact of elevated CO2 and temperature on growth, development and nutrient uptake of tomato. Horticulturae 2021, 7, 509. [Google Scholar] [CrossRef]
- Zhang, Z.P.; Li, T.; Sun, S.; Xing, G. Effect of CO2 enrichment on fruit quality of thin-skinned melons in greenhouses. Chin. Squash 2019, 32, 20–25. (In Chinese) [Google Scholar] [CrossRef]
- Leakey, A.D.; Ainsworth, E.A.; Bernacchi, C.J.; Rogers, A.; Long, S.P.; Ort, D.R. Elevated CO2 effects on plant carbon, nitrogen, and water relations: Six important lessons from FACE. J. Exp. Bot. 2009, 60, 2859–2876. [Google Scholar] [CrossRef]
- Serrato, A.J.; de Dios Barajas-Lopez, J.; Chueca, A.; Sahrawy, M. Changing sugar partitioning in FBPase-manipulated plants. J. Exp. Bot. 2009, 60, 2923–2931. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.R.; Liu, M.M.; Li, Y.H.; Song, S.S.; Bai, Y.L.; Wang, Y. Research progress on the regulatory mechanism of plant starch biosynthesis. J. Plant Physiol. 2013, 49, 1319–1325. (In Chinese) [Google Scholar] [CrossRef]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef]
- Feng, Y.L.; Yin, F.; Xu, K.; Jia, X.Y.; Zhou, S.; Ma, C. Role of sucrose metabolism and signaling in plant development and response to adversity. J. Nucl. Agri. Sci. 2021, 35, 2044–2055. (In Chinese) [Google Scholar] [CrossRef]
- Gan, C.X.; Wu, C. Research progress on three key enzymes in sucrose metabolism. J. Yangtze Univ. (Nat. Sci. Ed.) 2007, 01, 74–78. (In Chinese) [Google Scholar] [CrossRef]
- Aoki, N.; Ono, K.; Sasaki, H.; Seneweera, S.; Sakai, H.; Kobayashi, K.; Ishimaru, K. Effects of elevated CO2 concentration on photosynthetic carbon metabolism in flag-leaf blades of rice before and after heading. Plant Prod. Sci. 2003, 6, 52–58. [Google Scholar] [CrossRef]
- Li, J.Y.; Liu, X.H.; Cai, Q.S.; Gu, H.; Zhang, S.S.; Wu, Y.Y.; Wang, C.J. Effects of elevated CO2 on growth, carbon assimilation, photosynthate accumulation and related enzymes in rice leaves during sink-source transition. J. Integr. Plant Biol. 2008, 50, 723–732. [Google Scholar] [CrossRef]
- Wei, H.; Gou, J.; Yordanov, Y.; Zhang, H.; Thakur, R.; Jones, W.; Burton, A. Global transcriptomic profiling of aspen trees under elevated [CO2] to identify potential molecular mechanisms responsible for enhanced radial growth. J. Plant Res. 2013, 126, 305–320. [Google Scholar] [CrossRef]
- Ma, Y.; Devi, M.J.; R. Reddy, V.; Song, L.; Gao, H.; Cao, B. Cloning and characterization of three sugar metabolism genes (LBGAE, LBGALA, and LBMS) regulated in response to elevated CO2 in Goji Berry (Lycium barbarum L.). Plants 2021, 10, 321. [Google Scholar] [CrossRef]
- Vu, J.C.; Allen, L.H.; Gesch, R.W. Up-regulation of photosynthesis and sucrose metabolism enzymes in young expanding leaves of sugarcane under elevated growth CO2. Plant Sci. 2006, 171, 123–131. [Google Scholar] [CrossRef]
- Song, H.; Li, Y.; Xu, X.; Zhang, J.; Zheng, S.; Hou, L.; Xing, G.; Li, M. Analysis of genes related to chlorophyll metabolism under elevated CO2 in cucumber (Cucumis sativus L.). Sci. Hortic. 2020, 261, 108988. [Google Scholar] [CrossRef]
- Duan, J.Y.; Cui, S.M.; Song, Y.; Zhang, L.H.; Jin, L.P.; Pan, L.; Zhang, M. Effect of high temperature and CO2 enrichment on carbohydrate distribution mechanism in cucumber seedlings. Jiangsu Agric. Sci. 2023, 51, 187–197. (In Chinese) [Google Scholar] [CrossRef]
- Li, Y.P.; Song, H.X. Effects of CO2 enrichment on photosynthetic characteristics and seedling quality of mini-cucumber in greenhouse. Northeast Agric. Sci. 2023, 48, 117–120+140. (In Chinese) [Google Scholar] [CrossRef]
- Miller, T.E.; Beneyton, T.; Schwander, T.; Diehl, C.; Girault, M.; McLean, R.; Chotel, T.; Claus, P.; Cortina, N.S.; Baret, J.C.; et al. Light-powered CO2 fixation in a chloroplast mimic with natural and synthetic parts. Science 2020, 368, 649–654. [Google Scholar] [CrossRef]
- Ma, Y.; Reddy, V.R.; Devi, M.J.; Song, L.; Cao, B. De novo characterization of the Goji berry (Lycium barbarium L.) fruit transcriptome and analysis of candidate genes involved in sugar metabolism under different CO2 concentrations. Tree Physiol. 2019, 39, 1032–1045. [Google Scholar] [CrossRef]
- Aranjuelo, I.; Cabrera-Bosquet, L.; Morcuende, R.; Avice, J.C.; Nogués, S.; Araus, J.L.; Martínez-Carrasco, R.; Pérez, P. Does ear C sink strength contribute to overcoming photosynthetic acclimation of wheat plants exposed to elevated CO2. J. Exp. Bot. 2011, 62, 3957–3969. [Google Scholar] [CrossRef] [PubMed]
- Rathore, R.S.; Garg, N.; Garg, S.; Kumar, A. Starch phosphorylase: Role in starch metabolism and biotechnological applications. Crit. Rev. Biotechnol. 2009, 29, 214–224. [Google Scholar] [CrossRef]
- Qin, Y.; Xiao, Z.; Zhao, H.; Wang, J.; Wang, Y.; Qiu, F. Starch phosphorylase2 is essential for cellular carbohydrate partitioning in maize. Plant Biol. 2022, 64, 1755–1769. [Google Scholar] [CrossRef]
- Cai, T.; Sun, H.; Qiao, J.; Zhu, L.; Zhang, F.; Zhang, J.; Tang, Z.; Wei, X.; Yang, J.; Yuan, Q.; et al. Cell-free chemoenzymatic starch synthesis from carbon dioxide. Science 2021, 373, 1523–1527. [Google Scholar] [CrossRef]
- Sharkey, T.D. The end game(s) of photosynthetic carbon metabolism. Plant Physiol. 2024, 195, 67–78. [Google Scholar] [CrossRef]
- Li, C.R.; Zhang, X.; Liu, C.M. Carbon reaction core of photosynthesis–Calvin Cycle and Regulation. Chin. Bull. Life Sci. 2024, 1–16. Available online: http://kns.cnki.net/kcms/detail/31.1600.Q.20240328.1659.004.html (accessed on 26 July 2024). (In Chinese).
- Li, Q.P.; Zhang, X.F.; Liang, M. Research progress on fructose 1,6-diphosphate esterase. J. Liaocheng Univ. (Soc. Sci. Ed.) 2021, 34, 73–80. (In Chinese) [Google Scholar] [CrossRef]
- Uematsu, K.; Suzuki, N.; Iwamae, T.; Inui, M.; Yukawa, H. Increased fructose 1,6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. J. Exp. Bot. 2012, 63, 3001–3009. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.H.; Jang, A.; Bhoo, S.H.; Jeon, J.S.; Hahn, T.R. Manipulation of triose phosphate/phosphate translocator and cytosolic fructose-1,6-bisphosphatase, the key components in photosynthetic sucrose synthesis, enhances the source capacity of transgenic Arabidopsis plants. Photosynth. Res. 2012, 111, 261–268. [Google Scholar] [CrossRef]
- Sahrawy, M.; Avila, C.; Chueca, A.; Cánovas, F.M.; López-Gorgé, J. Increased sucrose level and altered nitrogen metabolism in Arabidopsis thaliana transgenic plants expressing antisense chloroplastic fructose-1,6-bisphosphatase. J. Exp. Bot. 2004, 55, 2495–2503. [Google Scholar] [CrossRef]
- Schormann, N.; Hayden, K.L.; Lee, P.; Banerjee, S.; Chattopadhyay, D. An overview of structure, function, and regulation of pyruvate kinases. Protein Sci. 2019, 28, 1771–1784. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X. Construction of pyruvate producing strain with intact pyruvate dehydrogenase and genome-wide transcription analysis. World J. Microb. Biot. 2017, 33, 59. [Google Scholar] [CrossRef]
- Tretter, L.; Adam-Vizi, V. Alpha-ketoglutarate dehydrogenase: A target and generator of oxidative stress. Philos. Trans. R. Soc. Lond. Ser. B-Contain. Pap. A Biol. Character 2005, 360, 2335–2345. [Google Scholar] [CrossRef]
- Ban, Q.; Han, Y.; He, Y.; Jin, M.; Han, S.; Suo, J.; Rao, J. Functional characterization of persimmon β-galactosidase gene DkGAL1 in tomato reveals cell wall modification related to fruit ripening and radicle elongation. Plant Sci. 2018, 274, 109–120. [Google Scholar] [CrossRef]
- María, M.S.; Alejandro, A.C.; Knox, J.P.; Dopico, B.; Labrador, E.; Martín, I. β-(1,4)-Galactan remodelling in Arabidopsis cell walls affects the xyloglucan structure during elongation. Planta 2019, 249, 351–362. [Google Scholar] [CrossRef]
- Baiya, S.; Pengthaisong, S.; Kitjaruwankul, S.; Ketudat Cairns, J.R. Structural analysis of rice Os4BGlu18 monolignol β-glucosidase. PLoS ONE 2021, 16, e0241325. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xie, F.; Huang, X.; Luo, Z. A Chromosome-level genome of ‘Xiaobaixing’ (Prunus armeniaca L.) provides clues to its domestication and identification of key bHLH genes in amygdalin biosynthesis. Plants 2023, 12, 2756. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, S.; Dong, Y.; Ren, R.; Chen, D.; Chen, X. Chloroplastic Os3BGlu6 contributes significantly to cellular ABA pools and impacts drought tolerance and photosynthesis in rice. New Phytol. 2020, 226, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Anees, M.; Gao, L.; Umer, M.J.; Yuan, P.; Zhu, H.; Lu, X.; He, N.; Gong, C.; Kaseb, M.O.; Zhao, S.; et al. Identification of Key Gene Networks Associated with Cell Wall Components Leading to Flesh Firmness in Watermelon. Front. Plant Sci. 2021, 12, 630243. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Niu, Y.; Ye, L.; Shi, Y.; Luo, A. Transcriptomic analysis reveals ozone treatment delays kiwifruit postharvest softening by modulating cell wall metabolism. J. Food Sci. 2024, 89, 2001–2016. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.S.; Pharr, D.M. Leaf area prediction models for cucumber from linear measurements. Hortscience 1987, 22, 1264–1266. [Google Scholar] [CrossRef]
- Garcês, H.; Sinha, N. Fixing and sectioning tissue from the plant Kalanchoë daigremontiana. Cold Spring Harb. Protoc. 2009, 2009, pdb.prot5301. [Google Scholar] [CrossRef]
- Shi, C.; Yang, X.Q.; Yan, H.B. Observation of white thorn leaves in Nitraria tangutorum under salt stress by scanning electron microscopy. J. Shanxi Agric. Univ. (Nat. Sci. Ed.) 2017, 37, 35–39. (In Chinese) [Google Scholar] [CrossRef]
- Liu, R.; Hu, G.; Wei, Q.; Wang, W.; Li, F.; Hu, B. Effects of low temperature on ultrastructure and physiological characteristics of chloroplast in transferring CBF3 gene tobacoseeding. Anhui Agric. Sci. 2010, 38, 9501–9503. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Liu, G.S.; Bi, Q.W.; Li, G.; Wang, H.M.; Cai, Z.C. Study on enzyme and carbohydrate activities in flue-cured tobacco during maturing stage. Acta Tabacaria Sin. 2010, 16, 45–48. (In Chinese) [Google Scholar]
- Tao, Z.Y.; Wu, S.B.; Yan, Q.S.; Yu, W.Q. Determination of monosaccharide components and content in Cassava residue by ion chromatography. Pap. Sci. Technol. 2012, 3, 25–27. (In Chinese) [Google Scholar] [CrossRef]
- Sahrawy, M.; Chueca, A.; Hermoso, R.; Lázaro, J.J.; Lopez gorg, J. Role of light in the biosynthesis and turnover of photosynthetic fructose-1,6-bisphosphatase in pea (Pisum sativum L.) seedlings. New Phytol. 2010, 115, 603–608. [Google Scholar] [CrossRef]
- Sun, M.; Qi, X.; Hou, L.; Xu, X.; Zhu, Z.; Li, M. Gene expression analysis of pak choi in response to vernalization. PLoS ONE 2015, 10, e0141446. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Bolger, M.; Schwacke, R.; Usadel, B. MapMan visualization of RNA-Seq data using mercator4 functional annotations. Methods Mol. Biol. 2021, 2354, 195–212. [Google Scholar] [CrossRef]
- Ye, L.; Bai, F.; Zhang, L.; Luo, M.; Gao, L.; Wang, Z. Transcriptome and metabolome analyses of anthocyanin biosynthesis in post-harvest fruits of a full red-type kiwifruit (Actinidia arguta) ‘Jinhongguan’. Front. Plant Sci. 2023, 14, 1280970. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]

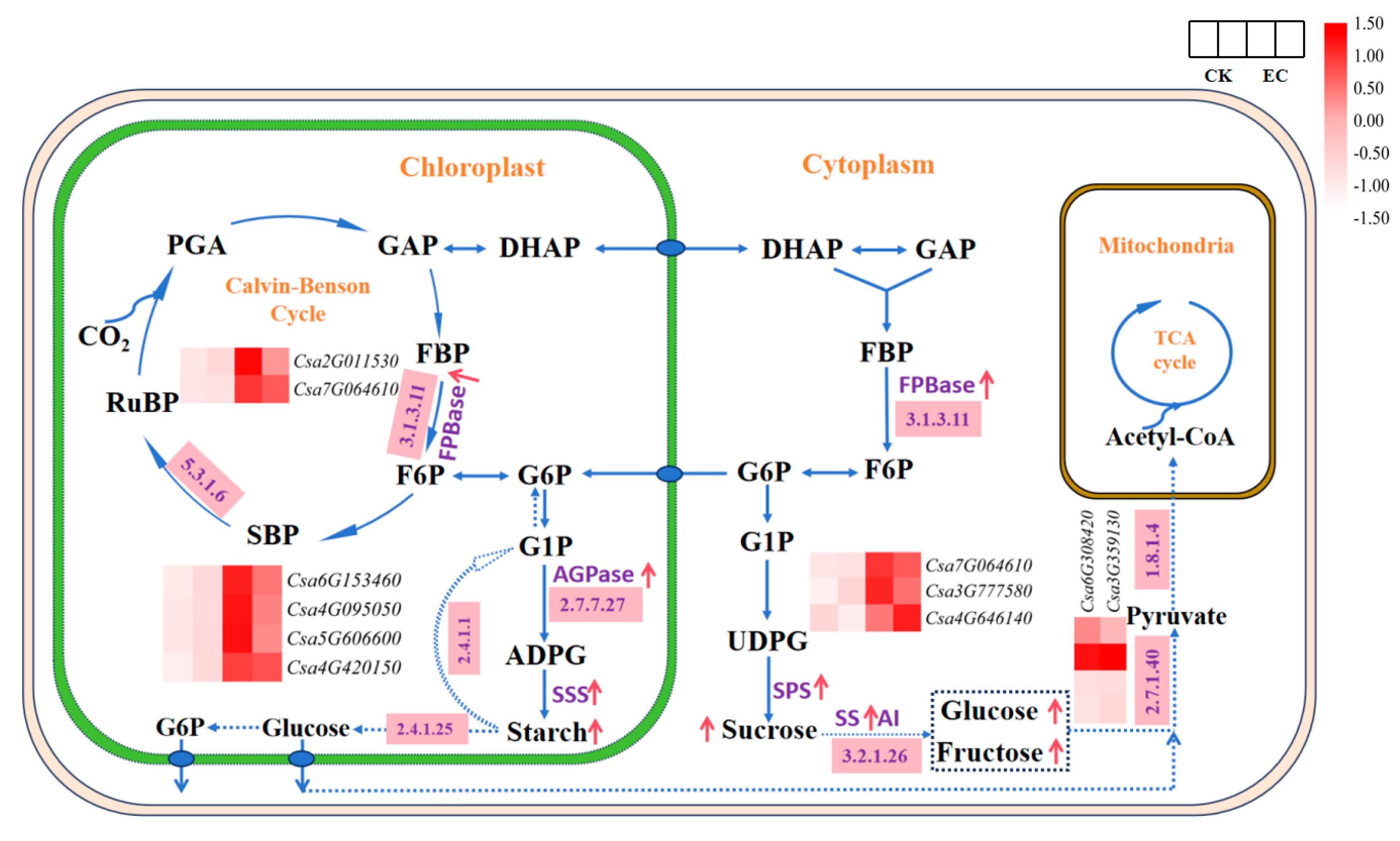
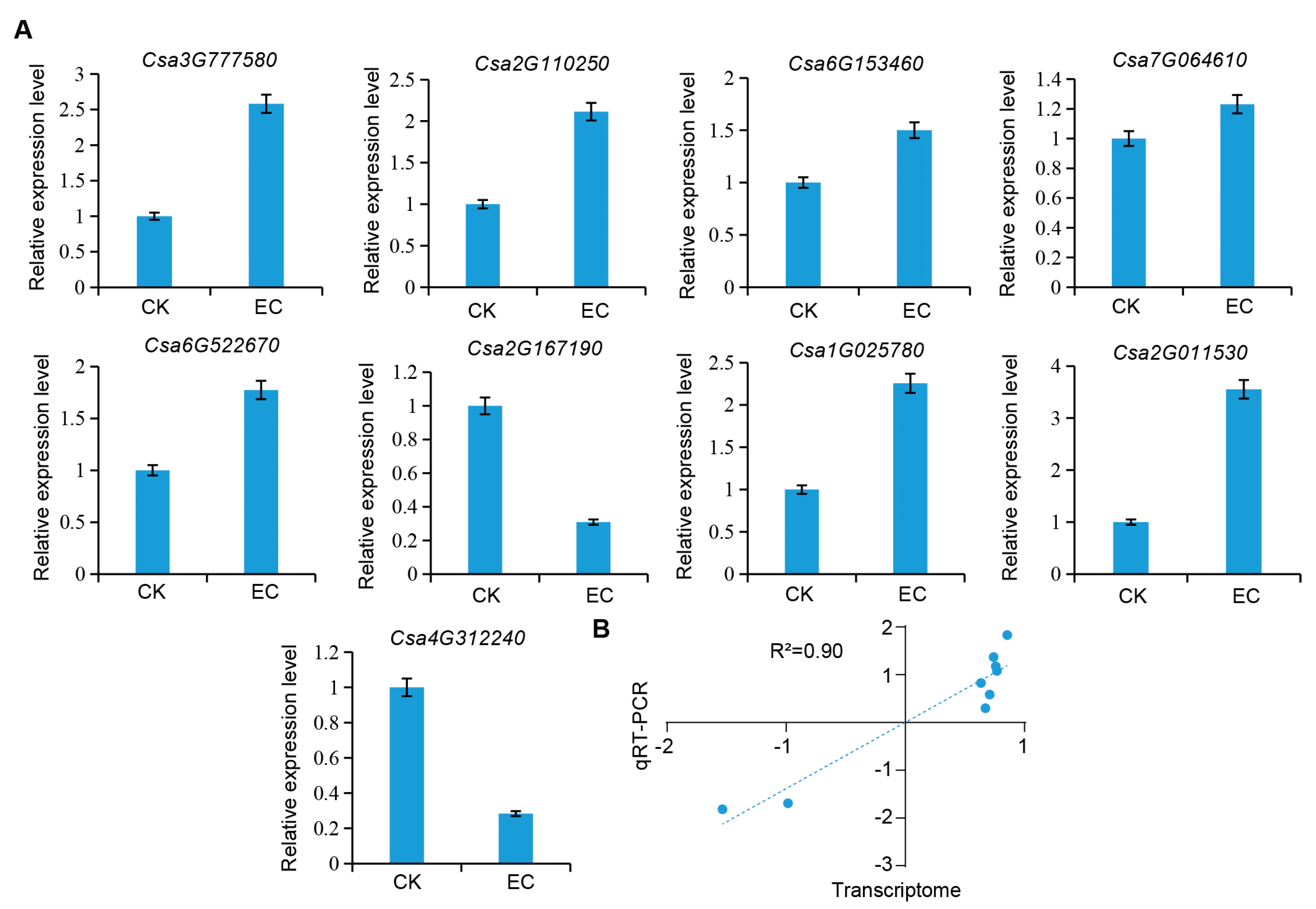
| Enzyme ID | Enzyme Name | Gene ID | FPKM | Expression Pattern | |
|---|---|---|---|---|---|
| 5.3.1.6 | ribose-5-phosphate isomerase | Csa2G011530 | 224.92 | 396.89 | Up |
| 3.1.3.11 | fructose-1,6-bisphosphatase | Csa7G064610 | 13.94 | 21.90 | Up |
| 2.7.7.27 | ADP-glucose pyrophosphorylase | Csa6G153460 | 64.82 | 104.42 | Up |
| 2.4.1.1 | starch phosphorylase | Csa4G095050 | 14.42 | 26.24 | Up |
| Csa5G606600 | 10.84 | 20.61 | |||
| 3.2.1.21 | β-glucosidase | Csa1G025780 | 10.33 | 17.38 | Up |
| Csa5G612840 | 2.13 | 4.25 | |||
| Csa1G042700 | 44.60 | 74.45 | |||
| 3.2.1.26 | β-fructofuranosidase | Csa3G777580 | 8.56 | 14.14 | Up |
| Csa4G646140 | 5.53 | 8.96 | |||
| 3.2.1.23 | β-galactosidase | Csa3G865330 | 6.51 | 12.97 | Up |
| Csa6G504610 | 17.43 | 28.51 | |||
| Csa2G337760 | 3.97 | 16.10 | |||
| 2.4.1.43 | α-1,4-galacturonosyltransferase | Csa6G075210 | 17.48 | 26.45 | Up |
| Csa1G179740 | 17.16 | 25.49 | |||
| 3.1.1.11 | pectinesterase | Csa7G343850 | 1.41 | 3.89 | Up |
| Csa6G514890 | 2.70 | 5.19 | |||
| Csa3G646640 | 32.93 | 60.06 | |||
| Csa7G414390 | 23.30 | 39.99 | |||
| 2.4.1.25 | 4-α-glucanotransferase | Csa4G420150 | 9.29 | 16.03 | Up |
| 2.7.1.40 | pyruvate kinase | Csa3G359130 | 19.47 | 30.87 | Up |
| 1.8.1.4 | dihydrolipoamide dehydrogenase | Csa6G308420 | 25.75 | 62.13 | Up |
| 1.1.1.40 | malate dehydrogenase | Csa2G373430 | 19.66 | 34.77 | Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Song, H.; Li, X.; Hou, L.; Li, M. An Analysis of the Mechanism About CO2 Enrichment Promoting Carbohydrate Metabolism in Cucumber (Cucumis sativus L.) Leaves. Int. J. Mol. Sci. 2024, 25, 11309. https://doi.org/10.3390/ijms252011309
Li Y, Song H, Li X, Hou L, Li M. An Analysis of the Mechanism About CO2 Enrichment Promoting Carbohydrate Metabolism in Cucumber (Cucumis sativus L.) Leaves. International Journal of Molecular Sciences. 2024; 25(20):11309. https://doi.org/10.3390/ijms252011309
Chicago/Turabian StyleLi, Yanling, Hongxia Song, Xuan Li, Leiping Hou, and Meilan Li. 2024. "An Analysis of the Mechanism About CO2 Enrichment Promoting Carbohydrate Metabolism in Cucumber (Cucumis sativus L.) Leaves" International Journal of Molecular Sciences 25, no. 20: 11309. https://doi.org/10.3390/ijms252011309
APA StyleLi, Y., Song, H., Li, X., Hou, L., & Li, M. (2024). An Analysis of the Mechanism About CO2 Enrichment Promoting Carbohydrate Metabolism in Cucumber (Cucumis sativus L.) Leaves. International Journal of Molecular Sciences, 25(20), 11309. https://doi.org/10.3390/ijms252011309






