Comparative Phylogenomic Study of Malaxidinae (Orchidaceae) Sheds Light on Plastome Evolution and Gene Divergence
Abstract
1. Introduction
2. Results
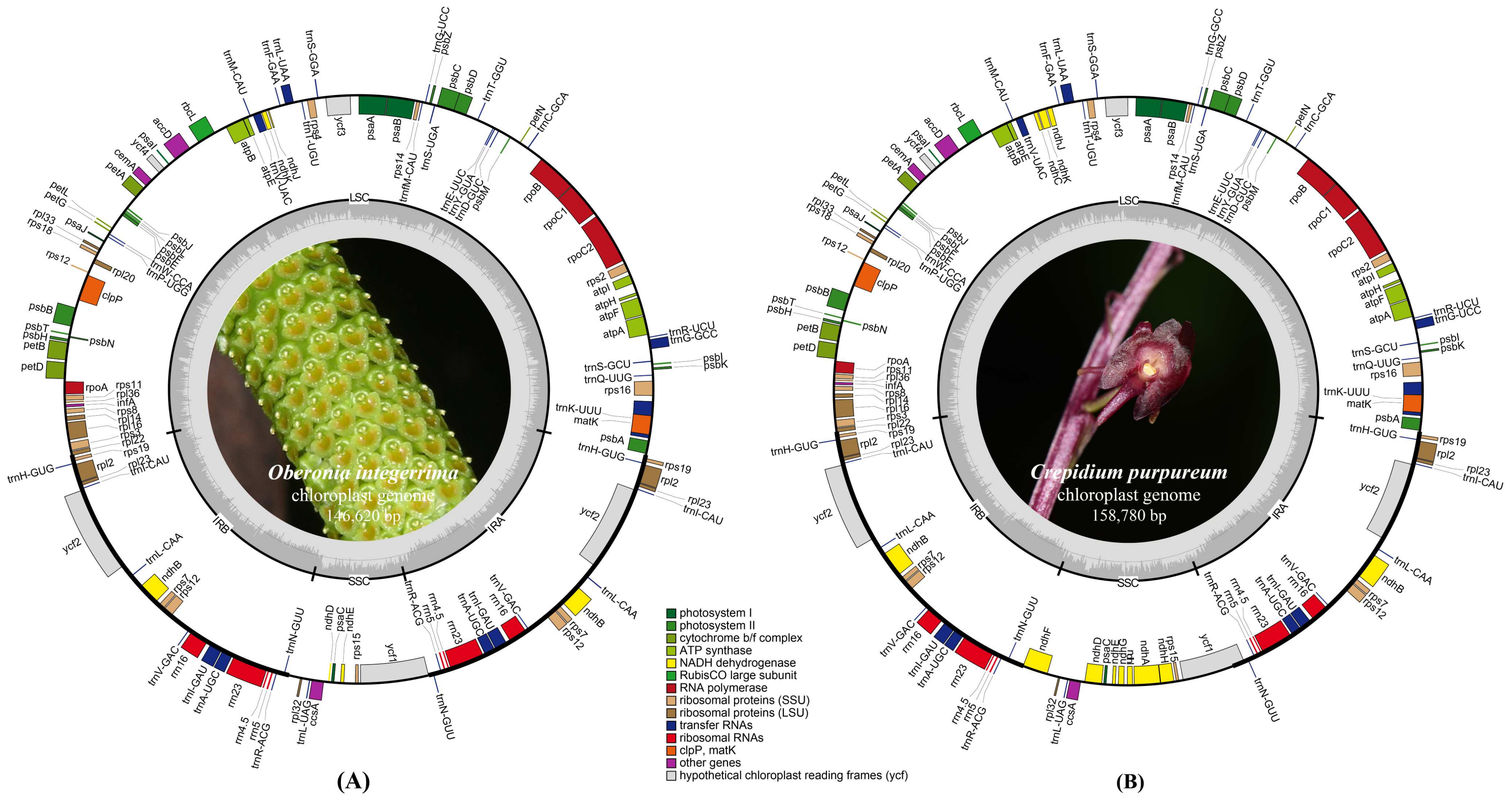
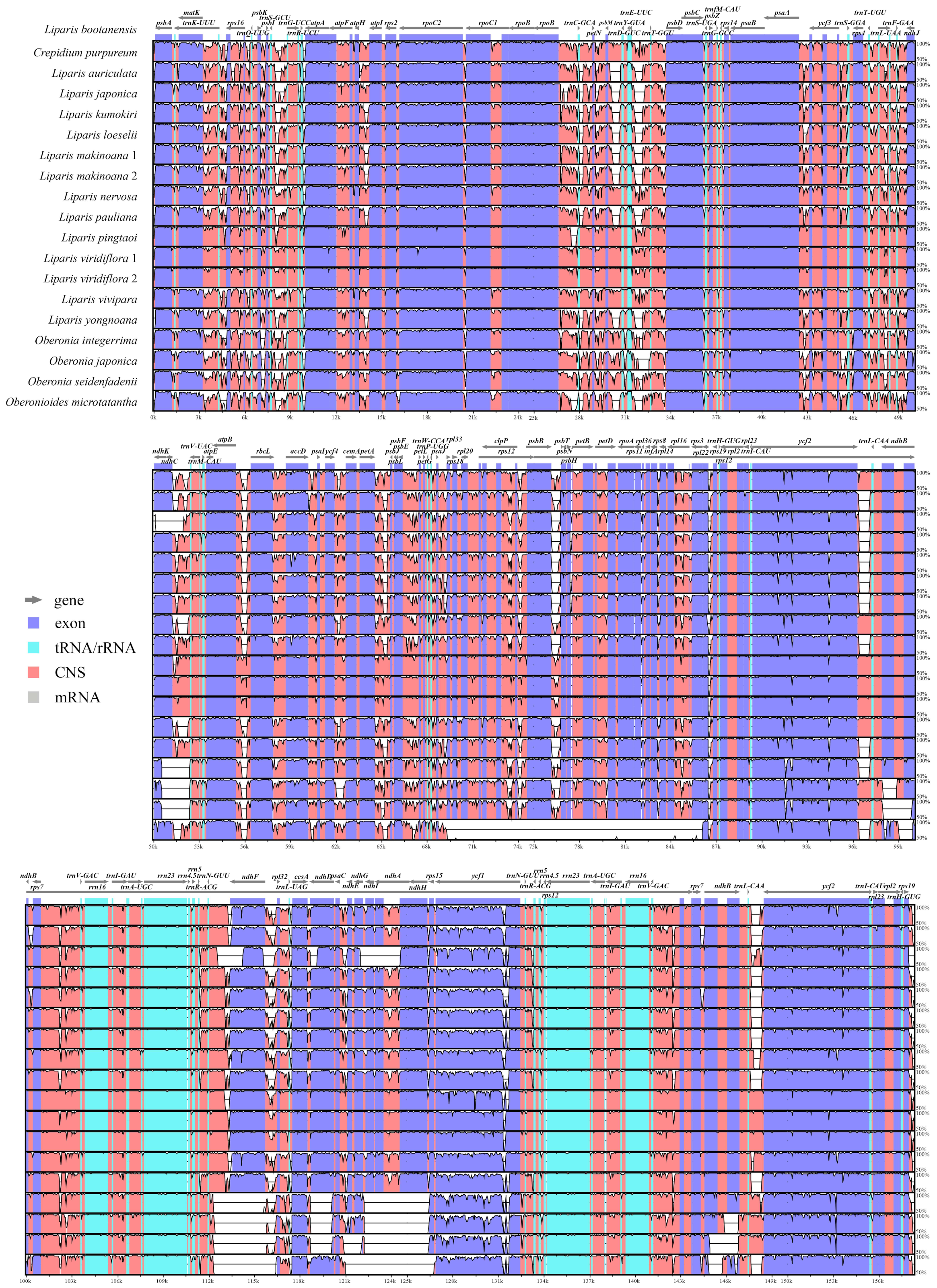
2.1. Plastome Features and Genome Rearrangement
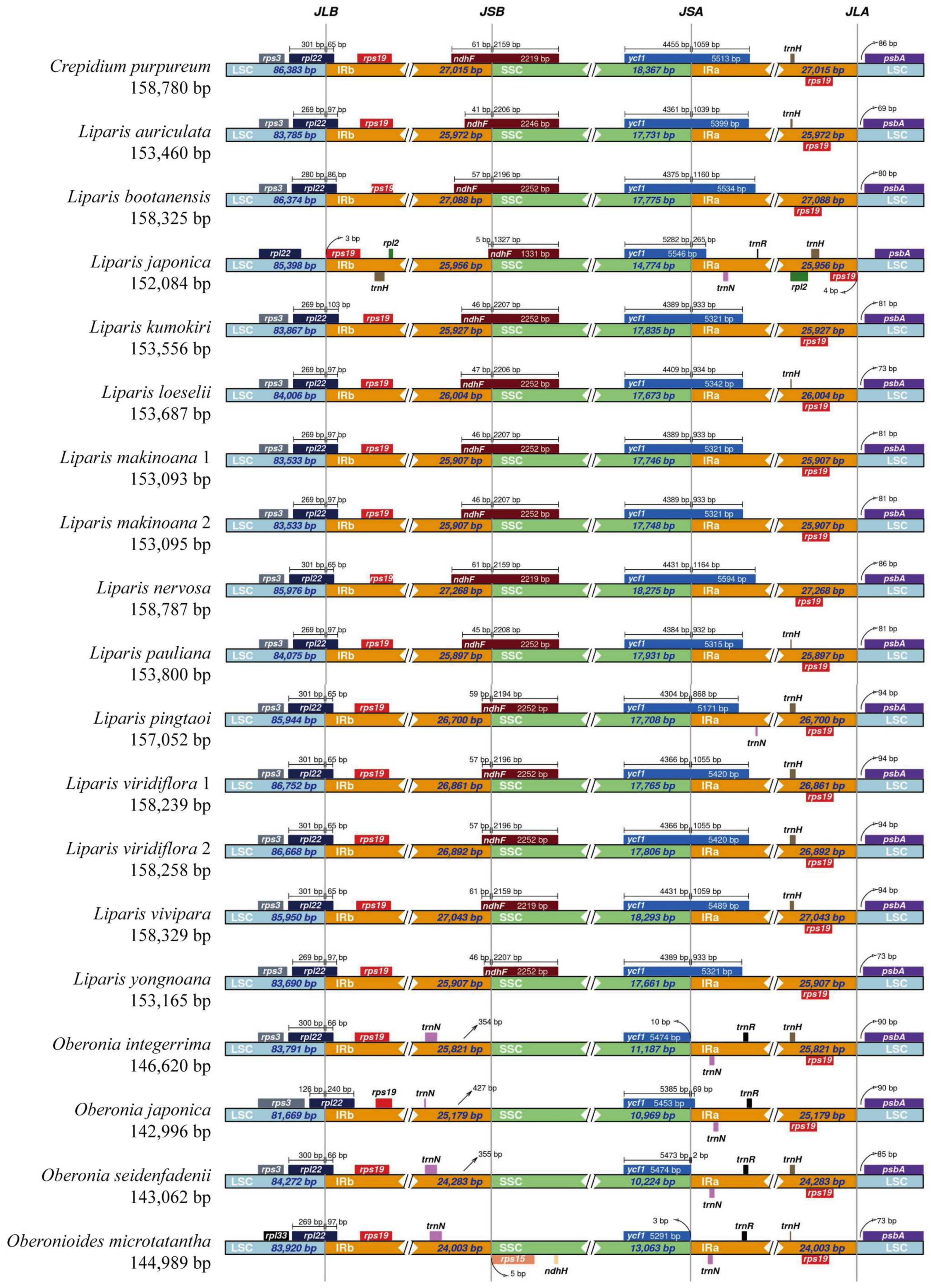
2.2. IR Region Border Analysis
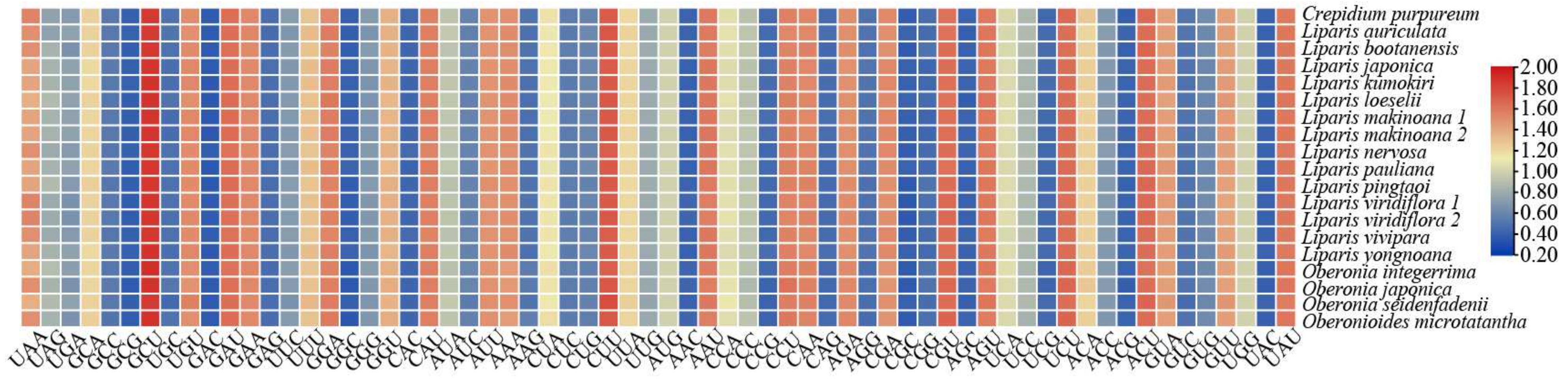
2.3. Codon Usage Analysis
2.4. Nucleotide Mutation Hotspots
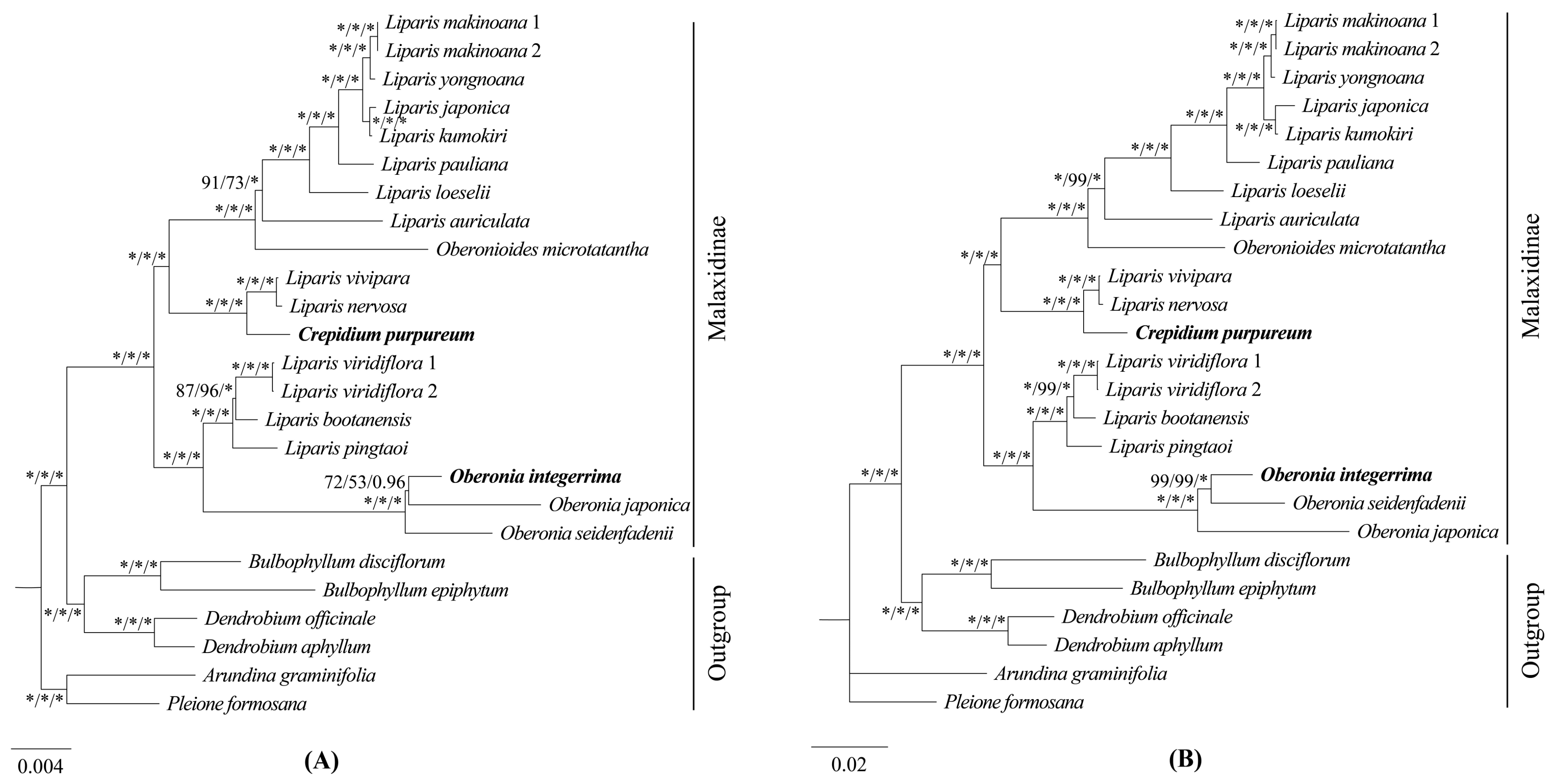
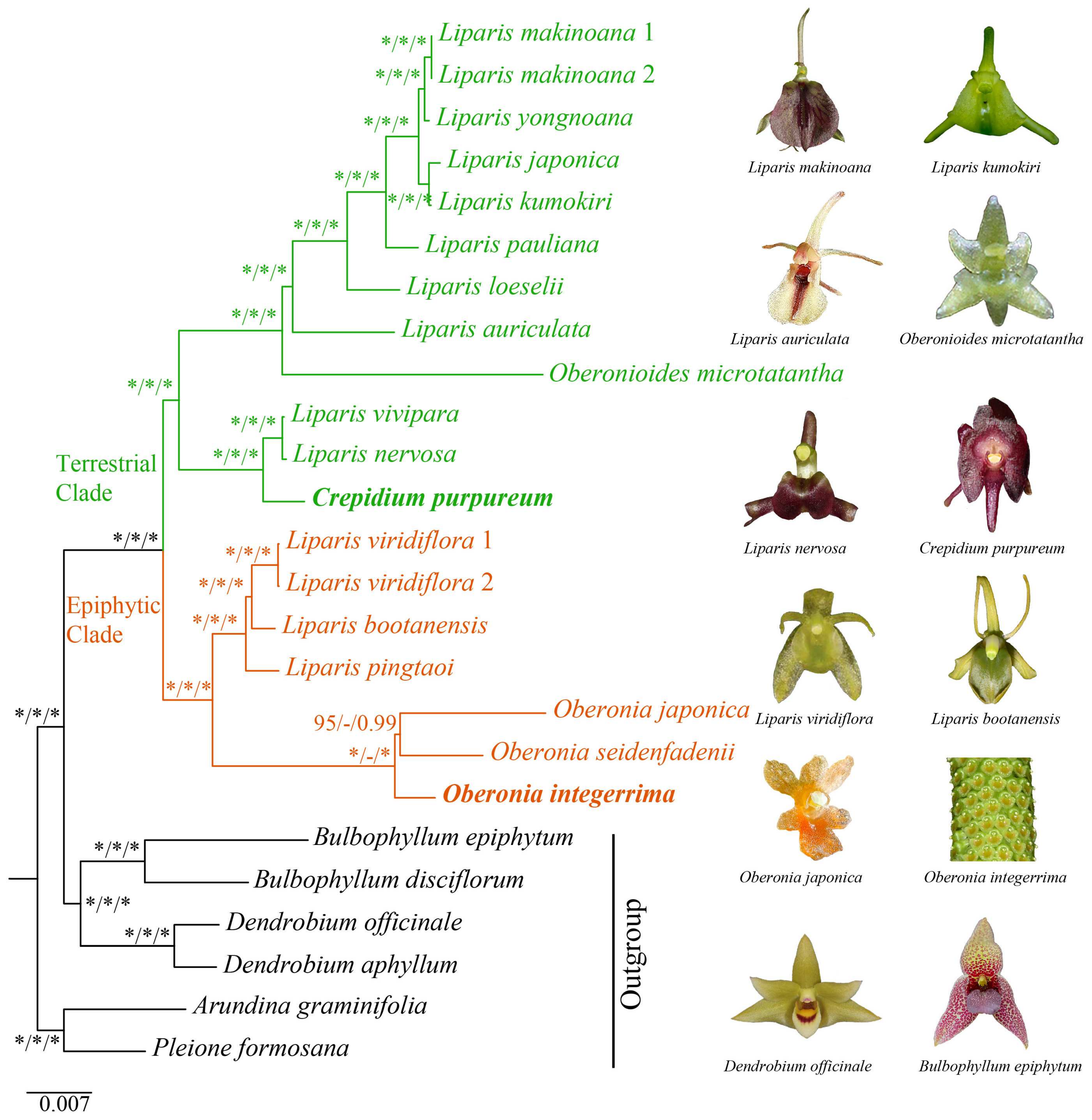
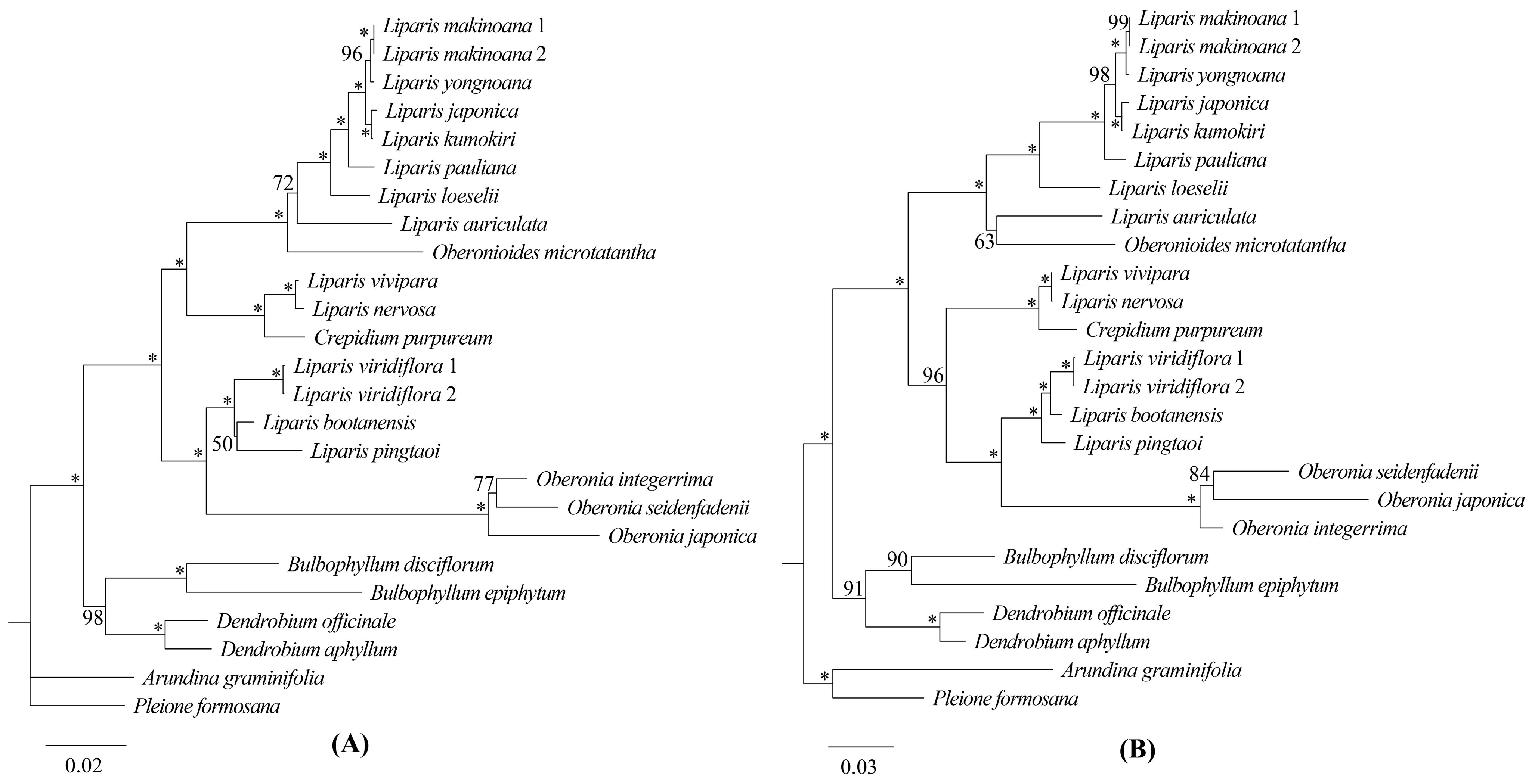
2.5. Phylogenetic Analysis
3. Discussion
3.1. Plastome Structure Conservation and Divergence
3.2. Phylogenetic Relationships
4. Materials and Methods
4.1. Sample Preparation, Sequencing, and Data Acquisition
4.2. Plastome DNA Assembly, Annotation, and Comparison
4.3. Plastome Features Analysis
4.4. Codon Usage and Nucleotide Mutation Hotspots Analysis
4.5. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pridgeon, A.M.; Cribb, P.J.; Chase, M.W.; Rasmussen, F.N. Genera Orchidacearum Volume 6: Epidendroideae (Part 1); Oxford University Press: Oxford, UK, 2005. [Google Scholar]
- Chase, M.W.; Cameron, K.M.; Freudestein, J.V.; Pridgeon, A.M.; Salazar, G.; Van den Berg, C.; Schuiteman, A. An update classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef]
- Govaerts, R.J.; Bernet, P.; Kratochvil, K.; Gerlach, G.; Carr, G.; Alrich, P.; Pridgeon, A.M.; Pfahl, J.; Campacci, M.A.; Holland Baptista, D.; et al. World Checklist of Orchidaceae. The Board of Trustees of the Royal Botanic Gardens, Kew. 2021. Available online: http://www.kew.org/wcsp/ (accessed on 15 November 2021).
- Cameron, K.M. Leave it to the leaves: A molecular phylogenetic study of Malaxideae (Epidendroideae, Orchidaceae). Am. J. Bot. 2005, 92, 1025–1032. [Google Scholar] [CrossRef]
- Liang, W.; Guo, X.; Nagle, D.G.; Zhang, W.D.; Tian, X.H. Genus Liparis: A review of its traditional uses in China, phytochemistry and pharmacology. J. Ethnopharmacol. 2019, 234, 154–171. [Google Scholar] [CrossRef]
- Ren, J.; Xie, Y.G.; Guo, Y.G.; Yan, S.K.; Jin, H.Z. Chemical constituents of Liparis viridiflora. Chem. Nat. Compd. 2019, 55, 552–554. [Google Scholar] [CrossRef]
- Szlachetko, D.L. Systema orchidalium. Fragm. Florist. Geobot. Pol. 1995, 3, 152. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. Int. J. Org. Evol. 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Margońska, H.B. Crossoliparis—A new genus of Malaxidinae (Orchidaceae, Malaxideae), from neotropic. Acta Soc. Bot. Pol. 2009, 78, 297–299. [Google Scholar] [CrossRef][Green Version]
- Margońska, H.B.; Kowalkowska, A.K.; Górniak, M.; Rutkowski, P. Taxonomic Redefinition of the Subtribe Malaxidinae (Orchidales, Malaxideae); Koeltz Scientific Books: Koenigstein, Germany, 2012; pp. 1–606. [Google Scholar]
- Tsutsumi, C.; Yukawa, T.; Lee, N.S.; Lee, C.S.; Kato, M. Phylogeny and comparative seed morphology of epiphytic and terrestrial species of Liparis (Orchidacea) in Japan. J. Plant Res. 2007, 120, 405–412. [Google Scholar] [CrossRef]
- Li, L.; Yan, H. A remarkable new species of Liparis (Orchidaceae) from China and its phylogenetic implications. PLoS ONE 2013, 8, e78112. [Google Scholar] [CrossRef]
- Salazar, J.R.; Salazar, G.A.; Cabrera, L.I.; Nez-Machorro, R.; Batista, J.A. A new paludicolous species of Malaxis (Orchidaceae) from Argentina and Uruguay. Phytotaxa 2014, 175, 121–132. [Google Scholar] [CrossRef][Green Version]
- Tang, G.D.; Zhang, G.Q.; Hong, W.J.; Liu, Z.J.; Zhuang, X.Y. Phylogenetic analysis of Malaxideae (Orchidaceae: Epidendroideae): Two new species based on the combined nrDNA ITS and chloroplast matK sequences. Guihaia 2015, 35, 447–463. [Google Scholar]
- Li, M.H.; Zhang, G.Q.; Lan, S.R.; Liu, Z.J. A molecular phylogeny of Chinese orchids. J. Syst. Evol. 2016, 54, 349–362. [Google Scholar] [CrossRef]
- Li, L.; Chung, S.W.; Li, B.; Zeng, S.J.; Yan, H.F.; Li, S.J. New insight into the molecular phylogeny of the genus Liparis s.l. (Orchidaceae: Malaxideae) with a new generic segregate: Blepharoglossum. Plant Syst. Evol. 2020, 306, 54. [Google Scholar] [CrossRef]
- Kumar, P.; Li, J.; Gale, S.W. Integrative analyses of Crepidium (Orchidaceae, Epidendroideae, Malaxideae) shed more light on its relationships with Dienia, Liparis and Malaxis and justify reinstatement of narrow endemic C. Allanii. Bot. J. Linn. Soc. 2022, 198, 285–305. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; Depamphilis, C.W.; Kai, F.M.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Liu, D.K.; Tu, X.D.; Zhao, Z.; Zeng, M.Y.; Zhang, S.; Ma, L.; Zhang, G.Q.; Wang, M.M.; Liu, Z.J.; Lan, S.R.; et al. Plastid phylogenomic data yield new and robust insights into the phylogeny of Cleisostoma–Gastrochilus clades (Orchidaceae, Aeridinae). Mol. Phylogenet. Evol. 2020, 145, 106729. [Google Scholar] [CrossRef]
- Tu, X.D.; Liu, D.K.; Xu, S.W.; Zhou, C.Y.; Gao, X.Y.; Zeng, M.Y.; Zhang, S.; Chen, J.L.; Ma, L.; Zhou, Z.; et al. Plastid phylogenomics improves resolution of phylogenetic relationship in the Cheirostylis and Goodyera clades of Goodyerinae (Orchidoideae, Orchidaceae). Mol. Phylogenet. Evol. 2021, 164, 107269. [Google Scholar] [CrossRef]
- Chen, J.L.; Wang, F.; Zhou, C.Y.; Ahmad, S.; Zhou, Y.Z.; Li, M.H.; Liu, Z.J.; Peng, D.H. Comparative phylogenetic analysis for Aerides (Aeridinae, Orchidaceae) based on six complete plastid genomes. Int. J. Mol. Sci. 2023, 24, 12473. [Google Scholar] [CrossRef]
- Yan, R.; Gu, L.; Qu, L.; Wang, X.; Hu, G. New Insights into phylogenetic relationship of Hydrocotyle (Araliaceae) based on plastid genomes. Int. J. Mol. Sci. 2023, 24, 16629. [Google Scholar] [CrossRef]
- Huang, H.X.; Chen, L.J.; Liu, Z.J.; Li, M.H. Liparis vivipara (Orchidaceae: Malaxideae), a new species from China: Evidence from morphological and molecular analyses. Phytotaxa 2018, 351, 289–295. [Google Scholar] [CrossRef]
- Ha, Y.H.; Gil, H.Y.; Lee, J.; Kim, D.K.; Choi, K.; Chang, K.S.; Oh, S.H. The complete chloroplast genome sequence of Liparis yongnoana, an endemic orchid of Korea. Mitochondrial DNA B Resour. 2019, 4, 2666–2667. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, J.F.; Chen, M.H. The complete chloroplast genome sequence of Oberonia seidenfadenii (Orchidaceae), a rare plant species endemic to China. Mitochondrial DNA B Resour. 2019, 4, 3362–3363. [Google Scholar] [CrossRef]
- Zhang, Q.H.; Wang, X.T.; Zheng, S.Z.; Zhu, W.Y.; Song, Z. The complete chloroplast genome of Liparis nervosa (Orchidaceae). Mitochondrial DNA B Resour. 2020, 5, 123–124. [Google Scholar] [CrossRef]
- Liu, C.Q.; Kang, N.; Liu, X.; Chen, Y.; Tao, Y.; Zhang, Y.; Zhang, Y.Y.; Li, Y.L.; Tang, G.D.; Li, Y.L. Complete plastid genome sequence of Oberonioides microtatantha (Schltr.) Szlach.(Orchidaceae), an endemic herb in China. Mitochondrial DNA B Resour. 2021, 6, 703–704. [Google Scholar] [CrossRef]
- Lin, C.S.; Chen, J.J.W.; Huang, Y.T.; Chan, M.T.; Daniell, H.; Chang, W.J.; Hsu, C.T.; Liao, D.C.; Wu, F.H.; Lin, S.Y.; et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 2015, 5, 9040. [Google Scholar] [CrossRef]
- Niu, Z.; Zhu, S.; Pan, J.; Li, L.; Sun, J.; Ding, X. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci. Rep. 2017, 7, 2073. [Google Scholar]
- Zhao, Z.; Zeng, M.Y.; Wu, Y.W.; Li, J.W.; Zhou, Z.; Liu, Z.J.; Li, M.H. Characterization and comparative analysis of the complete plastomes of five Epidendrum (Epidendreae, Orchidaceae) species. Int. J. Mol. Sci. 2023, 24, 14437. [Google Scholar] [CrossRef]
- Lin, C.S.; Chen, J.J.W.; Chiu, C.C.; Hsiao, H.C.W.; Yang, C.J.; Jin, X.H.; Leebens-Mack, J.; de Pamphilis, C.W.; Huang, Y.T.; Yang, L.H.; et al. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 2017, 90, 994–1006. [Google Scholar] [CrossRef]
- Zhou, C.Y.; Zeng, M.Y.; Gao, X.; Zhao, Z.; Li, R.; Wu, Y.; Liu, Z.J.; Zhang, D.; Li, M.H. Characteristics and comparative analysis of seven complete plastomes of Trichoglottis s.l. (Aeridinae, Orchidaceae). Int. J. Mol. Sci. 2023, 24, 14544. [Google Scholar] [CrossRef]
- Zavala-Páez, M.; Vieira, L.D.N.; Baura, V.A.D.; Balsanelli, E.; Souza, E.M.D.; Cevallos, M.C.; Smidt, E.D.C. Comparative plastid genomics of neotropical Bulbophyllum (Orchidaceae; Epidendroideae). Front. Plant Sci. 2020, 11, 799. [Google Scholar] [CrossRef]
- Yang, J.B.; Tang, M.; Li, H.T.; Zhang, Z.R.; Li, D.Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013, 13, 84. [Google Scholar] [CrossRef]
- Downie, S.R.; Jansen, R.K. A comparative analysis of whole plastid genomes from the Apiales: Expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 2015, 40, 336–351. [Google Scholar] [CrossRef]
- Weng, M.L.; Ruhlman, T.A.; Jansen, R.K. Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 2017, 214, 842–851. [Google Scholar] [CrossRef]
- Liu, Q.P.; Dou, S.J.; Ji, Z.J.; Xue, Q.Z. Synonymous codon usage and gene function are strongly related in Oryza sativa. Biosystems 2005, 80, 123–131. [Google Scholar] [CrossRef]
- Quax, T.E.F.; Claassens, N.J.; Söll, D.; van der Oost, J. Codon bias as a means to fine-tune gene expression. Molec. Cell 2015, 59, 149–161. [Google Scholar] [CrossRef]
- Liu, H.; Ye, H.; Zhang, N.; Ma, J.; Wang, J.; Hu, G.; Li, M.; Zhao, P. Comparative analyses of chloroplast genomes provide comprehensive insights into the adaptive evolution of Paphiopedilum (Orchidaceae). Horticulturae 2022, 8, 391. [Google Scholar] [CrossRef]
- Knill, T.; Reichelt, M.; Paetz, C.; Gershenzon, J.; Binder, S. Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation. Plant Mol. Biol. 2009, 71, 227–239. [Google Scholar] [CrossRef]
- Hildebrandt, T.; Nunes-Nesi, A.; Araujo, W.; Braun, H.-P. Amino Acid Catabolism in Plants. Mol. Plant 2015, 8, 1563–1579. [Google Scholar] [CrossRef]
- Xiao, T.; He, L.; Yue, L.; Zhang, Y.; Lee, S.Y. Comparative phylogenetic analysis of complete plastid genomes of Renanthera (Orchidaceae). Front. Genet. 2022, 13, 998575. [Google Scholar] [CrossRef]
- Smidt, E.C.; Páez, M.Z.; Vieira, L.D.N.; Viruel, J.; De Baura, V.A.; Balsanelli, E.; Maltempi de Souza, E.; Chase, M.W. Characterization of sequence variability hotspots in Cranichideae plastomes (Orchidaceae, Orchidoideae). PLoS ONE 2020, 15, e227991. [Google Scholar] [CrossRef]
- Andrews, S. FASTQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 12 March 2022).
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; de Pamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Qu, X.J.; Moore, M.J.; Li, D.Z.; Yi, T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Brudno, M.; Do, C.B.; Cooper, G.M.; Kim, M.F.; Davydov, E.; Green, E.D.; Sidow, A.; Batzoglou, S. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003, 13, 721–731. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Rissman, A.I.; Mau, B.; Biehl, B.S.; Darling, A.E.; Glasner, J.D.; Perna, N.T. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 2009, 25, 2071–2073. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Li, W.X.; Jakovlić, I.; Zou, H.; Zhang, J.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z. DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 2001, 92, 371–373. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools-an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 289660. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Freudenstein, J.V.; Rasmussen, F.N. What does morphology tell us about orchid relationships?—A cladistic analysis. Am. J. Bot. 1999, 86, 225–248. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP: Phylogenetic Analysis Using Parsimony and Other Methods; Version 4; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, M.-Y.; Li, M.-H.; Lan, S.; Yin, W.-L.; Liu, Z.-J. Comparative Phylogenomic Study of Malaxidinae (Orchidaceae) Sheds Light on Plastome Evolution and Gene Divergence. Int. J. Mol. Sci. 2024, 25, 11181. https://doi.org/10.3390/ijms252011181
Zeng M-Y, Li M-H, Lan S, Yin W-L, Liu Z-J. Comparative Phylogenomic Study of Malaxidinae (Orchidaceae) Sheds Light on Plastome Evolution and Gene Divergence. International Journal of Molecular Sciences. 2024; 25(20):11181. https://doi.org/10.3390/ijms252011181
Chicago/Turabian StyleZeng, Meng-Yao, Ming-He Li, Siren Lan, Wei-Lun Yin, and Zhong-Jian Liu. 2024. "Comparative Phylogenomic Study of Malaxidinae (Orchidaceae) Sheds Light on Plastome Evolution and Gene Divergence" International Journal of Molecular Sciences 25, no. 20: 11181. https://doi.org/10.3390/ijms252011181
APA StyleZeng, M.-Y., Li, M.-H., Lan, S., Yin, W.-L., & Liu, Z.-J. (2024). Comparative Phylogenomic Study of Malaxidinae (Orchidaceae) Sheds Light on Plastome Evolution and Gene Divergence. International Journal of Molecular Sciences, 25(20), 11181. https://doi.org/10.3390/ijms252011181






