Molecular Biomarkers in Prediction of High-Grade Transformation and Outcome in Patients with Follicular Lymphoma: A Comprehensive Systemic Review
Abstract
1. Introduction
2. Literature Search
- (i)
- None, if the biomarker was investigated but no statistically significant impact on prognosis or risk of transformation was reported,
- (ii)
- Favorable, if the biomarker was associated with superior prognosis or lower risk of transformation,
- (iii)
- Inferior, if the biomarker was associated with worse prognosis or higher risk of transformation.
3. Review of Studies of Putative Biomarkers
3.1. Genetic Abnormalities
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| Genomic or karyotypic changes | [15] | [16,17,18,19,20] | [21,22,23] | |||
| Increasing number of mutations | [24,25] | [21] | [21,26,27,28,29,30] | [21,31,32,33,34] | ||
| M7-FLIPI [35] | [24] | [26,34,35,36] | [27,32,33,37,38,39,40] | |||
| TNFRSF14-KMT2D-HIST1H1E-FLIPI [41] | [41] | |||||
| DLBCL-like mutational status [42] | [42] | |||||
| TT genetic subtype (NFκB members and TP53) [43] | [43] | |||||
3.1.1. Cytogenetic Abnormalities
| Reported Risk of Transformation | Reported Prognostic Value | Reported Risk of Transformation | Reported Prognostic Value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Losses on | Gains on | ||||||||||
| Favorable | Inferior | None | Favorable | Inferior | None | Favorable | Inferior | None | Favorable | Inferior | None | |
| 1p | [18,47,48] | [19] | [19] | |||||||||
| 1p36 | [46] | [24] | [33,45,46] | [53] | ||||||||
| 1q | [18,19] | [49] | [46] | [46,47] | [17,19] | |||||||
| 2 | [49] | [47] | ||||||||||
| 2p | [18,47] | [64] | [20,49] | [64] | ||||||||
| 2q | [20] | |||||||||||
| 3p | [47] | |||||||||||
| 3q | [47] | [49] | [49] | |||||||||
| 3q27 | [19] | [18] | ||||||||||
| 4 | [47] | |||||||||||
| 4p | [53] | |||||||||||
| 4q | [18] | [17] | ||||||||||
| 5 | [49] | [47,65] | ||||||||||
| 5p | [46] | [46,49,50] | ||||||||||
| 5q | [18,53] | [49] | ||||||||||
| 6 | ||||||||||||
| 6p | [49] | [46] | [33] | [46,47] | ||||||||
| 6q | [49] | [18,46] | [24] | [16,17,18,46,48,50,65] | [19,47,53] | |||||||
| 7 | [64] | [18,19,47,64,65] | ||||||||||
| 7p | [46] | [22,46] | [17,51] | |||||||||
| 7q | [46] | [17,46] | ||||||||||
| 8 | [47] | |||||||||||
| 8p | [51] | |||||||||||
| 8q | [66] | [20,66] | [46] | [64] | [16,46] | [64] | ||||||
| 9p | [16,50] | |||||||||||
| 9q | [53] | |||||||||||
| 10p | [47] | |||||||||||
| 10q | [46] | [18,19,46,47,53] | [19] | |||||||||
| 11q | [50] | [51] | ||||||||||
| 12 | [22,47] | [18,19,65] | ||||||||||
| 12p | [47] | [17] | ||||||||||
| 12q | [46,49] | [64] | [49] | [18,46,64] | ||||||||
| 13 | [65] | |||||||||||
| 13p | [47] | |||||||||||
| 13q | [17,18,19,53] | |||||||||||
| 15 | [47,65] | |||||||||||
| 16p | [20] | |||||||||||
| 17p | [18] | [46] | [18,20,46,47,48,53] | [17] | ||||||||
| 17q | [19,47,53] | [18] | [46] | [20,46] | [47] | |||||||
| 18 | [52] | [18,19,47] | ||||||||||
| 18p | [46] | [17,46] | ||||||||||
| 18q | [47,53] | [46,64] | [33,51] | [17,46,64] | ||||||||
| 19p | [46] | [46] | ||||||||||
| 21 | [49] | [19] | [47] | |||||||||
| 22 | [47] | |||||||||||
| 22q | [46] | [20] | [46] | |||||||||
| X | [16,47,49] | [18,19,65] | ||||||||||
| Xp | [16] | [17] | ||||||||||
| Xq | [17] | |||||||||||
3.1.2. Gene Variants
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| Uniparental disomy | ||||||
| Number of abnormalities | [64] | |||||
| 1p36 | [64] | |||||
| 6p | [64] | [64] | ||||
| 6q | [64] | [64] | ||||
| 10q | [64] | [64] | ||||
| 12q | [64] | [64] | ||||
| 16p | [64] | [64] | ||||
| Gene rearrangements | ||||||
| Number of structural rearrangements | [21] | [21] | ||||
| BCL6 | [60] | [58] | [61] | [59] | ||
| LAZ3 | [82] | |||||
| MYC | [66,83] | [62] | [66] | [58,62,84,85] | ||
| Copy number changes | ||||||
| BCL2 | [62] | [62] | ||||
| BCL6 | [62] | [62] | ||||
| IRF4 | [62] | [86] | ||||
| MYC | [62,66] | [62] | [66] | |||
| Reported Risk of Transformation | Reported Prognostic Value | Reported Risk of Transformation | Reported Prognostic Value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Favorable | Inferior | None | Favorable | Inferior | None | Gene | Favorable | Inferior | None | Favorable | Inferior | None |
| ABL2 | [51] | HSP27 | [73] | ||||||||||
| ACTA | [73] | HSP40 | [73] | ||||||||||
| ACTB | [21,73] | HTR2B | [73] | ||||||||||
| ADAM17 | [103] | HVCN1 | [38] | [21] | |||||||||
| APEX1 | [104] | ID2 | [73] | ||||||||||
| ARHGEF1 | [21] | IFNGR1 | [105] | [92] | |||||||||
| ARID1A | [81] | [35] | [69] | [21,26,27,33,51] | IGHV | [106] | |||||||
| ARID1B | [21,33] | IGHV1 | [107] | ||||||||||
| ATP6AP1 | [21] | IGHV3 | [107] | ||||||||||
| ATP6V1B2 | [21,33,38] | IGHV4 | [107] | ||||||||||
| B2M | [41] | [21] | [27,32] | IGHV5 | [107] | ||||||||
| BACH2 | [21] | IGHV6 | [107] | ||||||||||
| BCL2 | [72] | [69,70,71,72] | [21,26,33,38,51,52,73] | IGLL5 | [32,33] | ||||||||
| BCL6 | [75] | [21,73,74,75] | IKZF3 | [21] | |||||||||
| BCL7A | [24] | [21,38] | IL10 | [28,108] | |||||||||
| BCR | [21] | IL12A | [108] | ||||||||||
| BHMT | [104] | IL12B | [28,92] | ||||||||||
| BIM | [109] | IL13 | [28] | ||||||||||
| BMP6 | [73] | IL16 | [28] | ||||||||||
| BMP7 | [103] | IL17A | [108] | ||||||||||
| BRCA1 | [104] | IL17F | [108] | ||||||||||
| BRCA2 | [104] | IL1RN | [28] | [92] | |||||||||
| BTG1 | [21] | [27] | IL2 | [92] | [28] | [108] | |||||||
| BTG2 | [21] | [21,38] | IL2RG | [73] | |||||||||
| BTK | [69] | [38] | IL3 | [28] | |||||||||
| C1QA | [110] | [110] | IL4R | [105] | [21,92,105] | ||||||||
| C1QB | [111] | IL5 | [28] | [105] | |||||||||
| C1QC | [111] | IL6 | [105] | ||||||||||
| C1QTNF7 | [111] | IL7R | [105] | ||||||||||
| C1RL | [111] | IL8 | [92] | [105] | [28,105] | ||||||||
| C1S | [111] | IL8RB | [28] | ||||||||||
| C2 | [111] | IRF4 | [21] | ||||||||||
| C3 | [111] | IRF8 | [69] | [24] | [21,26,27,33,38] | ||||||||
| C3AR1 | [111] | IL10 | [105] | ||||||||||
| C4BPA | [111] | IL12B | [105] | ||||||||||
| C5 | [111] | IL16 | [105] | ||||||||||
| C5AR1 | [111] | ITPKB | [21] | ||||||||||
| C6 | [111] | JUN | [73] | ||||||||||
| C6orf15 | [112] | [112] | [92] | KLHL6 | [21] | ||||||||
| C7 | [111] | KMT2C | [21] | ||||||||||
| C8B | [111] | KMT2D [MLL2] | [41] | [25] | [21,26,33,38,51,52] | ||||||||
| C9 | [111] | [111] | [92] | LGMN | [48] | ||||||||
| CARD11 | [35] | [21,26,27,33,38,52] | LIG4 | [104] | |||||||||
| CARD15 | [105] | LRRC7 | [21] | ||||||||||
| CBS | [104] | LRRN3 | [38] | ||||||||||
| CCDC129 | [38] | MAP3K11 | [73] | ||||||||||
| CCNB | [73] | MASP2 | [111] | ||||||||||
| CCND3 | [71] | [21] | MBD2 | [104] | |||||||||
| CCR2 | [105] | MBL2 | [111] | ||||||||||
| CCR4 | [37] | MDM2 | [113] | [113] | |||||||||
| CD46 | [92,111] | MEF2B | [21,27,38,52] | ||||||||||
| CD55 | [111] | [92] | MEF2C | [21] | |||||||||
| CD59 | [111] | MGMT | [104] | ||||||||||
| CD69 | [48] | MIF | [105] | [92] | |||||||||
| CD79B | [21,33,38] | MINOR | [73] | ||||||||||
| CD83 | [21] | MKI67 | [21] | ||||||||||
| CD8A | [37,48] | MLH1 | [97] | [104] | |||||||||
| CD8B | [37] | MSH2 | [104] | ||||||||||
| CD93 | [111] | MTHFD2 | [104] | ||||||||||
| CDC2 | [73] | MTHFR | [92] | [104] | |||||||||
| CDKN1A | [73] | MTHFS | [104] | ||||||||||
| CDKN2A | [69] | [69,89,90] | [90] | MTR | [104] | ||||||||
| CFB | [111] | MTRR | [104] | ||||||||||
| CFD | [111] | MUC4 | [38] | ||||||||||
| CFH | [111] | [111] | [92] | MYC | [21,73] | ||||||||
| CFHR1 | [111] | MYD88 | [21] | [32,52] | |||||||||
| CFHR5 | [111] | [92] | MYOM2 | [21] | |||||||||
| CHI3L1 | [114] | NBS1 | [104] | ||||||||||
| CHD8 | [21] | NCOR2 | [33] | ||||||||||
| CIITA | [21] | NLRC5 | [21] | ||||||||||
| CLU | [111] | NOTCH1 | [21] | ||||||||||
| COL3A1 | [73] | NOTCH2 | [24,41] | [21,33] | |||||||||
| CR1 | [111] | NPM3 | [73] | ||||||||||
| CR2 | [111] | NR2F6 | [73] | ||||||||||
| CREBBP | [25,41] | [76] | [25] | [32,38,76] | [21,26,27,33,51,52] | OVGL | [73] | ||||||
| CSMD3 | [41] | ||||||||||||
| CTLA4 | [105] | P2RY8 | [21] | ||||||||||
| CTSS | [115] | [21] | PDCD4 | [73] | |||||||||
| CX3CR1 | [114] | PIEZO1 | [73] | ||||||||||
| CXCR3 | [37] | PIM1 | [34] | [21,27,38] | |||||||||
| CXCR4 | [38] | PLAU | [73] | ||||||||||
| CXCR5 | [116] | [116] | [116] | [116] | POU2AF1 | [21,38] | |||||||
| CYBA | [97] | POU2F2 | [38] | ||||||||||
| CYHR1 | [33] | PRF1 | [37] | ||||||||||
| DEFB115 | [38] | PRKCB | [73] | ||||||||||
| DNAH9 | [21] | PRKCG | [73] | ||||||||||
| DTX1 | [24] | [21] | PSMB1 | [91] | |||||||||
| DUSP2 | [103] | PSMB5 | [91] | ||||||||||
| DUXA | [38] | PSMB8 | [91] | ||||||||||
| E2FS | [117] | PSMB9 | [91] | ||||||||||
| EBF1 | [21,27] | RAD23B | [104] | ||||||||||
| EBF3 | [21] | RAG1 | [104] | ||||||||||
| EEF1A1 | [21] | RFX5 | [21] | ||||||||||
| EIF2B | [73] | RHOA | [21] | ||||||||||
| EML6 | [117] | RHOH | [21] | ||||||||||
| EOMES | [37] | RPS9 | [73] | ||||||||||
| EP300 | [41] | [27,35] | [21,26,38,52] | RRAGC | [21] | ||||||||
| ERCC1 | [104] | S1PR2 | [21] | ||||||||||
| ERCC2 | [104] | SELE | [105] | [92] | |||||||||
| ERCC4 | [104] | SERPING1 | [111] | ||||||||||
| ERCC5 | [104] | SGK1 | [21,27] | ||||||||||
| EVI2A | [21] | SHMT1 | [104] | ||||||||||
| EZH2 | [41] | [25] | [25,79] | [26,35,52,77,78] | [21,27,33,38,79,80] | SLC19A1 | [104] | ||||||
| FAS | [21] | [27,52] | LC25A23 | [33] | |||||||||
| FAT4 | [21] | SMAD1 | [73] | ||||||||||
| FCGR2A | [28,91] | [91,92,93,94,95,96,97] | SMARCA4 | [21,26] | |||||||||
| FCGR2B | [95] | [95] | [95] | SOCS1 | [21,32] | [27,33] | |||||||
| FCGR3A | [91,96,118] | [96] | [93,94,95,97,98] | SORT1 | [33] | ||||||||
| FLT3LG | [37] | STAT3 | [21] | ||||||||||
| FOXO1 | [26,34,35] | [27] | STAT6 | [21,26,32,33,38,51] | |||||||||
| FPGS | [104] | TBX-21 | [37] | ||||||||||
| FTHFD | [104] | [92] | TCF3 | [21] | |||||||||
| GADD45B | [103] | TCN1 | [104] | ||||||||||
| GALNT12 | [103] | [92] | TGFB1 | [108] | |||||||||
| GAPDH | [73] | TGFBR1 | [108] | ||||||||||
| GGH | [104] | [92] | TGFBR2 | [108] | |||||||||
| GNA13 | [69] | [21,26,27,38] | TLE1 | [73] | |||||||||
| GNAI2 | [21] | TNF/LTA | [73] | [28] | |||||||||
| GSTA1 | [97] | TNFAIP3 | [21,52] | ||||||||||
| GSTM1 | [119] | TNFRSF14 | [41] | [45] | [120] | [45] | [21,26,33,38,48,52] | ||||||
| GSTT1 | [119] | TP53 | [41,69] | [113] | [21,24,32,35,57,69,74,121] | [27,33,38,104,113] | |||||||
| GZMM | [37] | TPTE2 | [38] | ||||||||||
| GZMK | [37] | TYMS | [104] | ||||||||||
| HIST1H2AC | [69] | UBE2A | [24,41] | ||||||||||
| HIST1H1B | [21] | UNC5C | [21] | ||||||||||
| HIST1H1C | [21,33,38] | USP44 | [117] | ||||||||||
| HIST1H1D | [33] | [38] | VEGFA | [122] | |||||||||
| HIST1H1E | [24,41] | [21,26,33,38] | VMA21 | [38] | |||||||||
| HIST1H2AM | [21,38] | WRN | [104] | ||||||||||
| HIST1H2BK | [38] | XBP1 | [21] | [27] | |||||||||
| HIST1H3G | [38] | XPB | [73] | ||||||||||
| HIST2H2AC | [38] | XPC | [104] | ||||||||||
| HLA-A | [123] | XRCC1 | [104] | ||||||||||
| HLA-B | [123] | XRCC2 | [104] | ||||||||||
| HLA-C | [123] | XRCC3 | [104] | ||||||||||
| HLA-DMB | [21] | XRCC4 | [104] | ||||||||||
| HLA-DRA | [73] | YY-1 | [73] | ||||||||||
| HLA-DRB | [123] | ZFP36L1 | [21] | ||||||||||
| HLA-DQA | [73] | ZFPC150 | [73] | ||||||||||
| HLA-DQB1 | [112] | [112] | ZFX | [73] | |||||||||
| HNRNPU | [38] | ZNF608 | [38] | ||||||||||
| HSF1 | [73] | ||||||||||||
3.2. Gene Expression
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| Pluripotency/embryonic stem cell-like signature [130] | [130] | [130] | ||||
| 23-GEP risk score [126] | [34,117,126,127] | [37,53] | ||||
| ICA13 [126] | [126] | |||||
| 33 gene-based ABC-like FL signature [131] | [131] | |||||
| 33 gene-based GCB-like FL signature [131] | [131] | |||||
| T-cell associated immune infiltration signature [37] | [37] | |||||
| T effector signature [132] | [132] | [53] | ||||
| T cell exhaustion signature [135] | [135] | |||||
| NFκB-linked signature [128] | [128,129] | [128,129] | ||||
| Somatic hypermutation signature [69] | [69] | [69] | ||||
| m6A score, low [136] | [136] | |||||
| FL loci risk score [133] | [133] | |||||
| MAP signature [32] | [32] | |||||
| STAT signature score [134] | [134] | |||||
| Immune-related 1 [124] | [124] | [53,125] | ||||
| Immune-related 2 [124] | [124,137] | [53,125] | ||||
| Localized-stage FL signature [125] | [125] | |||||
| TH17-axis related [138] | [138] | |||||
| Reported Risk of Transformation | Reported Prognostic Value | Reported Risk of Transformation | Reported Prognostic Value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Favorable | Inferior | None | Favorable | Inferior | None | Gene | Favorable | Inferior | None | Favorable | Inferior | None |
| ABHD6 | [137] | KLRB1 | [140] | ||||||||||
| ACTB | [142] | KNT2C | [143] | ||||||||||
| ACTN1 | [142] | KIAA0100 | [137] | ||||||||||
| AKAP12 | [142] | KIAA0101 | [143] | ||||||||||
| AKIRIN1 | [137] | KIAA0317 | [144] | ||||||||||
| AKT | [145] | KIAA1223 | [144] | ||||||||||
| ALDH1L1 | [137] | KRT19 | [137] | ||||||||||
| ANP32E | [143] | LAG3 | [139] | ||||||||||
| ARFGEF1 | [137] | LEF1 | [142] | ||||||||||
| ARPC2 | [137] | LGMN | [142] | ||||||||||
| ASAP2 | [137] | LIPA | [140] | ||||||||||
| ATPAF2 | [137] | LPP | [137] | ||||||||||
| BCL-XL | [146] | [145] | LYN | [140] | |||||||||
| BCL6 | [145] | MAL2 | [140] | ||||||||||
| BLCAP | [144] | MALAT1 | [141] | [141] | |||||||||
| BLNK | [144] | MAP3K7 | [128] | ||||||||||
| BNIP2 | [137] | MAPK1 | [142] | ||||||||||
| BRI3BP | [144] | MARCO | [147] | ||||||||||
| BTD | [137] | MED6 | [137] | ||||||||||
| BTK | [128] | MED8 | [137] | ||||||||||
| BTN2A3P | [137] | MLPH | [144] | ||||||||||
| BUB1B | [143] | MMP9 | [148] | ||||||||||
| C1S | [144] | MOX2 | [144] | ||||||||||
| C1QR1 | [144] | MYC | [145] | ||||||||||
| C3AR1 | [142] | MZB1 | [149] | ||||||||||
| C4B | [144] | NCF4 | [144] | ||||||||||
| CAV1 | [144] | NEK2 | [142] | ||||||||||
| CBFA2T2 | [137] | NFIB | [148] | ||||||||||
| CCL19 | [140] | [148] | NFκB | [145] | |||||||||
| CCL20 | [140] | NGFRAP1 | [144] | ||||||||||
| CCL3 | [142] | NK4 | [144] | ||||||||||
| CCL5 | [142] | NPDC1 | [144] | ||||||||||
| CCL8 | [142] | NSDHL | [137] | ||||||||||
| CCNA2 | [143] | PAICS | [143] | ||||||||||
| CCNB1 | [143] | PBX1 | [140] | ||||||||||
| CCNB2 | [143] | PD-1 | [139] | ||||||||||
| CCND1 | [144] | PD-L1 | [139] | ||||||||||
| CCR1 | [142] | PD-L2 | [139] | [53] | |||||||||
| CD101 | [140] | PIM1 | [134] | ||||||||||
| CD11d | [142] | PLA2G2D | [148] | ||||||||||
| CD137 | [139] | PPP4R1 | [128] | ||||||||||
| CD138 | [140] | PRB1 | [137] | ||||||||||
| CD19 | [142] | PRH1 | [137] | ||||||||||
| CD2 | [140] | [142] | PSMF1 | [137] | |||||||||
| CD3 | [142,150] | PTAFR | [137] | ||||||||||
| CD31/ PECAM1 | [151] | PTEN | [145] | ||||||||||
| CD34 | [151] | PTGDS | [148] | ||||||||||
| CD3D | [142] | PTP4A2 | [137] | ||||||||||
| CD4 | [139] | [142] | PTPRB | [140] | |||||||||
| CD47 | [142] | PTPRC | [140] | ||||||||||
| CD5 | [142] | PTPRF | [140] | ||||||||||
| CD6 | [142] | PTPRM | [144] | ||||||||||
| CD68 | [139] | [142] | RAB27B | [140] | |||||||||
| CD69 | [140] | [142] | RAB38 | (144) | |||||||||
| CD7 | [139,142] | RANBP9 | [137] | ||||||||||
| CD8 | [150] | RET | [144] | ||||||||||
| CD8A | [139] | RGL1 | [148] | ||||||||||
| CD8B | [142] | RPS9 | [142] | ||||||||||
| CD9 | [140] | ROCK1 | [128] | ||||||||||
| CDC2 | [143] | RRM2B | [144] | ||||||||||
| CDC40 | [137] | SEP-10 | [142] | ||||||||||
| CDC42BPK | [140] | SH2D1A | [140] | ||||||||||
| CDK2 | [142] | SIRT5 | [137] | ||||||||||
| CDKN3 | [143] | SLC21A9 | [144] | ||||||||||
| CKS1B | [143] | SLC24A2 | [137] | ||||||||||
| CRY1 | [144] | SLC7A11 | [137] | ||||||||||
| CXCL1 | [140] | SLP1 | [140] | ||||||||||
| CXCR6 | [140] | SMAD1 | [147] | ||||||||||
| CXCL12 | [142] | SNX9 | [144] | ||||||||||
| DAAM2 | [144] | SOCS1 | [134] | ||||||||||
| DNAAF1 | [137] | SOCS3 | [134] | ||||||||||
| DUSP6 | [144] | SPP1 | [144] | ||||||||||
| ELF3 | [140] | SSI-3 | [144] | ||||||||||
| EPHA1 | [147] | ST14 | [144] | ||||||||||
| EVA1B | [137] | STAT2 | [134] | ||||||||||
| EZH2 | [77] | STAT3 | [134] | ||||||||||
| FASTKD1 | [128] | STAT4 | [140] | [134,142] | |||||||||
| FCGR1A | [142] | STAT5a | [134] | ||||||||||
| FOXP3 | [139] | STAT6 | [134] | ||||||||||
| FREB | [144] | TAB2 | [128] | ||||||||||
| FRYL | [137] | TAF12 | [137] | ||||||||||
| GAPDH | [142] | TBK1 | [128] | ||||||||||
| GEM | [142] | TCP10L | [137] | ||||||||||
| GLE1L | [144] | TDRD12 | [137] | ||||||||||
| GMDS | [143] | TIA-1 | [140] | ||||||||||
| GZM-K | [140] | TIM3 | [139] | ||||||||||
| H2BFB | [144] | TIMP3 | [148] | ||||||||||
| H2BFG | [144] | TLR5 | [142] | ||||||||||
| HMGB2 | [143] | TM4SF1 | [140] | ||||||||||
| HMMR (RHAMM) | [143] | TMED7-TICAM2 | [128] | ||||||||||
| HSF2 | [137] | TMEM70 | [137] | ||||||||||
| IDH3A | [137] | TMP3 | [140] | ||||||||||
| IDO1 | [132] | TNF-alfa | [139] | ||||||||||
| IF2B | [142] | TNFSF13B | [144] | [142] | |||||||||
| IFITM1 | [144] | TNFRSF14 | [152] | ||||||||||
| IFN-γ | [140] | TNFSF10 | [144] | ||||||||||
| IGBP1 | [128] | TOP2A | [143] | ||||||||||
| IKBKG | [128] | TOP2B | [137] | ||||||||||
| IL1R | [140] | TRBα | [144] | ||||||||||
| IL2 | [134] | TRIM37 | [128] | ||||||||||
| IL2Rα (CD25) | [134] | [142] | TRPM4 | [137] | |||||||||
| IL4 | [134] | TSC22D3 | [128] | ||||||||||
| IL4R | [134] | TSPAN7 | [137] | [142] | |||||||||
| IL7 | [134] | TTLL3 | [137] | ||||||||||
| IL7R | [134] | TYROBP | [144] | ||||||||||
| ILF3 | [142] | UACA | [144] | ||||||||||
| INPP5B | [53] | UBQLN1 | [144] | ||||||||||
| IRAK1 | [128] | USP11 | [128] | ||||||||||
| ITK | [137] | [142] | VEGF | [151] | |||||||||
| JAK2 | [134] | YAP1 | [148] | ||||||||||
| JUNB | [144] | ZNF230 | [137] | ||||||||||
| KLK10 | [137] | ||||||||||||
3.3. MicroRNAs
3.4. B Cells/FL Tumor Cells
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| B cells | ||||||
| BCL2 | [168] | [169] | [48,61,170,171,172,173,174,175,176,177] | |||
| pBCL2 | [184] | |||||
| CD19 | [185] | |||||
| CD20 | [169] | [142,186,187] | ||||
| Interfollicular CD20 | [140] | [58,188] | ||||
| CD21 | [177] | |||||
| CD37 | [189] | [189] | ||||
| CD69 | [190] | |||||
| CD79a | [169] | |||||
| FOXP1 | [162,163] | [171,179] | ||||
| HVEM (TNFRSF14) | [152] | [152] | ||||
| Follicular HVEM | [152] | |||||
| Interfollicular HVEM | [152] | |||||
| MUM1 (IRF4) | [60] | [182,183] | [61,171,172,175] | |||
| OCT2 | [169] | |||||
| PAX5 | [181] | [181] | ||||
| Germinal center cells | ||||||
| BCL6 | [60] | [178] | [61,169,172,177] | |||
| Follicular BCL6 | [171] | |||||
| Interfollicular BCL6 | [171] | [179] | ||||
| CD10 | [60] | [172,178] | [61,142,169,175,177,191] | |||
| Follicular CD10 | [171,177] | |||||
| Interfollicular CD10 | [188] | [171,179] | ||||
| CD10 negative | [192] | |||||
| CD75 | [169] | |||||
| HGAL | [54] | |||||
| Serpin A9/GCET1 | [54] | |||||
| Immunoglobulins | ||||||
| IgA | [180] | |||||
| IgD | [172] | |||||
| IgG | [180] | |||||
| IgM | [180] | |||||
| κ Ig light chain | [61] | |||||
| λ Ig light chain | [61] | |||||
| Tumor phenotype | ||||||
| FL with features of pre-CSR, IgM+IgG− memory B-cells | [193] | |||||
| FL with features of normal GC B-cells | [193] | |||||
| Phenotypic diversity among malignant B-cells | [193] | |||||
| CSR, class switch recombination; GC, germinal center. | ||||||
3.5. The Tumor Microenvironment
3.5.1. T Cells
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| CD3 | [140,196,201] | [142,150,185,213,232,233,234] | [52,108,177,186,187,190,196,201,206,235,236] | |||
| Follicular CD3 | [201,237] | [196] | [52,188,190,196,201,206] | |||
| Interfollicular CD3 | [237] | [196] | [52,188,190,196,206] | |||
| CD5 | [238,239] | |||||
| CD7 | [196] | [142] | [177,196,203] | |||
| Follicular CD7 | [196] | [196] | ||||
| Interfollicular CD7 | [196] | [196] | ||||
| ZAP-70 | [187] | |||||
| CD4/CD8 ratio | [205] | [195,196] | [205] | |||
| CCR7 | [232] | |||||
| GATA3 | [195] | |||||
CD8+ T cells | ||||||
| CD8 | [201] | [140,185,196] | [52,185,196,197,198] | [240] | [142,150,177,187,190,201,203,212] | |
| Follicular CD8 | [201] | [196] | [117,187] | [108] | [52,190,196,201,215] | |
| Interfollicular CD8 | [196] | [52,196] | [117,215] | |||
| CD8+CXCR5+ | [241] | |||||
| Granzyme B | [196] | [198,199] | [171,242] | [108,196,203,216] | ||
| Follicular GrzB | [196] | [200] | [196] | |||
| Interfollicular GrzB | [196] | [200] | [196] | |||
| Granulysin | [216] | |||||
| Perforin | [196] | [108,196] | ||||
| Follicular perforin | [196] | [196] | ||||
| Interfollicular perforin | [196] | [196] | ||||
| TIA-1 | [201] | [196] | [177,196,197,201,203] | |||
| Follicular TIA-1 | [196,201] | [196,201] | ||||
| Interfollicular TIA-1 | [196] | [196] | ||||
| Tryptase | [196] | [196] | ||||
| Follicular tryptase | [196] | [196] | ||||
| Interfollicular tryptase | [196] | [196] | ||||
CD8+ Tregs | ||||||
| CD8+FOXP3+ | [216] | |||||
CD4+ T cells | ||||||
| CD4 | [201] | [196,205] | [203] | [142,196,202,207] | [52,108,150,177,185,190,197,201,204,205,206,207] | |
| Follicular CD4 | [140,201] | [196] | [117] | [196] | [52,190,201,206] | |
| Interfollicular CD4 | [140] | [196] | [117] | [52,190,196,206] | ||
CD4+FOXP3+ Tregs | ||||||
| FOXP3 | [201] | [196,205,209] | [152,206,212,213,214] | [211] | [52,150,168,171,175,183,190,196,197,201,203,205,209,210] | |
| Follicular FoxP3 | [205] | [196] | [196,206] | [108,175,205] | [52,117,190,215] | |
| Interfollicular FoxP3 | [201] | [140,196] | [117,190,203,215] | [52,196,201,206,211,216,217] | ||
| CD8/FOXP3 ratio | [215] | |||||
Activated Tregs | ||||||
| CD4+FOXP3+PD1+TIGIT+ | [218] | |||||
| CD25 | [205] | [219] | [203,205,220] | |||
| Follicular CD25 | [205] | [205] | ||||
T helper 1 cells | ||||||
| T-bet | [140] | |||||
T helper 17 cells | ||||||
| RORγt | [138] | |||||
T cell activation | ||||||
| CD27 | [220] | [169,172] | ||||
| CD28 | [220] | |||||
| CD69 | [140] | [213] | [202] | [190] | ||
| CD70 | [243] | |||||
| CD80 | [244] | |||||
| CD86 | [244] | |||||
| CD137 | [244] | |||||
| GITR | [244] | |||||
| GITRL | [244] | |||||
| ICOS | [150] | |||||
| OX40 | [244] | |||||
| OX40L | [244] | |||||
T cell phenotypes | ||||||
| CD4+CD8+ | [193] | [193,216] | ||||
| CD4+CD57+ | [232] | |||||
| CD4+CD57+PD-1low | [232] | |||||
| CD8+CD57+ | [232] | |||||
| CD4+PD1+ | [232] | |||||
| CD4+PD-1low | [245] | |||||
| CD4+PD-1high | [246] | [245] | ||||
| CD8+PD-1low | [245] | |||||
| LAG3+TIM3+ | [247] | |||||
| LAG3+PD1+ | [247] | |||||
| PD1+CXCR5−CD27+CD28+ | [220] | |||||
| PD1+CXCR5+CD27+CD28+ | [220] | |||||
| PD1+CCR4−CD27−CD28− | [220] | |||||
| PD1+CCR4+CD27−CD28− | [220] | |||||
| CD8EM/Th1-rich [193] | [193] | [193] | ||||
| Tfh-rich [193] | [193] | [193] | ||||
| Exhausted immunophenotypes [240,248] | [240,248] | |||||
Early-stage differentiation | ||||||
| Naïve CD4+ T cells | [249] | |||||
| Naïve CD8+ T cells | [249] | |||||
| CD45RA | [232] | |||||
| CD4+CD45RA+CCR7+ | [232] | |||||
| CD8+CD45RA+CCR7+ | [232] | |||||
| CD45RO−CCR7+ | [220] | |||||
Late-stage differentiation | ||||||
| CD4+CD45RA−CCR7+ | [232] | |||||
| CD45RA−CCR7+ T memory | [117] | |||||
| CD127 | [248] | |||||
| CD127+KLRG1+ | [248] | |||||
3.5.2. Immune Activation and Exhaustion
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| PD-1 | [214] | [251] | [196,209] | [196,214,250] | [168,195] | [52,108,150,209,212,220,244,247,251,252] |
| Follicular PD-1 | [209] | [201] | [196,251] | [117,150,152,196,209,246] | [251,252] | [52,171] |
| Interfollicular PD-1 | [209] | [196] | [235] | [204,209] | [52,117,171,196,216,217,246] | |
| PD-L1 | [204,244,256] | |||||
| Follicular PD-L1 | [201] | [201] | ||||
| Interfollicular PD-L1 | [216] | |||||
| PD-L2 | [244] | |||||
| TIM3 | [199] | [244] | ||||
| Follicular TIM3 | [117] | |||||
| Interfollicular TIM3 | [117] | |||||
| LAG3 | [253] | [247,253] | [244,256] | |||
| LAG3+PD-1+ | [253] | |||||
| TIGIT | [254] | |||||
| CTLA4 | [244] | |||||
| IDO1 | [255] | [255] | ||||
| Trp | [257] | |||||
| Kyn | [257] | |||||
| Galectin-9 | [244] | |||||
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| CD56 | [196] | [196] | ||||
| Follicular CD56 | [196] | [196] | ||||
| Interfollicular CD56 | [196] | [196] | ||||
| CD56/MS4A4A ratio | [246] | |||||
| CD57 | [201] | [140,196] | [202,232] | [177,196,197,201] | ||
| Follicular CD57 | [201] | [196] | [108] | [196,201] | ||
| Interfollicular CD57 | [196] | [196] | ||||
3.5.3. Follicular Dendritic Cells
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| CD21 | [258] | [140,201] | [209] | [201] | [142,190,209,258] | |
| Follicular CD21 | [201] | |||||
| CD23 | [140] | [259] | [108,190] | [61,188,212,239,260] | ||
| CD11c | [209] | [209,211,264] | ||||
| Follicular CD11c | [211] | |||||
| CD1a | [211] | |||||
| CD83 | [211] | |||||
| Ki-M4p | [260] | |||||
| PU.1 | [169] | [172] | ||||
Plasmacytoid dendritic cells | ||||||
| CD123 | [233] | [197,211] | ||||
3.5.4. Tumor-Associated Macrophages
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| CD68 | [140,177,196,209,269] | [91,212,236] | [142,168,177,266,267,268,269] | [52,108,171,190,196,197,203,204,209,235,264,268,270] | ||
| Follicular CD68 | [201] | [196] | [91,246] | [266] | [52,117,175,183,188,190,196,201,215] | |
| Interfollicular CD68 | [196] | [175,196,200] | [52,91,117,183,188,190,215,216,246,266] | |||
| CD14 | [209] | [86,202] | [209] | |||
| Follicular CD14 | [209] | [117,209] | ||||
| Interfollicular CD14 | [209] | [117,209] | ||||
| SIRPα | [86] | |||||
| Follicular SIRPα | [117] | |||||
| Interfollicular SIRPα | [117] | |||||
| CD14+SIRPa+ | [86] | |||||
Pro-inflammatory/ M1-like macrophages | ||||||
| iNOS | [108] | |||||
Anti-inflammatory/ M2-like macrophages | ||||||
| CD163 | [52] | [264,268] | [108,204] | |||
| Follicular CD163 | [52] | |||||
| Interfollicular CD163 | [52] | |||||
| CD163/CD8 ratio | [246] | |||||
| CD206 | [240] | |||||
| Interfollicular CD206 | [216] | |||||
| CSF-1R | [271] | [271] | ||||
| Follicular CSF-1R | [271] | [271] | ||||
3.5.5. Angiogenesis
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| Increased microvessel density | ||||||
| CD31 | [151,264] | [147] | ||||
| CD34 | [269] | [273] | [269] | [264,274] | ||
| Follicular CD34 | [275] | [275] | ||||
| Interfollicular CD34 | [275] | [275] | ||||
Angiogenesis | ||||||
| Estrogen receptor α | [276] | |||||
| FLT-1 | [277] | |||||
| FLT-4 | [277] | |||||
| KDR | [277] | [277] | ||||
| LYVE-1 | [264] | |||||
| Podoplanin | [264] | |||||
| PROX1 | [264] | |||||
| VEGF | [277] | |||||
| VEGF-C | [277] | |||||
| VWF | [264] | |||||
3.5.6. Energy Metabolism and Vitamin D Insufficiency
3.5.7. Cell Death
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| Other leukocyte markers | ||||||
| AID | [72] | [262] | ||||
| BAFF | [263] | |||||
| BAFFR | [263] | |||||
| BLIMP1 | [171] | |||||
| BTLA | [152] | [152] | ||||
| Follicular BTLA | [152] | |||||
| Interfollicular BTLA | [152] | |||||
| CD30 | [172] | |||||
| CD32B (FcγRIIB) | ||||||
| Follicular CD32B | [117] | |||||
| Interfollicular CD32B | [117] | |||||
| CD38 | [172] | |||||
| CD44 | [293] | |||||
| Follicular CD44 | [293] | [293] | ||||
| CD44s | [274] | |||||
| CD70 | ||||||
| Follicular CD70 | [117] | |||||
| Interfollicular CD70 | [117] | |||||
| CD9 | [294] | |||||
| ETV1 | ||||||
| Follicular ETV1 | [295] | [295] | ||||
| Interfollicular ETV1 | [295] | [295] | ||||
| HLA-DR | [242] | [190,211] | ||||
| Follicular HLA-DR | [211] | |||||
| LMO2 | [54] | |||||
| NAMPT | ||||||
| Follicular NAMPT | [295] | [295] | ||||
| Interfollicular NAMPT | [295] | [295] | ||||
| PI3Kδ | [294] | |||||
| PMCH | ||||||
| Follicular PMCH | [295] | [295] | ||||
| Interfollicular PMCH | [295] | [295] | ||||
T cell immunological synapse | ||||||
| Filamin A | ||||||
| Follicular Filamin A | [234] | |||||
| Interfollicular Filamin A | [234] | |||||
| Itk | ||||||
| Follicular Itk | [234] | |||||
| Interfollicular Itk | [234] | |||||
| RAB27A | [234] | |||||
| Follicular RAB27A | [234] | |||||
| Interfollicular RAB27A | [234] | |||||
Complement inhibitors | ||||||
| CD46 | [186] | |||||
| CD55 | [186] | |||||
| CD59 | [186] | |||||
| NFκB activity | ||||||
| p65 (RelA) | [91] | [296] | ||||
| pRB | [172] | |||||
Cytokines/chemokines | ||||||
| CCR1 | [297] | |||||
| CXCL13 | [209] | [209] | ||||
| IL-10 | [108] | |||||
| IL-12A | [108] | |||||
| IL-17A | [108] | |||||
| IL-17F | [108] | |||||
| IL-2 | [108] | |||||
| IL-21R | [298] | |||||
| TGFB1 | [108] | |||||
| TGFBR1 | [108] | |||||
The cytoskeleton and cellular migration | ||||||
| CDK6 | [172] | |||||
| FilGAP | [299] | |||||
| Integrin B2 | [299] | |||||
| RHAMM | [293] | [293] | ||||
| Follicular RHAMM | [293] | [293] | ||||
| CD44/RHAMM ratio, low | [293] | [293] | ||||
| Vimentin | [300] | [181] | [181] | |||
G protein-coupled signals | ||||||
| GNA13 | [301] | |||||
| Rac1 | [299] | |||||
Metalloproteinases | ||||||
| MMP2 | [302] | [302] | ||||
| MMP9 | [302] | [302] | ||||
| TIMP1 | [302] | [302] | ||||
| TIMP2 | [302] | [302] | ||||
Signal transduction | ||||||
| EPHA1 | [147] | |||||
| pJAK2 | [303] | |||||
| SOCS3 | [304] | [304] | ||||
| STAT5a | [134] | |||||
| pSTAT5 | [303] | |||||
| STAT1 | [270] | |||||
| CD68−STAT1+ | [270] | |||||
Peroxiredoxins | ||||||
| Peroxiredoxin, total | [305] | |||||
| PRDX1 | [305] | |||||
| PRDX2 | [305] | |||||
| PRDX3 | [305] | |||||
| PRDX4 | [305] | |||||
| PRDX5 | [305] | |||||
| PRDX6 | [305] | |||||
Oxidative stress | ||||||
| OHdG | [306] | |||||
| Gamma-GCS | [306] | |||||
| Thioredoxine | [305] | |||||
| Nitrotyrosine | [305] | |||||
| Superoxide dismutase | [306] | |||||
Cell cycle | ||||||
| ACPI | [307] | |||||
| BMI1 | [308] | |||||
| ECT2 | [299] | |||||
| CDK2 | [172] | |||||
| Cyclin A | [143,172] | |||||
| Cyclin B1 | [143] | [172] | ||||
| Cyclin D3 | [172] | |||||
| Cyclin E | [172] | |||||
| E2F6 | [172] | |||||
| EZH2 | [77] | |||||
| MDM2 | [61,172] | |||||
| MYC | [85] | [162] | ||||
| p18 | [172] | |||||
| p21 | [172] | |||||
| p27 | [91,172] | |||||
| p53 | [302] | [258] | [302,309] | [52,172,174] | ||
| Follicular p53 | [52] | |||||
| Interfollicular p53 | [52] | |||||
| P-glycoprotein | [258] | [309] | ||||
| S100 | [211] | |||||
| SKP2 | [172] | |||||
Glucose metabolism | ||||||
| GLUT1 | [240] | |||||
| Aldolase A | [300] | [281] | [281] | |||
| GAPDH | [300] | [281] | [281] | |||
| ATP synthase δ | [300] | |||||
Cell death | ||||||
| 14-3-3γ | [184] | |||||
| Akt | [184] | |||||
| pAkt | [184] | |||||
| Aurora A | [184] | |||||
| BAD | [184] | |||||
| BAK | [184] | |||||
| BAX | [289] | [172,173,184] | ||||
| BCL-rambo | [289] | [184] | ||||
| BCL-x | [173] | |||||
| BCL-xL | [289] | [172] | [177,184] | |||
| CASP3 | [289] | [184] | ||||
| CASP3a | [172] | |||||
| cCASP3 | [184] | |||||
| MCL1 | [289] | [176] | [184] | |||
| PARP | [184] | |||||
| cPARP | [184] | |||||
| SMAC | [184] | |||||
| Survivin | [172,184] | |||||
| XIAP | [184] | |||||
| BCL2/BAK ratio | [184] | |||||
| BCL2/BAX ratio | [184] | |||||
| YY1 | [290] | |||||
| YY1/PLK1 interaction | [310] | |||||
Metabolomics | ||||||
| Metabolomic profile [311] | [311] | |||||
3.6. Soluble Protein Measurements
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| APRIL | [324] | |||||
| BAFF | [324] | |||||
| CA-125 | [327] | |||||
| CCL17 | [325] | |||||
| CCL19 | [148] | |||||
| CCL22 | [325] | |||||
| CCL3 | [318] | |||||
| CCL4 | [318] | |||||
| CCL5 | [318] | |||||
| CFH | [326] | |||||
| CFHR1 | [326] | |||||
| CFHR3 | [326] | |||||
| CXCL9 | [318] | |||||
| CXCL10 | [318] | |||||
| EGF | [318] | |||||
| Eotaxin | [318] | |||||
| HGF | [318] | |||||
| IFN-α | [318] | |||||
| Ig free light chains | [328] | |||||
| IL-1RA | [318] | |||||
| IL-2 | [318] | |||||
| IL-2R | [319] | [315] | [219,312,313,314,315,316,317,318] | [329,330] | ||
| IL-2Ra | [320,321] | |||||
| IL-4 | [322] | [318] | ||||
| IL-6 | [315] | [315] | ||||
| IL-8 | [318] | |||||
| IL-10 | [318] | |||||
| IL-12 | [318,323] | |||||
| IL-13 | [318] | |||||
| LR11 | [331] | |||||
| MCP1 | [318] | |||||
| Selenium | [332] | |||||
| Thymidine kinase 1 (TK1) | [333] | |||||
| TNF-α | [315] | [315] | ||||
| Triiodothyronine (T3) | [334] | |||||
| Vitamin D insufficiency | ||||||
| Low vitamin D | [284,285] | |||||
| Cholesterols | ||||||
| High-density lipoprotein cholesterol | [335] | |||||
| Low-density lipoprotein cholesterol | [335] | |||||
Circulating Tumor DNA
| Reported Risk of Transformation | Reported Prognostic Value | |||||
|---|---|---|---|---|---|---|
| Favorable | Inferior | None | Favorable | Inferior | None | |
| ctDNA | ||||||
| High proportion of ctDNA | [30,31,336,337] | |||||
| Detectable ctDNA mutations | [338] | |||||
| Specific genetic mutations in | ||||||
| BCL2 | [71] | |||||
| CARD11 | [338] | |||||
| CREBBP | [30,338] | |||||
| EP300 | [338] | |||||
| KMT2D | [338] | |||||
| PCLO | [338] | |||||
| STAT6 | [338] | |||||
| TP53 | [30] | |||||
cfDNA | ||||||
| High proportion of cfDNA | [31,71] | [339] | ||||
| cfDNA, cell-free DNA; ctDNA, circulating tumor DNA. | ||||||
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Friedberg, J.W. Update on follicular lymphoma. Hematol. Oncol. 2023, 41 (Suppl. S1), 43–47. [Google Scholar] [CrossRef]
- Carbone, A.; Roulland, S.; Gloghini, A.; Younes, A.; von Keudell, G.; López-Guillermo, A.; Fitzgibbon, J. Follicular lymphoma. Nat. Rev. Dis. Primers. 2019, 5, 83. [Google Scholar] [CrossRef]
- Huet, S.; Sujobert, P.; Salles, G. From genetics to the clinic: A translational perspective on follicular lymphoma. Nat. Rev. Cancer 2018, 18, 224–239. [Google Scholar] [CrossRef]
- Kridel, R.; Sehn, L.H.; Gascoyne, R.D. Can histologic transformation of follicular lymphoma be predicted and prevented? Blood 2017, 130, 258–266. [Google Scholar] [CrossRef]
- Kumar, E.A.; Okosun, J.; Fitzgibbon, J. The Biological Basis of Histologic Transformation. Hematol. Oncol. Clin. North. Am. 2020, 34, 771–784. [Google Scholar] [CrossRef]
- Jacobsen, E. Follicular lymphoma: 2023 update on diagnosis and management. Am. J. Hematol. 2022, 97, 1638–1651. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Burrows, P.D.; Wang, J.Y. B Cell Development and Maturation. Adv. Exp. Med. Biol. 2020, 1254, 1–22. [Google Scholar]
- Fischer, T.; Zing, N.P.C.; Chiattone, C.S.; Federico, M.; Luminari, S. Transformed follicular lymphoma. Ann. Hematol. 2018, 97, 17–29. [Google Scholar] [CrossRef]
- Montoto, S.; Fitzgibbon, J. Transformation of indolent B-cell lymphomas. J. Clin. Oncol. 2011, 29, 1827–1834. [Google Scholar] [CrossRef]
- Lackraj, T.; Goswami, R.; Kridel, R. Pathogenesis of follicular lymphoma. Best. Pract. Res. Clin. Haematol. 2018, 31, 2–14. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Bmj 2021, 372, n71. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Moher, D. Updating guidance for reporting systematic reviews: Development of the PRISMA 2020 statement. J. Clin. Epidemiol. 2021, 134, 103–112. [Google Scholar] [CrossRef]
- Mamessier, E.; Broussais-Guillaumot, F.; Chetaille, B.; Bouabdallah, R.; Xerri, L.; Jaffe, E.S.; Nadel, B. Nature and importance of follicular lymphoma precursors. Haematologica 2014, 99, 802–810. [Google Scholar] [CrossRef]
- Levine, E.G.; Arthur, D.C.; Frizzera, G.; Peterson, B.A.; Hurd, D.D.; Bloomfield, C.D. Cytogenetic abnormalities predict clinical outcome in non-Hodgkin lymphoma. Ann. Intern. Med. 1988, 108, 14–20. [Google Scholar] [CrossRef]
- Bouska, A.; McKeithan, T.W.; Deffenbacher, K.E.; Lachel, C.; Wright, G.W.; Iqbal, J.; Smith, L.M.; Zhang, W.; Kucuk, C.; Rinaldi, A.; et al. Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood 2014, 123, 1681–1690. [Google Scholar] [CrossRef]
- Viardot, A.; Möller, P.; Högel, J.; Werner, K.; Mechtersheimer, G.; Ho, A.D.; Ott, G.; Barth, T.F.; Siebert, R.; Gesk, S.; et al. Clinicopathologic correlations of genomic gains and losses in follicular lymphoma. J. Clin. Oncol. 2002, 20, 4523–4530. [Google Scholar] [CrossRef]
- Tilly, H.; Rossi, A.; Stamatoullas, A.; Lenormand, B.; Bigorgne, C.; Kunlin, A.; Monconduit, M.; Bastard, C. Prognostic value of chromosomal abnormalities in follicular lymphoma. Blood 1994, 84, 1043–1049. [Google Scholar] [CrossRef]
- Mitsui, T.; Yokohama, A.; Koiso, H.; Saito, A.; Toyama, K.; Shimizu, H.; Ishizaki, T.; Irisawa, H.; Takizawa, M.; Saitoh, T.; et al. Prognostic impact of trisomy 21 in follicular lymphoma. Br. J. Haematol. 2019, 184, 570–577. [Google Scholar] [CrossRef]
- Qu, X.; Li, H.; Braziel, R.M.; Passerini, V.; Rimsza, L.M.; Hsi, E.D.; Leonard, J.P.; Smith, S.M.; Kridel, R.; Press, O.; et al. Genomic alterations important for the prognosis in patients with follicular lymphoma treated in SWOG study S0016. Blood 2019, 133, 81–93. [Google Scholar] [CrossRef]
- Kridel, R.; Chan, F.C.; Mottok, A.; Boyle, M.; Farinha, P.; Tan, K.; Meissner, B.; Bashashati, A.; McPherson, A.; Roth, A.; et al. Histological Transformation and Progression in Follicular Lymphoma: A Clonal Evolution Study. PLoS Med. 2016, 13, e1002197. [Google Scholar] [CrossRef]
- Juneja, S.; Matthews, J.; Lukeis, R.; Laidlaw, C.; Cooper, I.; Wolf, M.; Ironside, P.; Garson, O.M. Prognostic value of cytogenetic abnormalities in previously untreated patients with non-Hodgkin’s lymphoma. Leuk. Lymphoma 1997, 25, 493–501. [Google Scholar] [CrossRef]
- Suguro, M.; Yoshida, N.; Umino, A.; Kato, H.; Tagawa, H.; Nakagawa, M.; Fukuhara, N.; Karnan, S.; Takeuchi, I.; Hocking, T.D.; et al. Clonal heterogeneity of lymphoid malignancies correlates with poor prognosis. Cancer Sci. 2014, 105, 897–904. [Google Scholar] [CrossRef]
- González-Rincón, J.; Méndez, M.; Gómez, S.; García, J.F.; Martín, P.; Bellas, C.; Pedrosa, L.; Rodríguez-Pinilla, S.M.; Camacho, F.I.; Quero, C.; et al. Unraveling transformation of follicular lymphoma to diffuse large B-cell lymphoma. PLoS ONE 2019, 14, e0212813. [Google Scholar] [CrossRef]
- Bai, B.; Wise, J.F.; Vodák, D.; Nakken, S.; Sharma, A.; Blaker, Y.N.; Brodtkorb, M.; Hilden, V.; Trøen, G.; Ren, W.; et al. Multi-omics profiling of longitudinal samples reveals early genomic changes in follicular lymphoma. Blood Cancer J. 2024, 14, 147. [Google Scholar] [CrossRef]
- García-Álvarez, M.; Alonso-Álvarez, S.; Prieto-Conde, I.; Jiménez, C.; Sarasquete, M.E.; Chillón, M.C.; Medina, A.; Balanzategui, A.; Maldonado, R.; Antón, A.; et al. Genetic complexity impacts the clinical outcome of follicular lymphoma patients. Blood Cancer J. 2021, 11, 11. [Google Scholar] [CrossRef]
- Sorigue, M.; Oliveira, A.; Mercadal, S.; Tapia, G.; Climent, F.; Perez-Roca, L.; Lorences, I.; Domingo-Domenech, E.; Cabezon, M.; Navarro, J.T.; et al. m7FLIPI and targeted sequencing in high-risk follicular lymphoma. Hematol. Oncol. 2019, 37, 564–568. [Google Scholar] [CrossRef]
- Cerhan, J.R.; Wang, S.; Maurer, M.J.; Ansell, S.M.; Geyer, S.M.; Cozen, W.; Morton, L.M.; Davis, S.; Severson, R.K.; Rothman, N.; et al. Prognostic significance of host immune gene polymorphisms in follicular lymphoma survival. Blood 2007, 109, 5439–5446. [Google Scholar] [CrossRef]
- Tsukamoto, T.; Nakano, M.; Sato, R.; Adachi, H.; Kiyota, M.; Kawata, E.; Uoshima, N.; Yasukawa, S.; Chinen, Y.; Mizutani, S.; et al. High-risk follicular lymphomas harbour more somatic mutations including those in the AID-motif. Sci. Rep. 2017, 7, 14039. [Google Scholar] [CrossRef]
- Yoon, S.E.; Shin, S.H.; Nam, D.K.; Cho, J.; Kim, W.S.; Kim, S.J. Feasibility of Circulating Tumor DNA Analysis in Patients with Follicular Lymphoma. Cancer Res. Treat. 2024, 56, 920–935. [Google Scholar] [CrossRef]
- Fernández-Miranda, I.; Pedrosa, L.; Llanos, M.; Franco, F.F.; Gómez, S.; Martín-Acosta, P.; García-Arroyo, F.R.; Gumá, J.; Horcajo, B.; Ballesteros, A.K.; et al. Monitoring of Circulating Tumor DNA Predicts Response to Treatment and Early Progression in Follicular Lymphoma: Results of a Prospective Pilot Study. Clin. Cancer Res. 2023, 29, 209–220. [Google Scholar] [CrossRef]
- Russler-Germain, D.A.; Krysiak, K.; Ramirez, C.; Mosior, M.; Watkins, M.P.; Gomez, F.; Skidmore, Z.L.; Trani, L.; Gao, F.; Geyer, S.; et al. Mutations associated with progression in follicular lymphoma predict inferior outcomes at diagnosis: Alliance A151303. Blood Adv. 2023, 7, 5524–5539. [Google Scholar] [CrossRef]
- Gao, F.; Liu, H.; Meng, X.; Liu, J.; Wang, J.; Yu, J.; Liu, X.; Liu, X.; Li, L.; Qiu, L.; et al. Integrative genomic and transcriptomic analysis reveals genetic alterations associated with the early progression of follicular lymphoma. Br. J. Haematol. 2023, 202, 1151–1164. [Google Scholar] [CrossRef]
- Mozas, P.; López, C.; Grau, M.; Nadeu, F.; Clot, G.; Valle, S.; Kulis, M.; Navarro, A.; Ramis-Zaldivar, J.E.; González-Farré, B.; et al. Genomic landscape of follicular lymphoma across a wide spectrum of clinical behaviors. Hematol. Oncol. 2023, 41, 631–643. [Google Scholar] [CrossRef]
- Pastore, A.; Jurinovic, V.; Kridel, R.; Hoster, E.; Staiger, A.M.; Szczepanowski, M.; Pott, C.; Kopp, N.; Murakami, M.; Horn, H.; et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: A retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015, 16, 1111–1122. [Google Scholar] [CrossRef]
- Jurinovic, V.; Kridel, R.; Staiger, A.M.; Szczepanowski, M.; Horn, H.; Dreyling, M.H.; Rosenwald, A.; Ott, G.; Klapper, W.; Zelenetz, A.D.; et al. Clinicogenetic risk models predict early progression of follicular lymphoma after first-line immunochemotherapy. Blood 2016, 128, 1112–1120. [Google Scholar] [CrossRef]
- Rai, S.; Inoue, H.; Sakai, K.; Hanamoto, H.; Matsuda, M.; Maeda, Y.; Haeno, T.; Watatani, Y.; Kumode, T.; Serizawa, K.; et al. Decreased expression of T-cell-associated immune markers predicts poor prognosis in patients with follicular lymphoma. Cancer Sci. 2022, 113, 660–673. [Google Scholar] [CrossRef]
- Krysiak, K.; Gomez, F.; White, B.S.; Matlock, M.; Miller, C.A.; Trani, L.; Fronick, C.C.; Fulton, R.S.; Kreisel, F.; Cashen, A.F.; et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood 2017, 129, 473–483. [Google Scholar] [CrossRef]
- Lockmer, S.; Ren, W.; Brodtkorb, M.; Østenstad, B.; Wahlin, B.E.; Pan-Hammarström, Q.; Kimby, E. M7-FLIPI is not prognostic in follicular lymphoma patients with first-line rituximab chemo-free therapy. Br. J. Haematol. 2020, 188, 259–267. [Google Scholar] [CrossRef]
- Rodríguez-Sevilla, J.J.; Fernández-Rodríguez, C.; Bento, L.; Diez-Feijóo, R.; Pinzón, S.; Gibert, J.; Fernández-Ibarrondo, L.; Lafuente, M.; Ferrer, A.; Sánchez-González, B.; et al. Evaluation of 4 prognostic indices in follicular lymphoma treated in first line with immunochemotherapy. Blood Adv. 2023, 7, 1606–1614. [Google Scholar] [CrossRef]
- Fernández-Miranda, I.; Pedrosa, L.; González-Rincón, J.; Espinet, B.; de la Cruz Vicente, F.; Climent, F.; Gómez, S.; Royuela, A.; Camacho, F.I.; Martín-Acosta, P.; et al. Generation and External Validation of a Histologic Transformation Risk Model for Patients with Follicular Lymphoma. Mod. Pathol. 2024, 37, 100516. [Google Scholar] [CrossRef]
- Dreval, K.; Hilton, L.K.; Cruz, M.; Shaalan, H.; Ben-Neriah, S.; Boyle, M.; Collinge, B.; Coyle, K.M.; Duns, G.; Farinha, P.; et al. Genetic subdivisions of follicular lymphoma defined by distinct coding and noncoding mutation patterns. Blood 2023, 142, 561–573. [Google Scholar] [CrossRef]
- Shelton, V.; Detroja, R.; Liu, T.; Isaev, K.; Silva, A.; Passerini, V.; Bakhtiari, M.; Calvente, L.; Hong, M.; He, M.Y.; et al. Identification of genetic subtypes in follicular lymphoma. Blood Cancer J. 2024, 14, 128. [Google Scholar] [CrossRef]
- Pasqualucci, L. Molecular pathogenesis of germinal center-derived B cell lymphomas. Immunol. Rev. 2019, 288, 240–261. [Google Scholar] [CrossRef]
- Cheung, K.J.; Johnson, N.A.; Affleck, J.G.; Severson, T.; Steidl, C.; Ben-Neriah, S.; Schein, J.; Morin, R.D.; Moore, R.; Shah, S.P.; et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010, 70, 9166–9174. [Google Scholar] [CrossRef]
- Cheung, K.J.; Shah, S.P.; Steidl, C.; Johnson, N.; Relander, T.; Telenius, A.; Lai, B.; Murphy, K.P.; Lam, W.; Al-Tourah, A.J.; et al. Genome-wide profiling of follicular lymphoma by array comparative genomic hybridization reveals prognostically significant DNA copy number imbalances. Blood 2009, 113, 137–148. [Google Scholar] [CrossRef]
- Höglund, M.; Sehn, L.; Connors, J.M.; Gascoyne, R.D.; Siebert, R.; Säll, T.; Mitelman, F.; Horsman, D.E. Identification of cytogenetic subgroups and karyotypic pathways of clonal evolution in follicular lymphomas. Genes Chromosomes Cancer 2004, 39, 195–204. [Google Scholar] [CrossRef]
- Horn, H.; Jurinovic, V.; Leich, E.; Kalmbach, S.; Bausinger, J.; Staiger, A.M.; Kurz, K.S.; Möller, P.; Bernd, H.W.; Feller, A.C.; et al. Molecular Cytogenetic Profiling Reveals Similarities and Differences Between Localized Nodal and Systemic Follicular Lymphomas. Hemasphere 2022, 6, e767. [Google Scholar] [CrossRef]
- Eide, M.B.; Liestøl, K.; Lingjaerde, O.C.; Hystad, M.E.; Kresse, S.H.; Meza-Zepeda, L.; Myklebost, O.; Trøen, G.; Aamot, H.V.; Holte, H.; et al. Genomic alterations reveal potential for higher grade transformation in follicular lymphoma and confirm parallel evolution of tumor cell clones. Blood 2010, 116, 1489–1497. [Google Scholar] [CrossRef]
- Schwaenen, C.; Viardot, A.; Berger, H.; Barth, T.F.; Bentink, S.; Döhner, H.; Enz, M.; Feller, A.C.; Hansmann, M.L.; Hummel, M.; et al. Microarray-based genomic profiling reveals novel genomic aberrations in follicular lymphoma which associate with patient survival and gene expression status. Genes Chromosomes Cancer 2009, 48, 39–54. [Google Scholar] [CrossRef]
- Kalmbach, S.; Grau, M.; Zapukhlyak, M.; Leich, E.; Jurinovic, V.; Hoster, E.; Staiger, A.M.; Kurz, K.S.; Weigert, O.; Gaitzsch, E.; et al. Novel insights into the pathogenesis of follicular lymphoma by molecular profiling of localized and systemic disease forms. Leukemia 2023, 37, 2058–2065. [Google Scholar] [CrossRef]
- Stevens, W.B.C.; Mendeville, M.; Redd, R.; Clear, A.J.; Bladergroen, R.; Calaminici, M.; Rosenwald, A.; Hoster, E.; Hiddemann, W.; Gaulard, P.; et al. Prognostic relevance of CD163 and CD8 combined with EZH2 and gain of chromosome 18 in follicular lymphoma: A study by the Lunenburg Lymphoma Biomarker Consortium. Haematologica 2017, 102, 1413–1423. [Google Scholar] [CrossRef]
- Leich, E.; Brodtkorb, M.; Schmidt, T.; Altenbuchinger, M.; Lingjærde, O.C.; Lockmer, S.; Holte, H.; Nedeva, T.; Grieb, T.; Sander, B.; et al. Gene expression and copy number profiling of follicular lymphoma biopsies from patients treated with first-line rituximab without chemotherapy. Leuk. Lymphoma 2023, 64, 1927–1937. [Google Scholar] [CrossRef]
- Menter, T.; Gasser, A.; Juskevicius, D.; Dirnhofer, S.; Tzankov, A. Diagnostic Utility of the Germinal Center-associated Markers GCET1, HGAL, and LMO2 in Hematolymphoid Neoplasms. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 491–498. [Google Scholar] [CrossRef]
- Isobe, M.; Emanuel, B.S.; Givol, D.; Oren, M.; Croce, C.M. Localization of gene for human p53 tumour antigen to band 17p13. Nature 1986, 320, 84–85. [Google Scholar] [CrossRef]
- Lo Cunsolo, C.; Casciano, I.; Banelli, B.; Tonini, G.P.; Romani, M. Refined chromosomal localization of the putative tumor suppressor gene TP73. Cytogenet. Cell Genet. 1998, 82, 199–201. [Google Scholar] [CrossRef]
- O’Shea, D.; O’Riain, C.; Taylor, C.; Waters, R.; Carlotti, E.; Macdougall, F.; Gribben, J.; Rosenwald, A.; Ott, G.; Rimsza, L.M.; et al. The presence of TP53 mutation at diagnosis of follicular lymphoma identifies a high-risk group of patients with shortened time to disease progression and poorer overall survival. Blood 2008, 112, 3126–3129. [Google Scholar] [CrossRef]
- Ikoma, H.; Miyaoka, M.; Hiraiwa, S.; Yukie Kikuti, Y.; Shiraiwa, S.; Hara, R.; Kojima, M.; Ohmachi, K.; Ando, K.; Carreras, J.; et al. Clinicopathological analysis of follicular lymphoma with BCL2, BCL6, and MYC rearrangements. Pathol. Int. 2022, 72, 321–331. [Google Scholar] [CrossRef]
- Watanabe, R.; Tomita, N.; Matsumoto, C.; Hattori, Y.; Matsuura, S.; Takasaki, H.; Hashimoto, C.; Fujita, H.; Fujisawa, S.; Ishigatsubo, Y. The 3q27 and 18q21 translocations for follicular lymphoma and diffuse large B-cell lymphoma in the rituximab era. J. Clin. Exp. Hematop. 2013, 53, 107–114. [Google Scholar] [CrossRef][Green Version]
- Kridel, R.; Mottok, A.; Farinha, P.; Ben-Neriah, S.; Ennishi, D.; Zheng, Y.; Chavez, E.A.; Shulha, H.P.; Tan, K.; Chan, F.C.; et al. Cell of origin of transformed follicular lymphoma. Blood 2015, 126, 2118–2127. [Google Scholar] [CrossRef]
- Duarte, I.X.; Domeny-Duarte, P.; Wludarski, S.C.; Natkunam, Y.; Bacchi, C.E. Follicular lymphoma in young adults: A clinicopathological and molecular study of 200 patients. Mod. Pathol. 2013, 26, 1183–1196. [Google Scholar] [CrossRef]
- Kim, M.; Hwang, H.S.; Son, E.M.; Cho, H.; Yoon, D.H.; Suh, C.; Park, C.S.; Go, H.; Huh, J. Clinicopathological and prognostic significance of BCL2, BCL6, MYC, and IRF4 copy number gains and translocations in follicular lymphoma: A study by FISH analysis. Leuk. Lymphoma 2020, 61, 3342–3350. [Google Scholar] [CrossRef]
- Pasqualucci, L. The germinal center in the pathogenesis of B cell lymphomas. Hematol. Oncol. 2023, 41 (Suppl. S1), 62–69. [Google Scholar] [CrossRef]
- O’Shea, D.; O’Riain, C.; Gupta, M.; Waters, R.; Yang, Y.; Wrench, D.; Gribben, J.; Rosenwald, A.; Ott, G.; Rimsza, L.M.; et al. Regions of acquired uniparental disomy at diagnosis of follicular lymphoma are associated with both overall survival and risk of transformation. Blood 2009, 113, 2298–2301. [Google Scholar] [CrossRef]
- Tsukamoto, T.; Kiyota, M.; Kawata, E.; Uoshima, N.; Tatekawa, S.; Chinen, Y.; Nagoshi, H.; Mizutani, S.; Shimura, Y.; Yamamoto-Sugitani, M.; et al. Detection of chromosomal abnormalities by G-banding and prognostic impact in follicular lymphoma in the rituximab era. Int. J. Hematol. 2017, 105, 658–667. [Google Scholar] [CrossRef]
- Bussot, L.; Chevalier, S.; Cristante, J.; Grange, B.; Tesson, B.; Deteix-Santana, C.; Orsini-Piocelle, F.; Leyronnas, C.; Dupire, S.; Gressin, R.; et al. Adverse outcome in follicular lymphoma is associated with MYC rearrangements but not MYC extra copies. Br. J. Haematol. 2021, 194, 382–392. [Google Scholar] [CrossRef]
- Okosun, J.; Bödör, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014, 46, 176–181. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Khiabanian, H.; Fangazio, M.; Vasishtha, M.; Messina, M.; Holmes, A.B.; Ouillette, P.; Trifonov, V.; Rossi, D.; Tabbò, F.; et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014, 6, 130–140. [Google Scholar] [CrossRef]
- Crouch, S.; Painter, D.; Barrans, S.L.; Roman, E.; Beer, P.A.; Cooke, S.L.; Glover, P.; Van Hoppe, S.J.L.; Webster, N.; Lacy, S.E.; et al. Molecular subclusters of follicular lymphoma: A report from the United Kingdom’s Haematological Malignancy Research Network. Blood Adv. 2022, 6, 5716–5731. [Google Scholar] [CrossRef]
- Correia, C.; Maurer, M.J.; McDonough, S.J.; Schneider, P.A.; Ross, P.E.; Novak, A.J.; Feldman, A.L.; Cerhan, J.R.; Slager, S.L.; Witzig, T.E.; et al. Relationship between BCL2 mutations and follicular lymphoma outcome in the chemoimmunotherapy era. Blood Cancer J. 2023, 13, 81. [Google Scholar] [CrossRef]
- Hatipoğlu, T.; Esmeray Sönmez, E.; Hu, X.; Yuan, H.; Danyeli, A.E.; Şeyhanlı, A.; Önal-Süzek, T.; Zhang, W.; Akman, B.; Olgun, A.; et al. Plasma Concentrations and Cancer-Associated Mutations in Cell-Free Circulating DNA of Treatment-Naive Follicular Lymphoma for Improved Non-Invasive Diagnosis and Prognosis. Front. Oncol. 2022, 12, 870487. [Google Scholar] [CrossRef]
- Correia, C.; Schneider, P.A.; Dai, H.; Dogan, A.; Maurer, M.J.; Church, A.K.; Novak, A.J.; Feldman, A.L.; Wu, X.; Ding, H.; et al. BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma. Blood 2015, 125, 658–667. [Google Scholar] [CrossRef]
- Sakhinia, E.; Glennie, C.; Hoyland, J.A.; Menasce, L.P.; Brady, G.; Miller, C.; Radford, J.A.; Byers, R.J. Clinical quantitation of diagnostic and predictive gene expression levels in follicular and diffuse large B-cell lymphoma by RT-PCR gene expression profiling. Blood 2007, 109, 3922–3928. [Google Scholar] [CrossRef]
- Bellido, M.; Capello, D.; Altés, A.; Estivill, C.; Gaidano, G.; Pujol, R.; Bordes, R.; Baiget, M.; Saglio, G.; Sierra, J.; et al. Bcl-6 p53 mutations in lymphomas carrying the bcl-2/Jh rearrangement. Haematologica 2002, 87, 908–917. [Google Scholar]
- Jardin, F.; Ruminy, P.; Parmentier, F.; Picquenot, J.M.; Courel, M.N.; Bertrand, P.; Buchonnet, G.; Tilly, H.; Bastard, C. Clinical and biological relevance of single-nucleotide polymorphisms and acquired somatic mutations of the BCL6 first intron in follicular lymphoma. Leukemia 2005, 19, 1824–1830. [Google Scholar] [CrossRef]
- Hershenfeld, S.A.; Tobin, J.W.D.; Shelton, V.; Calvente, L.; Lajkosz, K.; Liu, T.; Brodtkorb, M.; d’Amore, F.A.; Ludvigsen, M.; Baetz, T.; et al. Single gene mutations and prognosis in limited-stage follicular lymphoma treated with radiation therapy. Br. J. Haematol. 2024. [Google Scholar] [CrossRef]
- Huet, S.; Xerri, L.; Tesson, B.; Mareschal, S.; Taix, S.; Mescam-Mancini, L.; Sohier, E.; Carrère, M.; Lazarovici, J.; Casasnovas, O.; et al. EZH2 alterations in follicular lymphoma: Biological and clinical correlations. Blood Cancer J. 2017, 7, e555. [Google Scholar] [CrossRef]
- Martínez-Laperche, C.; Sanz-Villanueva, L.; Díaz Crespo, F.J.; Muñiz, P.; Martín Rojas, R.; Carbonell, D.; Chicano, M.; Suárez-González, J.; Menárguez, J.; Kwon, M.; et al. EZH2 mutations at diagnosis in follicular lymphoma: A promising biomarker to guide frontline treatment. BMC Cancer 2022, 22, 982. [Google Scholar] [CrossRef]
- Bödör, C.; O’Riain, C.; Wrench, D.; Matthews, J.; Iyengar, S.; Tayyib, H.; Calaminici, M.; Clear, A.; Iqbal, S.; Quentmeier, H.; et al. EZH2 Y641 mutations in follicular lymphoma. Leukemia 2011, 25, 726–729. [Google Scholar] [CrossRef]
- Szumera-CieĆkiewicz, A.; Poleszczuk, J.; Paszkiewicz-Kozik, E.; Rymkiewicz, G.; SokÓŁ, K.; Borysiuk, A.; Kotarska, M.; Owczarek, D.; Kawecka, M.; Pytlak, B.; et al. EZH2 Expression in Follicular Lymphoma Is Variable and Independent from the Progression of Disease Within 24 Months of First Treatment. Anticancer. Res. 2020, 40, 6685–6697. [Google Scholar] [CrossRef]
- Barisic, D.; Chin, C.R.; Meydan, C.; Teater, M.; Tsialta, I.; Mlynarczyk, C.; Chadburn, A.; Wang, X.; Sarkozy, M.; Xia, M.; et al. ARID1A orchestrates SWI/SNF-mediated sequential binding of transcription factors with ARID1A loss driving pre-memory B cell fate and lymphomagenesis. Cancer Cell 2024, 42, 583–604.e511. [Google Scholar] [CrossRef]
- Bastard, C.; Deweindt, C.; Kerckaert, J.P.; Lenormand, B.; Rossi, A.; Pezzella, F.; Fruchart, C.; Duval, C.; Monconduit, M.; Tilly, H. LAZ3 rearrangements in non-Hodgkin’s lymphoma: Correlation with histology, immunophenotype, karyotype, and clinical outcome in 217 patients. Blood 1994, 83, 2423–2427. [Google Scholar] [CrossRef]
- Yano, T.; Jaffe, E.S.; Longo, D.L.; Raffeld, M. MYC rearrangements in histologically progressed follicular lymphomas. Blood 1992, 80, 758–767. [Google Scholar] [CrossRef]
- Chaudhary, S.; Brown, N.; Song, J.Y.; Yang, L.; Skrabek, P.; Nasr, M.R.; Wong, J.T.; Bedell, V.; Murata-Collins, J.; Kochan, L.; et al. Relative frequency and clinicopathologic characteristics of MYC-rearranged follicular lymphoma. Hum. Pathol. 2021, 114, 19–27. [Google Scholar] [CrossRef]
- Aukema, S.M.; van Pel, R.; Nagel, I.; Bens, S.; Siebert, R.; Rosati, S.; van den Berg, E.; Bosga-Bouwer, A.G.; Kibbelaar, R.E.; Hoogendoorn, M.; et al. MYC expression and translocation analyses in low-grade and transformed follicular lymphoma. Histopathology 2017, 71, 960–971. [Google Scholar] [CrossRef]
- Chen, Y.P.; Kim, H.J.; Wu, H.; Price-Troska, T.; Villasboas, J.C.; Jalali, S.; Feldman, A.L.; Novak, A.J.; Yang, Z.Z.; Ansell, S.M. SIRPα expression delineates subsets of intratumoral monocyte/macrophages with different functional and prognostic impact in follicular lymphoma. Blood Cancer J. 2019, 9, 84. [Google Scholar] [CrossRef]
- Bouska, A.; Zhang, W.; Gong, Q.; Iqbal, J.; Scuto, A.; Vose, J.; Ludvigsen, M.; Fu, K.; Weisenburger, D.D.; Greiner, T.C.; et al. Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma. Leukemia 2017, 31, 83–91. [Google Scholar] [CrossRef]
- Sander, C.A.; Yano, T.; Clark, H.M.; Harris, C.; Longo, D.L.; Jaffe, E.S.; Raffeld, M. p53 mutation is associated with progression in follicular lymphomas. Blood 1993, 82, 1994–2004. [Google Scholar] [CrossRef]
- Alhejaily, A.; Day, A.G.; Feilotter, H.E.; Baetz, T.; Lebrun, D.P. Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin. Cancer Res. 2014, 20, 1676–1686. [Google Scholar] [CrossRef]
- Taniguchi, T.; Chikatsu, N.; Takahashi, S.; Fujita, A.; Uchimaru, K.; Asano, S.; Fujita, T.; Motokura, T. Expression of p16INK4A and p14ARF in hematological malignancies. Leukemia 1999, 13, 1760–1769. [Google Scholar] [CrossRef][Green Version]
- Coiffier, B.; Li, W.; Henitz, E.D.; Karkera, J.D.; Favis, R.; Gaffney, D.; Shapiro, A.; Theocharous, P.; Elsayed, Y.A.; van de Velde, H.; et al. Prespecified candidate biomarkers identify follicular lymphoma patients who achieved longer progression-free survival with bortezomib-rituximab versus rituximab. Clin. Cancer Res. 2013, 19, 2551–2561. [Google Scholar] [CrossRef]
- Baecklund, F.; Foo, J.N.; Bracci, P.; Darabi, H.; Karlsson, R.; Hjalgrim, H.; Rosenquist, R.; Adami, H.O.; Glimelius, B.; Melbye, M.; et al. A comprehensive evaluation of the role of genetic variation in follicular lymphoma survival. BMC Med. Genet. 2014, 15, 113. [Google Scholar] [CrossRef]
- Carlotti, E.; Palumbo, G.A.; Oldani, E.; Tibullo, D.; Salmoiraghi, S.; Rossi, A.; Golay, J.; Pulsoni, A.; Foà, R.; Rambaldi, A. FcgammaRIIIA and FcgammaRIIA polymorphisms do not predict clinical outcome of follicular non-Hodgkin’s lymphoma patients treated with sequential CHOP and rituximab. Haematologica 2007, 92, 1127–1130. [Google Scholar] [CrossRef]
- Ghesquières, H.; Cartron, G.; Seymour, J.F.; Delfau-Larue, M.H.; Offner, F.; Soubeyran, P.; Perrot, A.; Brice, P.; Bouabdallah, R.; Sonet, A.; et al. Clinical outcome of patients with follicular lymphoma receiving chemoimmunotherapy in the PRIMA study is not affected by FCGR3A and FCGR2A polymorphisms. Blood 2012, 120, 2650–2657. [Google Scholar] [CrossRef]
- Strefford, J.C.; Nowicka, M.; Hargreaves, C.E.; Burton, C.; Davies, A.; Ganderton, R.; Hiddemann, W.; Iriyama, C.; Klapper, W.; Latham, K.V.; et al. Single-nucleotide Fcγ receptor polymorphisms do not impact obinutuzumab/rituximab outcome in patients with lymphoma. Blood Adv. 2021, 5, 2935–2944. [Google Scholar] [CrossRef]
- Cartron, G.; Dacheux, L.; Salles, G.; Solal-Celigny, P.; Bardos, P.; Colombat, P.; Watier, H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 2002, 99, 754–758. [Google Scholar] [CrossRef]
- Rossi, D.; Bruscaggin, A.; La Cava, P.; Galimberti, S.; Ciabatti, E.; Luminari, S.; Rigacci, L.; Tucci, A.; Pulsoni, A.; Bertoldero, G.; et al. The genotype of MLH1 identifies a subgroup of follicular lymphoma patients who do not benefit from doxorubicin: FIL-FOLL study. Haematologica 2015, 100, 517–524. [Google Scholar] [CrossRef]
- Prochazka, V.; Papajik, T.; Gazdova, J.; Divoka, M.; Rozmanova, S.; Faber, E.; Raida, L.; Kucerova, L.; Langova, K.; Jarosova, M.; et al. FcγRIIIA receptor genotype does not influence an outcome in patients with follicular lymphoma treated with risk-adapted immunochemotherapy. Neoplasma 2011, 58, 263–270. [Google Scholar] [CrossRef][Green Version]
- Zhou, X.; Hu, W.; Qin, X. The role of complement in the mechanism of action of rituximab for B-cell lymphoma: Implications for therapy. Oncologist 2008, 13, 954–966. [Google Scholar] [CrossRef]
- Weiner, G.J. Rituximab: Mechanism of action. Semin. Hematol. 2010, 47, 115–123. [Google Scholar] [CrossRef]
- Salles, G.; Barrett, M.; Foà, R.; Maurer, J.; O’Brien, S.; Valente, N.; Wenger, M.; Maloney, D.G. Rituximab in B-Cell Hematologic Malignancies: A Review of 20 Years of Clinical Experience. Adv. Ther. 2017, 34, 2232–2273. [Google Scholar] [CrossRef]
- Bezombes, C.; Fournié, J.J.; Laurent, G. Direct effect of rituximab in B-cell-derived lymphoid neoplasias: Mechanism, regulation, and perspectives. Mol. Cancer Res. 2011, 9, 1435–1442. [Google Scholar] [CrossRef]
- Gibson, T.M.; Wang, S.S.; Cerhan, J.R.; Maurer, M.J.; Hartge, P.; Habermann, T.M.; Davis, S.; Cozen, W.; Lynch, C.F.; Severson, R.K.; et al. Inherited genetic variation and overall survival following follicular lymphoma. Am. J. Hematol. 2012, 87, 724–726. [Google Scholar] [CrossRef]
- Wang, S.S.; Maurer, M.J.; Morton, L.M.; Habermann, T.M.; Davis, S.; Cozen, W.; Lynch, C.F.; Severson, R.K.; Rothman, N.; Chanock, S.J.; et al. Polymorphisms in DNA repair and one-carbon metabolism genes and overall survival in diffuse large B-cell lymphoma and follicular lymphoma. Leukemia 2009, 23, 596–602. [Google Scholar] [CrossRef][Green Version]
- Aschebrook-Kilfoy, B.; Zheng, T.; Foss, F.; Ma, S.; Han, X.; Lan, Q.; Holford, T.; Chen, Y.; Leaderer, B.; Rothman, N.; et al. Polymorphisms in immune function genes and non-Hodgkin lymphoma survival. J. Cancer Surviv. 2012, 6, 102–114. [Google Scholar] [CrossRef][Green Version]
- García-Álvarez, M.; Alonso-Álvarez, S.; Prieto-Conde, I.; Jiménez, C.; Sarasquete, M.E.; Chillón, M.C.; Medina, A.; Balanzategui, A.; Maldonado, R.; Antón, A.; et al. Immunoglobulin gene rearrangement IGHV3-48 is a predictive marker of histological transformation into aggressive lymphoma in follicular, l.y.m.p.h.o.m.a.s. Blood Cancer J. 2019, 9, 52. [Google Scholar] [CrossRef]
- Berget, E.; Molven, A.; Løkeland, T.; Helgeland, L.; Vintermyr, O.K. IGHV gene usage and mutational status in follicular lymphoma: Correlations with prognosis and patient age. Leuk. Res. 2015, 39, 702–708. [Google Scholar] [CrossRef]
- Assis-Mendonça, G.R.; Fattori, A.; Rocha, R.M.; Lourenço, G.J.; Delamain, M.T.; Nonogaki, S.; de Lima, V.C.C.; Colleoni, G.W.B.; de Souza, C.A.; Soares, F.A.; et al. An integrative microenvironment approach for follicular lymphoma: Roles of inflammatory cell subsets and immune-response polymorphisms on disease clinical course. Oncotarget 2020, 11, 3153–3173. [Google Scholar] [CrossRef]
- Ito, Y.; Umezu, T.; Tadokoro, K.; Saito, Y.; Katagiri, S.; Suguro, T.; Asano, M.; Yoshizawa, S.; Akahane, D.; Tanaka, Y.; et al. BIM deletion polymorphism accounts for lack of favorable outcome in Japanese females with follicular lymphoma. Leuk. Lymphoma 2019, 60, 1283–1288. [Google Scholar] [CrossRef]
- Racila, E.; Link, B.K.; Weng, W.K.; Witzig, T.E.; Ansell, S.; Maurer, M.J.; Huang, J.; Dahle, C.; Halwani, A.; Levy, R.; et al. A polymorphism in the complement component C1qA correlates with prolonged response following rituximab therapy of follicular lymphoma. Clin. Cancer Res. 2008, 14, 6697–6703. [Google Scholar] [CrossRef]
- Charbonneau, B.; Maurer, M.J.; Fredericksen, Z.S.; Zent, C.S.; Link, B.K.; Novak, A.J.; Ansell, S.M.; Weiner, G.J.; Wang, A.H.; Witzig, T.E.; et al. Germline variation in complement genes and event-free survival in follicular and diffuse large B-cell lymphoma. Am. J. Hematol. 2012, 87, 880–885. [Google Scholar] [CrossRef]
- Wrench, D.; Leighton, P.; Skibola, C.F.; Conde, L.; Cazier, J.B.; Matthews, J.; Iqbal, S.; Carlotti, E.; Bödör, C.; Montoto, S.; et al. SNP rs6457327 in the HLA region on chromosome 6p is predictive of the transformation of follicular lymphoma. Blood 2011, 117, 3147–3150. [Google Scholar] [CrossRef]
- Wrench, D.; Waters, R.; Carlotti, E.; Iqbal, S.; Matthews, J.; Calaminici, M.; Gribben, J.; Lister, T.A.; Fitzgibbon, J. Clinical relevance of MDM2 SNP 309 and TP53 Arg72Pro in follicular lymphoma. Haematologica 2009, 94, 148–150. [Google Scholar] [CrossRef][Green Version]
- Nielsen, K.R.; Steffensen, R.; Bendtsen, M.D.; Rodrigo-Domingo, M.; Baech, J.; Haunstrup, T.M.; Bergkvist, K.S.; Schmitz, A.; Boedker, J.S.; Johansen, P.; et al. Inherited Inflammatory Response Genes Are Associated with B-Cell Non-Hodgkin’s Lymphoma Risk and Survival. PLoS ONE 2015, 10, e0139329. [Google Scholar] [CrossRef]
- Bararia, D.; Hildebrand, J.A.; Stolz, S.; Haebe, S.; Alig, S.; Trevisani, C.P.; Osorio-Barrios, F.; Bartoschek, M.D.; Mentz, M.; Pastore, A.; et al. Cathepsin S Alterations Induce a Tumor-Promoting Immune Microenvironment in Follicular Lymphoma. Cell Rep. 2020, 31, 107522. [Google Scholar] [CrossRef]
- Charbonneau, B.; Wang, A.H.; Maurer, M.J.; Asmann, Y.W.; Zent, C.S.; Link, B.K.; Ansell, S.M.; Weiner, G.J.; Ozsan, N.; Feldman, A.L.; et al. CXCR5 polymorphisms in non-Hodgkin lymphoma risk and prognosis. Cancer Immunol. Immunother. 2013, 62, 1475–1484. [Google Scholar] [CrossRef][Green Version]
- Mondello, P.; Fama, A.; Larson, M.C.; Feldman, A.L.; Villasboas, J.C.; Yang, Z.Z.; Galkin, I.; Svelolkin, V.; Postovalova, E.; Bagaev, A.; et al. Lack of intrafollicular memory CD4 + T cells is predictive of early clinical failure in newly diagnosed follicular lymphoma. Blood Cancer J. 2021, 11, 130. [Google Scholar] [CrossRef]
- Ghielmini, M.; Rufibach, K.; Salles, G.; Leoncini-Franscini, L.; Léger-Falandry, C.; Cogliatti, S.; Fey, M.; Martinelli, G.; Stahel, R.; Lohri, A.; et al. Single agent rituximab in patients with follicular or mantle cell lymphoma: Clinical and biological factors that are predictive of response and event-free survival as well as the effect of rituximab on the immune system: A study of the Swiss Group for Clinical Cancer Research (SAKK). Ann. Oncol. 2005, 16, 1675–1682. [Google Scholar]
- Hohaus, S.; Mansueto, G.; Massini, G.; D’Alo, F.; Giachelia, M.; Martini, M.; Larocca, L.M.; Voso, M.T.; Leone, G. Glutathione-S-transferase genotypes influence prognosis in follicular non-Hodgkin’s Lymphoma. Leuk. Lymphoma 2007, 48, 564–569. [Google Scholar] [CrossRef]
- Launay, E.; Pangault, C.; Bertrand, P.; Jardin, F.; Lamy, T.; Tilly, H.; Tarte, K.; Bastard, C.; Fest, T. High rate of TNFRSF14 gene alterations related to 1p36 region in de novo follicular lymphoma and impact on prognosis. Leukemia 2012, 26, 559–562. [Google Scholar] [CrossRef]
- Burack, W.R.; Li, H.; Adlowitz, D.; Spence, J.M.; Rimsza, L.M.; Shadman, M.; Spier, C.M.; Kaminski, M.S.; Leonard, J.P.; Leblanc, M.L.; et al. Subclonal TP53 mutations are frequent and predict resistance to radioimmunotherapy in follicular lymphoma. Blood Adv. 2023, 7, 5082–5090. [Google Scholar] [CrossRef]
- De Mendonça, G.R.; Brito, A.B.; Rocha, R.M.; Delamain, M.T.; de Andrade Natal, R.; Soares, F.A.; Colleoni, G.W.; Souza, C.A.; Vassallo, J.; Lima, C.S. Association of VEGFA-2578 C>A polymorphism with clinicopathological aspects and outcome in follicular lymphoma patients. Blood Cancer J. 2016, 6, e464. [Google Scholar] [CrossRef]
- Lu, Y.; Abdou, A.M.; Cerhan, J.R.; Morton, L.M.; Severson, R.K.; Davis, S.; Cozen, W.; Rothman, N.; Bernstein, L.; Chanock, S.; et al. Human leukocyte antigen class I and II alleles and overall survival in diffuse large B-cell lymphoma and follicular lymphoma. Sci. World J. 2011, 11, 2062–2070. [Google Scholar] [CrossRef]
- Dave, S.S.; Wright, G.; Tan, B.; Rosenwald, A.; Gascoyne, R.D.; Chan, W.C.; Fisher, R.I.; Braziel, R.M.; Rimsza, L.M.; Grogan, T.M.; et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N. Engl. J. Med. 2004, 351, 2159–2169. [Google Scholar] [CrossRef]
- Staiger, A.M.; Hoster, E.; Jurinovic, V.; Winter, S.; Leich, E.; Kalla, C.; Möller, P.; Bernd, H.W.; Feller, A.C.; Koch, K.; et al. Localized- and advanced-stage follicular lymphomas differ in their gene expression profiles. Blood 2020, 135, 181–190. [Google Scholar] [CrossRef]
- Huet, S.; Tesson, B.; Jais, J.P.; Feldman, A.L.; Magnano, L.; Thomas, E.; Traverse-Glehen, A.; Albaud, B.; Carrère, M.; Xerri, L.; et al. A gene-expression profiling score for prediction of outcome in patients with follicular lymphoma: A retrospective training and validation analysis in three international cohorts. Lancet Oncol. 2018, 19, 549–561. [Google Scholar] [CrossRef]
- Silva, A.; Bassim, S.; Sarkozy, C.; Mottok, A.; Lackraj, T.; Jurinovic, V.; Brodtkorb, M.; Lingjaerde, O.C.; Sehn, L.H.; Gascoyne, R.D.; et al. Convergence of risk prediction models in follicular lymphoma. Haematologica 2019, 104, e252–e255. [Google Scholar] [CrossRef]
- Brodtkorb, M.; Lingjærde, O.C.; Huse, K.; Trøen, G.; Hystad, M.; Hilden, V.I.; Myklebust, J.H.; Leich, E.; Rosenwald, A.; Delabie, J.; et al. Whole-genome integrative analysis reveals expression signatures predicting transformation in follicular lymphoma. Blood 2014, 123, 1051–1054. [Google Scholar] [CrossRef]
- Steen, C.B.; Leich, E.; Myklebust, J.H.; Lockmer, S.; Wise, J.F.; Wahlin, B.E.; Østenstad, B.; Liestøl, K.; Kimby, E.; Rosenwald, A.; et al. A clinico-molecular predictor identifies follicular lymphoma patients at risk of early transformation after first-line immunotherapy. Haematologica 2019, 104, e460–e464. [Google Scholar] [CrossRef]
- Gentles, A.J.; Alizadeh, A.A.; Lee, S.I.; Myklebust, J.H.; Shachaf, C.M.; Shahbaba, B.; Levy, R.; Koller, D.; Plevritis, S.K. A pluripotency signature predicts histologic transformation and influences survival in follicular lymphoma patients. Blood 2009, 114, 3158–3166. [Google Scholar] [CrossRef]
- Tsukamoto, T.; Tokuda, Y.; Nakano, M.; Tashiro, K.; Kuroda, J. Expression of activated B-cell gene signature is predictive of the outcome of follicular lymphoma. Blood Adv. 2022, 6, 1932–1936. [Google Scholar] [CrossRef]
- Bolen, C.R.; McCord, R.; Huet, S.; Frampton, G.M.; Bourgon, R.; Jardin, F.; Dartigues, P.; Punnoose, E.A.; Szafer-Glusman, E.; Xerri, L.; et al. Mutation load and an effector T-cell gene signature may distinguish immunologically distinct and clinically relevant lymphoma subsets. Blood Adv. 2017, 1, 1884–1890. [Google Scholar] [CrossRef]
- Zhong, C.; Chao, C.R.; Song, J.Y.; Weisenburger, D.D.; Luo, J.; Ding, Y.C.; Neuhausen, S.L.; Bernstein, L.; Cozen, W.; Wang, S.S. Follicular lymphoma polygenic risk score is associated with increased disease risk but improved overall survival among women in a population based case-control in Los Angeles County California. Cancer Epidemiol. 2020, 65, 101688. [Google Scholar] [CrossRef]
- Taskinen, M.; Valo, E.; Karjalainen-Lindsberg, M.L.; Hautaniemi, S.; Meri, S.; Leppä, S. Signal transducers and activators of transcription 5a-dependent cross-talk between follicular lymphoma cells and tumor microenvironment characterizes a group of patients with improved outcome after R-CHOP. Clin. Cancer Res. 2010, 16, 2615–2623. [Google Scholar] [CrossRef]
- Roider, T.; Baertsch, M.A.; Fitzgerald, D.; Vöhringer, H.; Brinkmann, B.J.; Czernilofsky, F.; Knoll, M.; Llaó-Cid, L.; Mathioudaki, A.; Faßbender, B.; et al. Multimodal and spatially resolved profiling identifies distinct patterns of T cell infiltration in nodal B cell lymphoma entities. Nat. Cell Biol. 2024, 26, 478–489. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, H.; Gao, F.; Gong, W.; Cui, Y.; He, J.; Li, L.; Qiu, L.; Qian, Z.; Zhou, S.; et al. m6A-Regulator Expression Signatures Identify a Subset of Follicular Lymphoma Harboring an Exhausted Tumor Microenvironment. Front. Immunol. 2022, 13, 922471. [Google Scholar] [CrossRef]
- Carreras, J.; Roncador, G.; Hamoudi, R. Artificial Intelligence Predicted Overall Survival and Classified Mature B-Cell Neoplasms Based on Immuno-Oncology and Immune Checkpoint Panels. Cancers 2022, 14, 5318. [Google Scholar] [CrossRef]
- Menter, T.; Hayoz, S.; Zucca, E.; Kimby, E.; Dirnhofer, S.; Tzankov, A. Immunomodulatory drugs may overcome the negative prognostic role of active Th17 axis in follicular lymphoma: Evidence from the SAKK35/10 trial. Br. J. Haematol. 2020, 190, e258–e261. [Google Scholar] [CrossRef]
- Tobin, J.W.D.; Keane, C.; Gunawardana, J.; Mollee, P.; Birch, S.; Hoang, T.; Lee, J.; Li, L.; Huang, L.; Murigneux, V.; et al. Progression of Disease within 24 Months in Follicular Lymphoma Is Associated With Reduced Intratumoral Immune Infiltration. J. Clin. Oncol. 2019, 37, 3300–3309. [Google Scholar] [CrossRef]
- Glas, A.M.; Knoops, L.; Delahaye, L.; Kersten, M.J.; Kibbelaar, R.E.; Wessels, L.A.; van Laar, R.; van Krieken, J.H.; Baars, J.W.; Raemaekers, J.; et al. Gene-expression and immunohistochemical study of specific T-cell subsets and accessory cell types in the transformation and prognosis of follicular lymphoma. J. Clin. Oncol. 2007, 25, 390–398. [Google Scholar] [CrossRef]
- Fernández-Garnacho, E.M.; Nadeu, F.; Martín, S.; Mozas, P.; Rivero, A.; Delgado, J.; Giné, E.; López-Guillermo, A.; Duran-Ferrer, M.; Salaverria, I.; et al. MALAT1 expression is associated with aggressive behavior in indolent B-cell neoplasms. Sci. Rep. 2023, 13, 16839. [Google Scholar] [CrossRef]
- Byers, R.J.; Sakhinia, E.; Joseph, P.; Glennie, C.; Hoyland, J.A.; Menasce, L.P.; Radford, J.A.; Illidge, T. Clinical quantitation of immune signature in follicular lymphoma by RT-PCR-based gene expression profiling. Blood 2008, 111, 4764–4770. [Google Scholar] [CrossRef]
- Björck, E.; Ek, S.; Landgren, O.; Jerkeman, M.; Ehinger, M.; Björkholm, M.; Borrebaeck, C.A.; Porwit-MacDonald, A.; Nordenskjöld, M. High expression of cyclin B1 predicts a favorable outcome in patients with follicular lymphoma. Blood 2005, 105, 2908–2915. [Google Scholar] [CrossRef]
- Bohen, S.P.; Troyanskaya, O.G.; Alter, O.; Warnke, R.; Botstein, D.; Brown, P.O.; Levy, R. Variation in gene expression patterns in follicular lymphoma and the response to rituximab. Proc. Natl. Acad. Sci. USA 2003, 100, 1926–1930. [Google Scholar] [CrossRef]
- Provencio, M.; Rodríguez, M.; Cantos, B.; Sabín, P.; Quero, C.; García-Arroyo, F.R.; Rueda, A.; Maximiano, C.; Rodríguez-Abreu, D.; Sánchez, A.; et al. mRNA in exosomas as a liquid biopsy in non-Hodgkin Lymphoma: A multicentric study by the Spanish Lymphoma Oncology Group. Oncotarget 2017, 8, 50949–50957. [Google Scholar] [CrossRef]
- Zhao, W.L.; Daneshpouy, M.E.; Mounier, N.; Brière, J.; Leboeuf, C.; Plassa, L.F.; Turpin, E.; Cayuela, J.M.; Ameisen, J.C.; Gisselbrecht, C.; et al. Prognostic significance of bcl-xL gene expression and apoptotic cell counts in follicular lymphoma. Blood 2004, 103, 695–697. [Google Scholar] [CrossRef]
- Harjunpää, A.; Taskinen, M.; Nykter, M.; Karjalainen-Lindsberg, M.L.; Nyman, H.; Monni, O.; Hemmer, S.; Yli-Harja, O.; Hautaniemi, S.; Meri, S.; et al. Differential gene expression in non-malignant tumour microenvironment is associated with outcome in follicular lymphoma patients treated with rituximab, C.H.O.P. Br. J. Haematol. 2006, 135, 33–42. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, S.; Tao, Y.; Chen, H.; Qin, Y.; He, X.; Zhou, S.; Liu, P.; Yang, J.; Yang, S.; et al. Low CCL19 expression is associated with adverse clinical outcomes for follicular lymphoma patients treated with chemoimmunotherapy. J. Transl. Med. 2021, 19, 399. [Google Scholar] [CrossRef]
- Herold, T.; Mulaw, M.A.; Jurinovic, V.; Seiler, T.; Metzeler, K.H.; Dufour, A.; Schneider, S.; Kakadia, P.M.; Spiekermann, K.; Mansmann, U.; et al. High expression of MZB1 predicts adverse prognosis in chronic lymphocytic leukemia, follicular lymphoma and diffuse large B-cell lymphoma and is associated with a unique gene expression signature. Leuk. Lymphoma 2013, 54, 1652–1657. [Google Scholar] [CrossRef]
- Laurent, C.; Flores, M.; Chartier, L.; Huet, S.; Bolen, C.R.; Venstrom, J.M.; Chassagne-Clément, C.; Dartigues-Cuilléres, P.; Charlotte, F.; Tesson, B.; et al. Long-term follow-up confirms the favourable prognostic impact of high numbers of tumour infiltrating CD3 T-cells in follicular lymphoma patients treated by rituximab-maintenance regimen. Br. J. Haematol. 2023, 202, 686–689. [Google Scholar] [CrossRef]
- Taskinen, M.; Jantunen, E.; Kosma, V.M.; Bono, P.; Karjalainen-Lindsberg, M.L.; Leppä, S. Prognostic impact of CD31-positive microvessel density in follicular lymphoma patients treated with immunochemotherapy. Eur. J. Cancer. 2010, 46, 2506–2512. [Google Scholar] [CrossRef]
- Carreras, J.; Lopez-Guillermo, A.; Kikuti, Y.Y.; Itoh, J.; Masashi, M.; Ikoma, H.; Tomita, S.; Hiraiwa, S.; Hamoudi, R.; Rosenwald, A.; et al. High TNFRSF14 and low BTLA are associated with poor prognosis in Follicular Lymphoma and in Diffuse Large B-cell Lymphoma transformation. J. Clin. Exp. Hematop. 2019, 59, 1–16. [Google Scholar] [CrossRef]
- Krajnović, M.; Radojković, M.; Davidović, R.; Dimitrijević, B.; Krtolica, K. Prognostic significance of epigenetic inactivation of p16, p15, MGMT and DAPK genes in follicular lymphoma. Med. Oncol. 2013, 30, 441. [Google Scholar] [CrossRef]
- Giachelia, M.; Bozzoli, V.; D’Alò, F.; Tisi, M.C.; Massini, G.; Maiolo, E.; Guidi, F.; Cupelli, E.; Martini, M.; Larocca, L.M.; et al. Quantification of DAPK1 promoter methylation in bone marrow and peripheral blood as a follicular lymphoma biomarker. J. Mol. Diagn. 2014, 16, 467–476. [Google Scholar] [CrossRef]
- Arzuaga-Mendez, J.; Lopez-Santillan, M.; Garcia-Ruiz, J.C.; Lopez-Lopez, E.; Martin-Guerrero, I. Systematic review of the potential of MicroRNAs in the management of patients with follicular lymphoma. Crit. Rev. Oncol. Hematol. 2021, 159, 103247. [Google Scholar] [CrossRef]
- Li, J.; Zou, J.; Wan, X.; Sun, C.; Peng, F.; Chu, Z.; Hu, Y. The Role of Noncoding RNAs in B-Cell Lymphoma. Front. Oncol. 2020, 10, 577890. [Google Scholar] [CrossRef]
- Larrabeiti-Etxebarria, A.; Lopez-Santillan, M.; Santos-Zorrozua, B.; Lopez-Lopez, E.; Garcia-Orad, A. Systematic Review of the Potential of MicroRNAs in Diffuse Large B Cell Lymphoma. Cancers 2019, 11, 144. [Google Scholar] [CrossRef]
- Lopez-Santillan, M.; Larrabeiti-Etxebarria, A.; Arzuaga-Mendez, J.; Lopez-Lopez, E.; Garcia-Orad, A. Circulating miRNAs as biomarkers in diffuse large B-cell lymphoma: A systematic review. Oncotarget 2018, 9, 22850–22861. [Google Scholar] [CrossRef]
- Musilova, K.; Mraz, M. MicroRNAs in B-cell lymphomas: How a complex biology gets more complex. Leukemia 2015, 29, 1004–1017. [Google Scholar] [CrossRef]
- Devan, J.; Janikova, A.; Mraz, M. New concepts in follicular lymphoma biology: From BCL2 to epigenetic regulators and non-coding RNAs. Semin. Oncol. 2018, 45, 291–302. [Google Scholar] [CrossRef]
- Fassina, A.; Marino, F.; Siri, M.; Zambello, R.; Ventura, L.; Fassan, M.; Simonato, F.; Cappellesso, R. The miR-17-92 microRNA cluster: A novel diagnostic tool in large B-cell malignancies. Lab. Investig. 2012, 92, 1574–1582. [Google Scholar] [CrossRef]
- Musilova, K.; Devan, J.; Cerna, K.; Seda, V.; Pavlasova, G.; Sharma, S.; Oppelt, J.; Pytlik, R.; Prochazka, V.; Prouzova, Z.; et al. miR-150 downregulation contributes to the high-grade transformation of follicular lymphoma by upregulating FOXP1 levels. Blood 2018, 132, 2389–2400. [Google Scholar] [CrossRef]
- Mottok, A.; Jurinovic, V.; Farinha, P.; Rosenwald, A.; Leich, E.; Ott, G.; Horn, H.; Klapper, W.; Boesl, M.; Hiddemann, W.; et al. FOXP1 expression is a prognostic biomarker in follicular lymphoma treated with rituximab and chemotherapy. Blood 2018, 131, 226–235. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Chi, J.; Taylor, S.; Tramonti, D.; Ballabio, E.; Palazzo, S.; Saunders, N.J.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Expression of microRNAs in diffuse large B cell lymphoma is associated with immunophenotype, survival and transformation from follicular lymphoma. J. Cell Mol. Med. 2009, 13, 1248–1260. [Google Scholar] [CrossRef]
- Lou, X.; Fu, J.; Zhao, X.; Zhuansun, X.; Rong, C.; Sun, M.; Niu, H.; Wu, L.; Zhang, Y.; An, L.; et al. MiR-7e-5p downregulation promotes transformation of low-grade follicular lymphoma to aggressive lymphoma by modulating an immunosuppressive stroma through the upregulation of FasL in M1 macrophages. J. Exp. Clin. Cancer Res. 2020, 39, 237. [Google Scholar] [CrossRef]
- Freedman, A.; Jacobsen, E. Follicular lymphoma: 2020 update on diagnosis and management. Am. J. Hematol. 2020, 95, 316–327. [Google Scholar] [CrossRef]
- López, C.; Mozas, P.; López-Guillermo, A.; Beà, S. Molecular Pathogenesis of Follicular Lymphoma: From Genetics to Clinical Practice. Hemato 2022, 3, 595–614. [Google Scholar] [CrossRef]
- Richendollar, B.G.; Pohlman, B.; Elson, P.; Hsi, E.D. Follicular programmed death 1-positive lymphocytes in the tumor microenvironment are an independent prognostic factor in follicular lymphoma. Hum. Pathol. 2011, 42, 552–557. [Google Scholar] [CrossRef]
- Torlakovic, E.E.; Bilalovic, N.; Golouh, R.; Zidar, A.; Angel, S. Prognostic significance of PU.1 in follicular lymphoma. J. Pathol. 2006, 209, 352–359. [Google Scholar] [CrossRef]
- Socha, D.S.; Zhao, X.; Bodo, J.; Durkin, L.; Hsi, E.D. Decreased BIM expression in BCL2-negative follicular lymphoma: A potential mechanism for resistance to apoptosis. Hum. Pathol. 2021, 107, 1–8. [Google Scholar] [CrossRef]
- Sohani, A.R.; Maurer, M.J.; Giri, S.; Pitcher, B.; Chadburn, A.; Said, J.W.; Bartlett, N.L.; Czuczman, M.S.; Martin, P.; Rosenbaum, C.A.; et al. Biomarkers for Risk Stratification in Patients With Previously Untreated Follicular Lymphoma Receiving Anti-CD20-based Biological Therapy. Am. J. Surg. Pathol. 2021, 45, 384–393. [Google Scholar] [CrossRef]
- Camacho, F.I.; Bellas, C.; Corbacho, C.; Caleo, A.; Arranz-Sáez, R.; Cannata, J.; Menárguez, J.; Sánchez-Verde, L.; González-Camacho, L.; Pérez-Martín, M.E.; et al. Improved demonstration of immunohistochemical prognostic markers for survival in follicular lymphoma cells. Mod. Pathol. 2011, 24, 698–707. [Google Scholar] [CrossRef]
- Logsdon, M.D.; Meyn, R.E.; Besa, P.C., Jr.; Pugh, W.C.; Stephens, L.C.; Peters, L.J.; Milas, L.; Cox, J.D.; Cabanillas, F.; Brisbay, S.; et al. Apoptosis and the Bcl-2 gene family -- patterns of expression and prognostic value in stage I and II follicular center lymphoma. Int. J. Radiat. Oncol. Biol. Phys. 1999, 44, 19–29. [Google Scholar] [CrossRef]
- Llanos, M.; Alvarez-Argüelles, H.; Alemán, R.; Oramas, J.; Diaz-Flores, L.; Batista, N. Prognostic significance of Ki-67 nuclear proliferative antigen, bcl-2 protein, and p53 expression in follicular and diffuse large B-cell lymphoma. Med. Oncol. 2001, 18, 15–22. [Google Scholar] [CrossRef]
- Kelley, T.; Beck, R.; Absi, A.; Jin, T.; Pohlman, B.; Hsi, E. Biologic predictors in follicular lymphoma: Importance of markers of immune response. Leuk. Lymphoma 2007, 48, 2403–2411. [Google Scholar] [CrossRef]
- Michels, J.; Foria, V.; Mead, B.; Jackson, G.; Mullee, M.; Johnson, P.W.; Packham, G. Immunohistochemical analysis of the antiapoptotic Mcl-1 and Bcl-2 proteins in follicular lymphoma. Br. J. Haematol. 2006, 132, 743–746. [Google Scholar] [CrossRef]
- Farinha, P.; Masoudi, H.; Skinnider, B.F.; Shumansky, K.; Spinelli, J.J.; Gill, K.; Klasa, R.; Voss, N.; Connors, J.M.; Gascoyne, R.D. Analysis of multiple biomarkers shows that lymphoma-associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL). Blood 2005, 106, 2169–2174. [Google Scholar] [CrossRef]
- Bilalovic, N.; Blystad, A.K.; Golouh, R.; Nesland, J.M.; Selak, I.; Trinh, D.; Torlakovic, E. Expression of bcl-6 and CD10 protein is associated with longer overall survival and time to treatment failure in follicular lymphoma. Am. J. Clin. Pathol. 2004, 121, 34–42. [Google Scholar] [CrossRef]
- Nichols, M.M.; Ondrejka, S.L.; Patil, S.; Durkin, L.; Hill, B.T.; Hsi, E.D. Ki67 proliferation index in follicular lymphoma is associated with favorable outcome in patients treated with R-CHOP. Leuk Lymphoma 2023, 64, 1433–1441. [Google Scholar] [CrossRef]
- Scherer, F.; van der Burgt, M.; Kiełbasa, S.M.; Bertinetti-Lapatki, C.; Dühren von Minden, M.; Mikesch, K.; Zirlik, K.; de Wreede, L.; Veelken, H.; Navarrete, M.A. Selection patterns of B-cell receptors and the natural history of follicular lymphoma. Br. J. Haematol. 2016, 175, 972–975. [Google Scholar] [CrossRef]
- Madsen, C.; Lauridsen, K.L.; Plesner, T.L.; Monrad, I.; Honoré, B.; Hamilton-Dutoit, S.; d’Amore, F.; Ludvigsen, M. High intratumoral expression of vimentin predicts histological transformation in patients with follicular lymphoma. Blood Cancer J. 2019, 9, 35. [Google Scholar] [CrossRef]
- Xerri, L.; Bachy, E.; Fabiani, B.; Canioni, D.; Chassagne-Clément, C.; Dartigues-Cuilléres, P.; Charlotte, F.; Brousse, N.; Rousselet, M.C.; Foussard, C.; et al. Identification of MUM1 as a prognostic immunohistochemical marker in follicular lymphoma using computerized image analysis. Hum. Pathol. 2014, 45, 2085–2093. [Google Scholar] [CrossRef]
- Sweetenham, J.W.; Goldman, B.; LeBlanc, M.L.; Cook, J.R.; Tubbs, R.R.; Press, O.W.; Maloney, D.G.; Fisher, R.I.; Rimsza, L.M.; Braziel, R.M.; et al. Prognostic value of regulatory T cells, lymphoma-associated macrophages, and MUM-1 expression in follicular lymphoma treated before and after the introduction of monoclonal antibody therapy: A Southwest Oncology Group Study. Ann. Oncol. 2010, 21, 1196–1202. [Google Scholar] [CrossRef]
- Gulmann, C.; Espina, V.; Petricoin, E.; 3rd Longo, D.L.; Santi, M.; Knutsen, T.; Raffeld, M.; Jaffe, E.S.; Liotta, L.A.; Feldman, A.L. Proteomic analysis of apoptotic pathways reveals prognostic factors in follicular lymphoma. Clin. Cancer Res. 2005, 11, 5847–5855. [Google Scholar] [CrossRef]
- Wahlin, B.E.; Sander, B.; Christensson, B.; Kimby, E. CD8+ T-cell content in diagnostic lymph nodes measured by flow cytometry is a predictor of survival in follicular lymphoma. Clin. Cancer Res. 2007, 13 Pt 1, 388–397. [Google Scholar] [CrossRef][Green Version]
- Weng, W.K.; Levy, R. Expression of complement inhibitors CD46, CD55, and CD59 on tumor cells does not predict clinical outcome after rituximab treatment in follicular non-Hodgkin lymphoma. Blood 2001, 98, 1352–1357. [Google Scholar] [CrossRef]
- Budau, L.; Wilhelm, C.; Moll, R.; Jäkel, J.; Hirt, C.; Dölken, G.; Maschmeyer, G.; Neubauer, E.; Strauch, K.; Burchert, A.; et al. Low number of intrafollicular T cells may predict favourable response to rituximab-based immuno-chemotherapy in advanced follicular lymphoma: A secondary analysis of a randomized clinical trial. J. Cancer Res. Clin. Oncol. 2019, 145, 2149–2156. [Google Scholar] [CrossRef]
- Klapper, W.; Hoster, E.; Rölver, L.; Schrader, C.; Janssen, D.; Tiemann, M.; Bernd, H.W.; Determann, O.; Hansmann, M.L.; Möller, P.; et al. Tumor sclerosis but not cell proliferation or malignancy grade is a prognostic marker in advanced-stage follicular lymphoma: The German Low Grade Lymphoma Study Group. J. Clin. Oncol. 2007, 25, 3330–3336. [Google Scholar] [CrossRef]
- Yoshimura, T.; Miyoshi, H.; Shimono, J.; Nakashima, K.; Takeuchi, M.; Yanagida, E.; Yamada, K.; Shimasaki, Y.; Moritsubo, M.; Furuta, T.; et al. CD37 expression in follicular lymphoma. Ann. Hematol. 2022, 101, 1067–1075. [Google Scholar] [CrossRef]
- De Jong, D.; Koster, A.; Hagenbeek, A.; Raemaekers, J.; Veldhuizen, D.; Heisterkamp, S.; de Boer, J.P.; van Glabbeke, M. Impact of the tumor microenvironment on prognosis in follicular lymphoma is dependent on specific treatment protocols. Haematologica 2009, 94, 70–77. [Google Scholar] [CrossRef]
- Szumera-Ciećkiewicz, A.; Rymkiewicz, G.; Sokół, K.; Paszkiewicz-Kozik, E.; Borysiuk, A.; Poleszczuk, J.; Bachnio, K.; Bystydzienski, Z.; Woroniecka, R.; Grygalewicz, B.; et al. Significance of CD10 protein expression in the diagnostics of follicular lymphoma: A comparison of conventional immunohistochemistry with flow cytometry supported by the establishment of BCL2 and BCL6 rearrangements. Int. J. Lab. Hematol. 2020, 42, 453–463. [Google Scholar] [CrossRef]
- Li, C.; Zhang, W.; Zhao, D.; Yang, P.; Wan, W.; Liu, S.; Jing, H. Development of a new risk scoring system based on spleen involvement and the lymphocyte/monocyte ratio for follicular lymphoma patients. Leuk. Res. 2022, 123, 106980. [Google Scholar] [CrossRef]
- Wang, X.; Nissen, M.; Gracias, D.; Kusakabe, M.; Simkin, G.; Jiang, A.; Duns, G.; Sarkozy, C.; Hilton, L.; Chavez, E.A.; et al. Single-cell profiling reveals a memory B cell-like subtype of follicular lymphoma with increased transformation risk. Nat. Commun. 2022, 13, 6772. [Google Scholar] [CrossRef]
- De Jong, D. Molecular pathogenesis of follicular lymphoma: A cross talk of genetic and immunologic factors. J. Clin. Oncol. 2005, 23, 6358–6363. [Google Scholar] [CrossRef]
- Menter, T.; Tzankov, A.; Zucca, E.; Kimby, E.; Hultdin, M.; Sundström, C.; Beiske, K.; Cogliatti, S.; Banz, Y.; Cathomas, G.; et al. Prognostic implications of the microenvironment for follicular lymphoma under immunomodulation therapy. Br. J. Haematol. 2020, 189, 707–717. [Google Scholar] [CrossRef]
- Wahlin, B.E.; Aggarwal, M.; Montes-Moreno, S.; Gonzalez, L.F.; Roncador, G.; Sanchez-Verde, L.; Christensson, B.; Sander, B.; Kimby, E. A unifying microenvironment model in follicular lymphoma: Outcome is predicted by programmed death-1--positive, regulatory, cytotoxic, and helper T cells and macrophages. Clin. Cancer Res. 2010, 16, 637–650. [Google Scholar] [CrossRef]
- Alvaro, T.; Lejeune, M.; Salvadó, M.T.; Lopez, C.; Jaén, J.; Bosch, R.; Pons, L.E. Immunohistochemical patterns of reactive microenvironment are associated with clinicobiologic behavior in follicular lymphoma patients. J. Clin. Oncol. 2006, 24, 5350–5357. [Google Scholar] [CrossRef]
- Laurent, C.; Müller, S.; Do, C.; Al-Saati, T.; Allart, S.; Larocca, L.M.; Hohaus, S.; Duchez, S.; Quillet-Mary, A.; Laurent, G.; et al. Distribution, function, and prognostic value of cytotoxic T lymphocytes in follicular lymphoma: A 3-D tissue-imaging study. Blood 2011, 118, 5371–5379. [Google Scholar] [CrossRef]
- Gravelle, P.; Do, C.; Franchet, C.; Mueller, S.; Oberic, L.; Ysebaert, L.; Larocca, L.M.; Hohaus, S.; Calmels, M.N.; Frenois, F.X.; et al. Impaired functional responses in follicular lymphoma CD8(+)TIM-3(+) T lymphocytes following TCR engagement. Oncoimmunology 2016, 5, e1224044. [Google Scholar] [CrossRef]
- Nesterova, E.S.; Kravchenko, S.K.; Gemdjian, E.G.; Osmanov, E.A.; Kovrigina, A.M. Tumor-associated cytotoxic lymphocytes and macrophages as predictive factors in follicular lymphoma. Biochem. (Mosc.) Suppl. Ser. A Membr. Cell Biol. 2014, 8, 198–205. [Google Scholar]
- Blaker, Y.N.; Spetalen, S.; Brodtkorb, M.; Lingjaerde, O.C.; Beiske, K.; Østenstad, B.; Sander, B.; Wahlin, B.E.; Melen, C.M.; Myklebust, J.H.; et al. The tumour microenvironment influences survival and time to transformation in follicular lymphoma in the rituximab era. Br. J. Haematol. 2016, 175, 102–114. [Google Scholar] [CrossRef]
- Gršković, P.; Hančić, S.; Dotlić, S.; Matulić, M.; Ostojić Kolonić, S.; Gašparov, S.; Dominis, M.; Korać, P. CD4+/CD57+/CD69+ T lymphocytes and CD14+ dendritic cells accumulate in advanced follicular lymphoma. Immunobiology 2022, 227, 152257. [Google Scholar] [CrossRef]
- Lee, A.M.; Clear, A.J.; Calaminici, M.; Davies, A.J.; Jordan, S.; MacDougall, F.; Matthews, J.; Norton, A.J.; Gribben, J.G.; Lister, T.A.; et al. Number of CD4+ cells and location of forkhead box protein P3-positive cells in diagnostic follicular lymphoma tissue microarrays correlates with outcome. J. Clin. Oncol. 2006, 24, 5052–5059. [Google Scholar] [CrossRef]
- Szumera-Ciećkiewicz, A.; Poleszczuk, J.; Kuczkiewicz-Siemion, O.; Paszkiewicz-Kozik, E.; Rymkiewicz, G.; Sokół, K.; Borysiuk, A.; Kotarska, M.; Kawecka, M.; Owczarek, D.; et al. PD1 distribution pattern, regardless of the cell origin, is an independent microenvironmental prognostic factor for progression-free survival in follicular lymphoma. Pathol. Res. Pract. 2020, 216, 153096. [Google Scholar] [CrossRef]
- Farinha, P.; Al-Tourah, A.; Gill, K.; Klasa, R.; Connors, J.M.; Gascoyne, R.D. The architectural pattern of FOXP3-positive T cells in follicular lymphoma is an independent predictor of survival and histologic transformation. Blood 2010, 115, 289–295. [Google Scholar] [CrossRef]
- Carreras, J.; Lopez-Guillermo, A.; Fox, B.C.; Colomo, L.; Martinez, A.; Roncador, G.; Montserrat, E.; Campo, E.; Banham, A.H. High numbers of tumor-infiltrating FOXP3-positive regulatory T cells are associated with improved overall survival in follicular lymphoma. Blood 2006, 108, 2957–2964. [Google Scholar] [CrossRef]
- Li, C.; Guo, N.; Han, S.; Yu, H.; Lei, T.; Chen, X.; Peng, S.; Yang, H.; Wu, M. Impact of positive CD4 cells on event-free survival in follicular lymphoma patients. Cancer Med. 2024, 13, e70117. [Google Scholar] [CrossRef]
- Fahlquist Hagert, C.; Degn, S.E. T follicular regulatory cells: Guardians of the germinal centre? Scand. J. Immunol. 2020, 92, e12942. [Google Scholar] [CrossRef]
- Smeltzer, J.P.; Jones, J.M.; Ziesmer, S.C.; Grote, D.M.; Xiu, B.; Ristow, K.M.; Yang, Z.Z.; Nowakowski, G.S.; Feldman, A.L.; Cerhan, J.R.; et al. Pattern of CD14+ follicular dendritic cells and PD1+ T cells independently predicts time to transformation in follicular lymphoma. Clin. Cancer Res. 2014, 20, 2862–2872. [Google Scholar] [CrossRef]
- Tzankov, A.; Meier, C.; Hirschmann, P.; Went, P.; Pileri, S.A.; Dirnhofer, S. Correlation of high numbers of intratumoral FOXP3+ regulatory T cells with improved survival in germinal center-like diffuse large B-cell lymphoma, follicular lymphoma and classical Hodgkin’s lymphoma. Haematologica 2008, 93, 193–200. [Google Scholar] [CrossRef]
- Chevalier, N.; Mueller, M.; Mougiakakos, D.; Ihorst, G.; Marks, R.; Schmitt-Graeff, A.; Veelken, H. Analysis of dendritic cell subpopulations in follicular lymphoma with respect to the tumor immune microenvironment. Leuk. Lymphoma 2016, 57, 2150–2160. [Google Scholar] [CrossRef]
- Tsakiroglou, A.M.; Astley, S.; Dave, M.; Fergie, M.; Harkness, E.; Rosenberg, A.; Sperrin, M.; West, C.; Byers, R.; Linton, K. Immune infiltrate diversity confers a good prognosis in follicular lymphoma. Cancer Immunol. Immunother. 2021, 70, 3573–3585. [Google Scholar] [CrossRef]
- Nelson, L.S.; Mansfield, J.R.; Lloyd, R.; Oguejiofor, K.; Salih, Z.; Menasce, L.P.; Linton, K.M.; Rose, C.J.; Byers, R.J. Automated prognostic pattern detection shows favourable diffuse pattern of FOXP3(+) Tregs in follicular lymphoma. Br. J. Cancer 2015, 113, 1197–1205. [Google Scholar] [CrossRef]
- Carreras, J.; Lopez-Guillermo, A.; Roncador, G.; Villamor, N.; Colomo, L.; Martinez, A.; Hamoudi, R.; Howat, W.J.; Montserrat, E.; Campo, E. High numbers of tumor-infiltrating programmed cell death 1-positive regulatory lymphocytes are associated with improved overall survival in follicular lymphoma. J. Clin. Oncol. 2009, 27, 1470–1476. [Google Scholar] [CrossRef]
- Saifi, M.; Maran, A.; Raynaud, P.; Picot, M.C.; Quittet, P.; Cartron, G.; Rossi, J.F.; Costes, V. High ratio of interfollicular CD8/FOXP3-positive regulatory T cells is associated with a high FLIPI index and poor overall survival in follicular lymphoma. Exp. Ther. Med. 2010, 1, 933–938. [Google Scholar] [CrossRef][Green Version]
- Hagos, Y.B.; Akarca, A.U.; Ramsay, A.; Rossi, R.L.; Pomplun, S.; Ngai, V.; Moioli, A.; Gianatti, A.; McNamara, C.; Rambaldi, A.; et al. High inter-follicular spatial co-localization of CD8+FOXP3+ with CD4+CD8+ cells predicts favorable outcome in follicular lymphoma. Hematol. Oncol. 2022, 40, 541–553. [Google Scholar] [CrossRef]
- Koch, K.; Hoster, E.; Unterhalt, M.; Ott, G.; Rosenwald, A.; Hansmann, M.L.; Engelhard, M.; Hiddemann, W.; Klapper, W. The composition of the microenvironment in follicular lymphoma is associated with the stage of the disease. Hum. Pathol. 2012, 43, 2274–2281. [Google Scholar] [CrossRef]
- Spasevska, I.; Sharma, A.; Steen, C.B.; Josefsson, S.E.; Blaker, Y.N.; Kolstad, A.; Rustad, E.H.; Meyer, S.; Isaksen, K.; Chellappa, S.; et al. Diversity of intratumoral regulatory T cells in B-cell non-Hodgkin lymphoma. Blood Adv. 2023, 7, 7216–7230. [Google Scholar] [CrossRef]
- Fujiwara, S.; Muroi, K.; Tatara, R.; Matsuyama, T.; Ohmine, K.; Suzuki, T.; Mori, M.; Nagai, T.; Tanaka, A.; Ozawa, K. Clinical features of de novo CD25-positive follicular lymphoma. Leuk. Lymphoma 2014, 55, 307–313. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Kim, H.J.; Villasboas, J.C.; Price-Troska, T.; Jalali, S.; Wu, H.; Luchtel, R.A.; Polley, M.C.; Novak, A.J.; Ansell, S.M. Mass Cytometry Analysis Reveals that Specific Intratumoral CD4(+) T Cell Subsets Correlate with Patient Survival in Follicular Lymphoma. Cell Rep. 2019, 26, 2178–2193.e2173. [Google Scholar] [CrossRef]
- Liston, A.; Aloulou, M. A fresh look at a neglected regulatory lineage: CD8(+)Foxp3(+) Regulatory T cells. Immunol. Lett. 2022, 247, 22–26. [Google Scholar] [CrossRef]
- Chung, Y.; Tanaka, S.; Chu, F.; Nurieva, R.I.; Martinez, G.J.; Rawal, S.; Wang, Y.H.; Lim, H.; Reynolds, J.M.; Zhou, X.H.; et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 2011, 17, 983–988. [Google Scholar] [CrossRef]
- Gonzalez-Figueroa, P.; Roco, J.A.; Papa, I.; Núñez Villacís, L.; Stanley, M.; Linterman, M.A.; Dent, A.; Canete, P.F.; Vinuesa, C.G. Follicular regulatory T cells produce neuritin to regulate B cells. Cell 2021, 184, 1775–1789.e1719. [Google Scholar] [CrossRef]
- Linterman, M.A.; Pierson, W.; Lee, S.K.; Kallies, A.; Kawamoto, S.; Rayner, T.F.; Srivastava, M.; Divekar, D.P.; Beaton, L.; Hogan, J.J.; et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 2011, 17, 975–982. [Google Scholar] [CrossRef]
- Wollenberg, I.; Agua-Doce, A.; Hernández, A.; Almeida, C.; Oliveira, V.G.; Faro, J.; Graca, L. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J. Immunol. 2011, 187, 4553–4560. [Google Scholar] [CrossRef]
- Lu, Y.; Craft, J. T Follicular Regulatory Cells: Choreographers of Productive Germinal Center Responses. Front. Immunol. 2021, 12, 679909. [Google Scholar] [CrossRef]
- Qi, J.; Liu, C.; Bai, Z.; Li, X.; Yao, G. T follicular helper cells and T follicular regulatory cells in autoimmune diseases. Front. Immunol. 2023, 14, 1178792. [Google Scholar] [CrossRef]
- Graca, L.; Jacobsen, J.; Kumar, S. The expanding family of T follicular regulatory cells. Sci. Immunol. 2023, 8, eadg7526. [Google Scholar] [CrossRef]
- Le Coz, C.; Oldridge, D.A.; Herati, R.S.; De Luna, N.; Garifallou, J.; Cruz Cabrera, E.; Belman, J.P.; Pueschl, D.; Silva, L.V.; Knox, A.V.C.; et al. Human T follicular helper clones seed the germinal center-resident regulatory pool. Sci. Immunol. 2023, 8, eade8162. [Google Scholar] [CrossRef]
- Fonseca, V.R.; Ribeiro, F.; Graca, L. T follicular regulatory (Tfr) cells: Dissecting the complexity of Tfr-cell compartments. Immunol. Rev. 2019, 288, 112–127. [Google Scholar] [CrossRef]
- Sage, P.T.; Sharpe, A.H. The multifaceted functions of follicular regulatory T cells. Curr. Opin. Immunol. 2020, 67, 68–74. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Kim, H.J.; Wu, H.; Tang, X.; Yu, Y.; Krull, J.; Larson, D.P.; Moore, R.M.; Maurer, M.J.; Pavelko, K.D.; et al. T-cell phenotype including CD57(+) T follicular helper cells in the tumor microenvironment correlate with a poor outcome in follicular lymphoma. Blood Cancer J. 2023, 13, 124. [Google Scholar] [CrossRef]
- Butsch, R.; Lukas Waelti, S.; Schaerer, S.; Braun, J.; Korol, D.; Probst-Hensch, N.; Moch, H.; Kurrer, M. Intratumoral plasmacytoid dendritic cells associate with increased survival in patients with follicular lymphoma. Leuk. Lymphoma 2011, 52, 1230–1238. [Google Scholar] [CrossRef]
- Ramsay, A.G.; Clear, A.J.; Kelly, G.; Fatah, R.; Matthews, J.; Macdougall, F.; Lister, T.A.; Lee, A.M.; Calaminici, M.; Gribben, J.G. Follicular lymphoma cells induce T-cell immunologic synapse dysfunction that can be repaired with lenalidomide: Implications for the tumor microenvironment and immunotherapy. Blood 2009, 114, 4713–4720. [Google Scholar] [CrossRef]
- Meirav, K.; Ginette, S.; Tamar, T.; Iris, B.; Arnon, N.; Abraham, A. Extrafollicular PD1 and Intrafollicular CD3 Expression Are Associated With Survival in Follicular Lymphoma. Clin. Lymphoma Myeloma Leuk. 2017, 17, 645–649. [Google Scholar] [CrossRef]
- Taskinen, M.; Karjalainen-Lindsberg, M.L.; Nyman, H.; Eerola, L.M.; Leppä, S. A high tumor-associated macrophage content predicts favorable outcome in follicular lymphoma patients treated with rituximab and cyclophosphamide-doxorubicin-vincristine-prednisone. Clin. Cancer Res. 2007, 13, 5784–5789. [Google Scholar] [CrossRef]
- Miyashita, H.; Taoka, K.; Kume, A.; Ushiku, A.; Ushiku, T.; Toyama, K.; Kurokawa, M. Clinical usefulness of a novel classification of T cell distribution patterns in the tumor microenvironment of follicular lymphoma to detect transformation. Ann. Hematol. 2022, 101, 2477–2483. [Google Scholar] [CrossRef]
- Li, Y.; Hu, S.; Zuo, Z.; Hong, M.; Lin, P.; Li, S.; Konoplev, S.; Wang, Z.; Khoury, J.D.; Young, K.H.; et al. CD5-positive follicular lymphoma: Clinicopathologic correlations and outcome in 88 cases. Mod. Pathol. 2015, 28, 787–798. [Google Scholar] [CrossRef]
- Miyoshi, H.; Sato, K.; Yoshida, M.; Kimura, Y.; Kiyasu, J.; Ichikawa, A.; Ishibashi, Y.; Arakawa, F.; Nakamura, Y.; Nakashima, S.; et al. CD5-positive follicular lymphoma characterized by CD25, MUM1, low frequency of t(14;18) and poor prognosis. Pathol. Int. 2014, 64, 95–103. [Google Scholar] [CrossRef]
- Deng, Y.; Ma, J.; Zhao, S.; Yang, M.; Sun, Y.; Zhang, Q. Expression of glucose transporter-1 in follicular lymphoma affected tumor-infiltrating immunocytes and was related to progression of disease within 24 months. Transl. Oncol. 2023, 28, 101614. [Google Scholar] [CrossRef]
- Chu, F.; Li, H.S.; Liu, X.; Cao, J.; Ma, W.; Ma, Y.; Weng, J.; Zhu, Z.; Cheng, X.; Wang, Z.; et al. CXCR5(+)CD8(+) T cells are a distinct functional subset with an antitumor activity. Leukemia 2019, 33, 2640–2653. [Google Scholar] [CrossRef]
- Frank, M.J.; Reagan, P.M.; Bartlett, N.L.; Gordon, L.I.; Friedberg, J.W.; Czerwinski, D.K.; Long, S.R.; Hoppe, R.T.; Janssen, R.; Candia, A.F.; et al. In Situ Vaccination with a TLR9 Agonist and Local Low-Dose Radiation Induces Systemic Responses in Untreated Indolent Lymphoma. Cancer Discov. 2018, 8, 1258–1269. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Grote, D.M.; Xiu, B.; Ziesmer, S.C.; Price-Troska, T.L.; Hodge, L.S.; Yates, D.M.; Novak, A.J.; Ansell, S.M. TGF-β upregulates CD70 expression and induces exhaustion of effector memory T cells in B-cell non-Hodgkin’s lymphoma. Leukemia 2014, 28, 1872–1884. [Google Scholar] [CrossRef]
- Yanagida, E.; Miyoshi, H.; Takeuchi, M.; Shimono, J.; Nakashima, K.; Yamada, K.; Kawamoto, K.; Moritsubo, M.; Shimasaki, Y.; Inoue, K.; et al. Clinicopathological analysis of immunohistochemical expression of immune checkpoint molecules in follicular lymphoma. Hematol. Oncol. 2022, 40, 530–540. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Grote, D.M.; Ziesmer, S.C.; Xiu, B.; Novak, A.J.; Ansell, S.M. PD-1 expression defines two distinct T-cell sub-populations in follicular lymphoma that differentially impact patient survival. Blood Cancer J. 2015, 5, e281. [Google Scholar] [CrossRef]
- Tumedei, M.M.; Piccinini, F.; Azzali, I.; Pirini, F.; Bravaccini, S.; De Matteis, S.; Agostinelli, C.; Castellani, G.; Zanoni, M.; Cortesi, M.; et al. Follicular Lymphoma Microenvironment Traits Associated with Event-Free Survival. Int. J. Mol. Sci. 2023, 24, 9909. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Kim, H.J.; Villasboas, J.C.; Chen, Y.P.; Price-Troska, T.; Jalali, S.; Wilson, M.; Novak, A.J.; Ansell, S.M. Expression of LAG-3 defines exhaustion of intratumoral PD-1(+) T cells and correlates with poor outcome in follicular lymphoma. Oncotarget 2017, 8, 61425–61439. [Google Scholar] [CrossRef]
- Wu, H.; Tang, X.; Kim, H.J.; Jalali, S.; Pritchett, J.C.; Villasboas, J.C.; Novak, A.J.; Yang, Z.Z.; Ansell, S.M. Expression of KLRG1 and CD127 defines distinct CD8(+) subsets that differentially impact patient outcome in follicular lymphoma. J. Immunother. Cancer 2021, 9. [Google Scholar] [CrossRef]
- Milcent, B.; Josseaume, N.; Petitprez, F.; Riller, Q.; Amorim, S.; Loiseau, P.; Toubert, A.; Brice, P.; Thieblemont, C.; Teillaud, J.L.; et al. Recovery of central memory and naive peripheral T cells in Follicular Lymphoma patients receiving rituximab-chemotherapy based regimen. Sci. Rep. 2019, 9, 13471. [Google Scholar] [CrossRef]
- Muenst, S.; Hoeller, S.; Willi, N.; Dirnhofera, S.; Tzankov, A. Diagnostic and prognostic utility of PD-1 in B cell lymphomas. Dis. Markers. 2010, 29, 47–53. [Google Scholar] [CrossRef]
- Beck Enemark, M.; Monrad, I.; Madsen, C.; Lystlund Lauridsen, K.; Honoré, B.; Plesner, T.L.; Hamilton-Dutoit, S.J.; d’Amore, F.; Ludvigsen, M. PD-1 Expression in Pre-Treatment Follicular Lymphoma Predicts the Risk of Subsequent High-Grade Transformation. Onco Targets Ther. 2021, 14, 481–489. [Google Scholar] [CrossRef]
- Takahashi, H.; Tomita, N.; Sakata, S.; Tsuyama, N.; Hashimoto, C.; Ohshima, R.; Matsuura, S.; Ogawa, K.; Yamamoto, W.; Kameda, Y.; et al. Prognostic significance of programmed cell death-1-positive cells in follicular lymphoma patients may alter in the rituximab era. Eur. J. Haematol. 2013, 90, 286–290. [Google Scholar] [CrossRef]
- Sarkozy, C.; Wu, S.; Takata, K.; Aoki, T.; Neriah, S.B.; Milne, K.; Goodyear, T.; Strong, C.; Rastogi, T.; Hilton, L.K.; et al. Integrated single cell analysis reveals co-evolution of malignant B cells and tumor micro-environment in transformed follicular lymphoma. Cancer Cell 2024, 42, 1003–1017.e1006. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Kim, H.J.; Wu, H.; Jalali, S.; Tang, X.; Krull, J.E.; Ding, W.; Novak, A.J.; Ansell, S.M. TIGIT Expression Is Associated with T-cell Suppression and Exhaustion and Predicts Clinical Outcome and Anti-PD-1 Response in Follicular Lymphoma. Clin. Cancer Res. 2020, 26, 5217–5231. [Google Scholar] [CrossRef]
- Enemark, M.B.H.; Sørensen, E.F.; Hybel, T.E.; Andersen, M.D.; Madsen, C.; Lauridsen, K.L.; Honoré, B.; d’Amore, F.; Plesner, T.L.; Hamilton-Dutoit, S.J.; et al. IDO1 Protein Is Expressed in Diagnostic Biopsies from Both Follicular and Transformed Follicular Patients. Int. J. Mol. Sci. 2023, 24, 7314. [Google Scholar] [CrossRef]
- Nasir, A.; Hegerova, L.; Yousaf, H.; Forster, C.L.; Shanley, R.; Linden, M.A.; Bachanova, V.; Yohe, S. Digital and manual interfollicular Ki-67 are associated with a progression-free survival in patients with low-grade follicular lymphoma. Am. J. Clin. Pathol. 2023. [CrossRef]
- Masaki, A.; Ishida, T.; Maeda, Y.; Ito, A.; Suzuki, S.; Narita, T.; Kinoshita, S.; Yoshida, T.; Ri, M.; Kusumoto, S.; et al. Clinical significance of tryptophan catabolism in follicular lymphoma. Hematol. Oncol. 2020, 38, 742–753. [Google Scholar] [CrossRef]
- Shiozawa, E.; Yamochi-Onizuka, T.; Yamochi, T.; Yamamoto, Y.; Naitoh, H.; Kawakami, K.; Nakamaki, T.; Tomoyasu, S.; Kushima, M.; Ota, H. Disappearance of CD21-positive follicular dendritic cells preceding the transformation of follicular lymphoma: Immunohistological study of the transformation using CD21, p53, Ki-67, and P-glycoprotein. Pathol. Res. Pract. 2003, 199, 293–302. [Google Scholar] [CrossRef]
- Olteanu, H.; Fenske, T.S.; Harrington, A.M.; Szabo, A.; He, P.; Kroft, S.H. CD23 expression in follicular lymphoma: Clinicopathologic correlations. Am. J. Clin. Pathol. 2011, 135, 46–53. [Google Scholar] [CrossRef]
- Jin, M.K.; Hoster, E.; Dreyling, M.; Unterhalt, M.; Hiddemann, W.; Klapper, W. Follicular dendritic cells in follicular lymphoma and types of non-Hodgkin lymphoma show reduced expression of CD23, CD35 and CD54 but no association with clinical outcome. Histopathology 2011, 58, 586–592. [Google Scholar] [CrossRef]
- Misiak, J.; Jean, R.; Rodriguez, S.; Deleurme, L.; Lamy, T.; Tarte, K.; Amé-Thomas, P. Human Lymphoid Stromal Cells Contribute to Polarization of Follicular T Cells Into IL-4 Secreting Cells. Front. Immunol. 2020, 11, 559866. [Google Scholar] [CrossRef]
- Willenbrock, K.; Renné, C.; Rottenkolber, M.; Klapper, W.; Dreyling, M.; Engelhard, M.; Küppers, R.; Hansmann, M.L.; Jungnickel, B. The expression of activation induced cytidine deaminase in follicular lymphoma is independent of prognosis and stage. Histopathology 2009, 54, 509–512. [Google Scholar] [CrossRef]
- Li, Y.J.; Jiang, W.Q.; Rao, H.L.; Huang, J.J.; Xia, Y.; Huang, H.Q.; Lin, T.Y.; Xia, Z.J.; Li, S.; Li, Z.M. Expression of BAFF and BAFF-R in follicular lymphoma: Correlation with clinicopathologic characteristics and survival outcomes. PLoS ONE. 2012, 7, e50936. [Google Scholar] [CrossRef]
- Clear, A.J.; Lee, A.M.; Calaminici, M.; Ramsay, A.G.; Morris, K.J.; Hallam, S.; Kelly, G.; Macdougall, F.; Lister, T.A.; Gribben, J.G. Increased angiogenic sprouting in poor prognosis FL is associated with elevated numbers of CD163+ macrophages within the immediate sprouting microenvironment. Blood 2010, 115, 5053–5056. [Google Scholar] [CrossRef]
- Gouni, S.; Marques-Piubelli, M.L.; Strati, P. Follicular lymphoma and macrophages: Impact of approved and novel therapies. Blood Adv. 2021, 5, 4303–4312. [Google Scholar] [CrossRef]
- Andjelic, B.; Mihaljevic, B.; Todorovic, M.; Bila, J.; Jakovic, L.; Jovanovic, M.P. The number of lymphoma-associated macrophages in tumor tissue is an independent prognostic factor in patients with follicular lymphoma. Appl. Immunohistochem. Mol. Morphol. 2012, 20, 41–46. [Google Scholar] [CrossRef]
- Canioni, D.; Salles, G.; Mounier, N.; Brousse, N.; Keuppens, M.; Morchhauser, F.; Lamy, T.; Sonet, A.; Rousselet, M.C.; Foussard, C.; et al. High numbers of tumor-associated macrophages have an adverse prognostic value that can be circumvented by rituximab in patients with follicular lymphoma enrolled onto the GELA-GOELAMS FL-2000 trial. J. Clin. Oncol. 2008, 26, 440–446. [Google Scholar] [CrossRef]
- Kridel, R.; Xerri, L.; Gelas-Dore, B.; Tan, K.; Feugier, P.; Vawda, A.; Canioni, D.; Farinha, P.; Boussetta, S.; Moccia, A.A.; et al. The Prognostic Impact of CD163-Positive Macrophages in Follicular Lymphoma: A Study from the BC Cancer Agency and the Lymphoma Study Association. Clin. Cancer Res. 2015, 21, 3428–3435. [Google Scholar] [CrossRef]
- Farinha, P.; Kyle, A.H.; Minchinton, A.I.; Connors, J.M.; Karsan, A.; Gascoyne, R.D. Vascularization predicts overall survival and risk of transformation in follicular lymphoma. Haematologica 2010, 95, 2157–2160. [Google Scholar] [CrossRef]
- Alvaro, T.; Lejeune, M.; Camacho, F.I.; Salvadó, M.T.; Sánchez, L.; García, J.F.; Lopez, C.; Jaén, J.; Bosch, R.; Pons, L.E.; et al. The presence of STAT1-positive tumor-associated macrophages and their relation to outcome in patients with follicular lymphoma. Haematologica 2006, 91, 1605–1612. [Google Scholar]
- Valero, J.G.; Matas-Céspedes, A.; Arenas, F.; Rodriguez, V.; Carreras, J.; Serrat, N.; Guerrero-Hernández, M.; Yahiaoui, A.; Balagué, O.; Martin, S.; et al. The receptor of the colony-stimulating factor-1 (CSF-1R) is a novel prognostic factor and therapeutic target in follicular lymphoma. Leukemia 2021, 35, 2635–2649. [Google Scholar] [CrossRef]
- Glennie, M.J.; French, R.R.; Cragg, M.S.; Taylor, R.P. Mechanisms of killing by anti-CD20 monoclonal antibodies. Mol. Immunol. 2007, 44, 3823–3837. [Google Scholar] [CrossRef]
- Koster, A.; van Krieken, J.H.; Mackenzie, M.A.; Schraders, M.; Borm, G.F.; van der Laak, J.A.; Leenders, W.; Hebeda, K.; Raemaekers, J.M. Increased vascularization predicts favorable outcome in follicular lymphoma. Clin. Cancer Res. 2005, 11, 154–161. [Google Scholar] [CrossRef]
- Jelicic, J.; Balint, M.T.; Jovanovic, M.P.; Boricic, N.; Micev, M.; Stojsic, J.; Antic, D.; Andjelic, B.; Bila, J.; Balint, B.; et al. The Role of Lymphocyte to Monocyte Ratio, Microvessel Density and HiGH CD44 Tumor Cell Expression in Non Hodgkin Lymphomas. Pathol. Oncol. Res. 2016, 22, 567–577. [Google Scholar] [CrossRef]
- Jørgensen, J.M.; Sørensen, F.B.; Bendix, K.; Nielsen, J.L.; Olsen, M.L.; Funder, A.M.; d’Amore, F. Angiogenesis in non-Hodgkin’s lymphoma: Clinico-pathological correlations and prognostic significance in specific subtypes. Leuk. Lymphoma 2007, 48, 584–595. [Google Scholar] [CrossRef]
- Ohe, R.; Meng, H.X.; Yamada, A.; Ye Aung, N.; Kabasawa, T.; Tamura, Y.; Utsunomiya, A.; Tamazawa, N.; Kawamura, I.; Kitaoka, T.; et al. Good prognosis for follicular lymphoma with estrogen receptor α-positive follicular dendritic cells. Hematol. Oncol. 2020, 38, 293–300. [Google Scholar] [CrossRef]
- Jørgensen, J.M.; Sørensen, F.B.; Bendix, K.; Nielsen, J.L.; Funder, A.; Karkkainen, M.J.; Tainola, T.; Sørensen, A.B.; Pedersen, F.S.; D’Amore, F. Expression level, tissue distribution pattern, and prognostic impact of vascular endothelial growth factors VEGF and VEGF-C and their receptors Flt-1, KDR, and Flt-4 in different subtypes of non-Hodgkin lymphomas. Leuk. Lymphoma 2009, 50, 1647–1660. [Google Scholar] [CrossRef]
- Bose, S.; Le, A. Glucose Metabolism in Cancer. Adv. Exp. Med. Biol. 2018, 1063, 3–12. [Google Scholar]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef]
- Monrad, I.; Madsen, C.; Lauridsen, K.L.; Honoré, B.; Plesner, T.L.; Hamilton-Dutoit, S.; d’Amore, F.; Ludvigsen, M. Glycolytic biomarkers predict transformation in patients with follicular lymphoma. PLoS ONE 2020, 15, e0233449. [Google Scholar] [CrossRef]
- El-Sharkawy, A.; Malki, A. Vitamin D Signaling in Inflammation and Cancer: Molecular Mechanisms and Therapeutic Implications. Molecules 2020, 25, 3219. [Google Scholar] [CrossRef]
- Sheeley, M.P.; Andolino, C.; Kiesel, V.A.; Teegarden, D. Vitamin D regulation of energy metabolism in cancer. Br. J. Pharmacol. 2022, 179, 2890–2905. [Google Scholar] [CrossRef]
- Kelly, J.L.; Salles, G.; Goldman, B.; Fisher, R.I.; Brice, P.; Press, O.; Casasnovas, O.; Maloney, D.G.; Soubeyran, P.; Rimsza, L.; et al. Low Serum Vitamin D Levels Are Associated With Inferior Survival in Follicular Lymphoma: A Prospective Evaluation in SWOG and LYSA Studies. J. Clin. Oncol. 2015, 33, 1482–1490. [Google Scholar] [CrossRef]
- Tracy, S.I.; Maurer, M.J.; Witzig, T.E.; Drake, M.T.; Ansell, S.M.; Nowakowski, G.S.; Thompson, C.A.; Inwards, D.J.; Johnston, P.B.; Micallef, I.N.; et al. Vitamin D insufficiency is associated with an increased risk of early clinical failure in follicular lymphoma. Blood Cancer J. 2017, 7, e595. [Google Scholar] [CrossRef]
- Federico, M.; Caballero Barrigón, M.D.; Marcheselli, L.; Tarantino, V.; Manni, M.; Sarkozy, C.; Alonso-Álvarez, S.; Wondergem, M.; Cartron, G.; Lopez-Guillermo, A.; et al. Rituximab and the risk of transformation of follicular lymphoma: A retrospective pooled analysis. Lancet Haematol. 2018, 5, e359–e367. [Google Scholar] [CrossRef]
- Madsen, C.; Clausen, M.R.; Plesner, T.L.; Pasanen, A.; Kuismanen, T.; Bentzen, H.H.; Jørgensen, J.M.; Sillesen, I.B.; Himmelstrup, B.M.; Rønnov-Jessen, D.; et al. Up-front rituximab maintenance improves outcome in patients with follicular lymphoma: A collaborative Nordic study. Blood Adv. 2018, 2, 1562–1571. [Google Scholar] [CrossRef]
- Neumann, F.; Acker, F.; Schormann, C.; Pfreundschuh, M.; Bittenbring, J.T. Determination of optimum vitamin D3 levels for NK cell-mediated rituximab- and obinutuzumab-dependent cellular cytotoxicity. Cancer Immunol. Immunother. 2018, 67, 1709–1718. [Google Scholar] [CrossRef]
- Enemark, M.B.H.; Wolter, K.; Campbell, A.J.; Andersen, M.D.; Sørensen, E.F.; Hybel, T.E.; Madsen, C.; Lauridsen, K.L.; Plesner, T.L.; Hamilton-Dutoit, S.J.; et al. Proteomics identifies apoptotic markers as predictors of histological transformation in patients with follicular lymphoma. Blood Adv. 2023, 7, 7418–7432. [Google Scholar] [CrossRef]
- Naidoo, K.; Clay, V.; Hoyland, J.A.; Swindell, R.; Linton, K.; Illidge, T.; Radford, J.A.; Byers, R.J. YY1 expression predicts favourable outcome in follicular lymphoma. J. Clin. Pathol. 2011, 64, 125–129. [Google Scholar] [CrossRef]
- Huang, Q.; Li, F.; Liu, X.; Li, W.; Shi, W.; Liu, F.F.; O’Sullivan, B.; He, Z.; Peng, Y.; Tan, A.C.; et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 2011, 17, 860–866. [Google Scholar] [CrossRef]
- Castillo Ferrer, C.; Berthenet, K.; Ichim, G. Apoptosis—Fueling the oncogenic fire. Febs J. 2021, 288, 4445–4463. [Google Scholar] [CrossRef]
- Enemark, M.B.; Hybel, T.E.; Madsen, C.; Lauridsen, K.L.; Honoré, B.; Plesner, T.L.; Hamilton-Dutoit, S.; d’Amore, F.; Ludvigsen, M. Tumor-Tissue Expression of the Hyaluronic Acid Receptor RHAMM Predicts Histological Transformation in Follicular Lymphoma Patients. Cancers 2022, 14, 1316. [Google Scholar] [CrossRef]
- Dong, T.; Liu, Z.; Zhao, S.; Hu, C.; Liu, Y.; Ma, W.; Zhang, Q. The Expression of CD9 and PIK3CD is Associated with Prognosis of Follicular Lymphoma. J. Cancer 2015, 6, 1222–1229. [Google Scholar] [CrossRef]
- Kiaii, S.; Clear, A.J.; Ramsay, A.G.; Davies, D.; Sangaralingam, A.; Lee, A.; Calaminici, M.; Neuberg, D.S.; Gribben, J.G. Follicular lymphoma cells induce changes in T-cell gene expression and function: Potential impact on survival and risk of transformation. J. Clin. Oncol. 2013, 31, 2654–2661. [Google Scholar] [CrossRef]
- Suzuki, Y.; Yoshida, T.; Horie, R.; Tsuruta, T.; Togano, T.; Ohsaka, M.; Miyazaki, K.; Danbara, M.; Ohtani, S.; Okayasu, I.; et al. Constitutive activity of nuclear transcription factor kappaB is observed in follicular lymphoma. J. Clin. Exp. Hematop. 2010, 50, 45–50. [Google Scholar] [CrossRef][Green Version]
- Anderson, M.W.; Zhao, S.; Ai, W.Z.; Tibshirani, R.; Levy, R.; Lossos, I.S.; Natkunam, Y. C-C chemokine receptor 1 expression in human hematolymphoid neoplasia. Am. J. Clin. Pathol. 2010, 133, 473–483. [Google Scholar] [CrossRef]
- Wood, B.; Sikdar, S.; Choi, S.J.; Virk, S.; Alhejaily, A.; Baetz, T.; LeBrun, D.P. Abundant expression of interleukin-21 receptor in follicular lymphoma cells is associated with more aggressive disease. Leuk. Lymphoma 2013, 54, 1212–1220. [Google Scholar] [CrossRef]
- Nishi, T.; Takahashi, H.; Hashimura, M.; Yoshida, T.; Ohta, Y.; Saegusa, M. FilGAP, a Rac-specific Rho GTPase-activating protein, is a novel prognostic factor for follicular lymphoma. Cancer Med. 2015, 4, 808–818. [Google Scholar] [CrossRef]
- Ludvigsen, M.; Madsen, C.; Kamper, P.; Hamilton-Dutoit, S.J.; Bendix, K.; d’Amore, F.; Honoré, B. Histologically transformed follicular lymphoma exhibits protein profiles different from both non-transformed follicular and de novo diffuse large B-cell lymphoma. Blood Cancer J. 2015, 5, e293. [Google Scholar] [CrossRef]
- Shimono, J.; Miyoshi, H.; Yoshida, N.; Kato, T.; Sato, K.; Sugio, T.; Miyawaki, K.; Kurita, D.; Sasaki, Y.; Kawamoto, K.; et al. Analysis of GNA13 Protein in Follicular Lymphoma and its Association With Poor Prognosis. Am. J. Surg. Pathol. 2018, 42, 1466–1471. [Google Scholar] [CrossRef]
- Pennanen, H.; Kuittinen, O.; Soini, Y.; Turpeenniemi-Hujanen, T. Prognostic significance of p53 and matrix metalloproteinase-9 expression in follicular lymphoma. Eur. J. Haematol. 2008, 81, 289–297. [Google Scholar] [CrossRef]
- Meier, C.; Hoeller, S.; Bourgau, C.; Hirschmann, P.; Schwaller, J.; Went, P.; Pileri, S.A.; Reiter, A.; Dirnhofer, S.; Tzankov, A. Recurrent numerical aberrations of JAK2 and deregulation of the JAK2-STAT cascade in lymphomas. Mod. Pathol. 2009, 22, 476–487. [Google Scholar] [CrossRef]
- Krishnadasan, R.; Bifulco, C.; Kim, J.; Rodov, S.; Zieske, A.W.; Vanasse, G.J. Overexpression of SOCS3 is associated with decreased survival in a cohort of patients with de novo follicular lymphoma. Br. J. Haematol. 2006, 135, 72–75. [Google Scholar] [CrossRef]
- Peroja, P.; Haapasaari, K.M.; Mannisto, S.; Miinalainen, I.; Koivunen, P.; Leppä, S.; Karjalainen-Lindsberg, M.L.; Kuusisto, M.E.; Turpeenniemi-Hujanen, T.; Kuittinen, O.; et al. Total peroxiredoxin expression is associated with survival in patients with follicular lymphoma. Virchows Arch. 2016, 468, 623–630. [Google Scholar] [CrossRef]
- Pasanen, A.K.; Kuitunen, H.; Haapasaari, K.M.; Karihtala, P.; Kyllönen, H.; Soini, Y.; Turpeenniemi-Hujanen, T.; Kuittinen, O. Expression and prognostic evaluation of oxidative stress markers in an immunohistochemical study of B-cell derived lymphomas. Leuk. Lymphoma 2012, 53, 624–631. [Google Scholar] [CrossRef]
- Alavaikko, M.; Aine, R.; Rinne, A.; Järvinen, M.; Blanco, G.; Apaja-Sarkkinen, M.; Hopsu-Havu, V.K. Behaviour of dendritic reticulum cells possessing immunoreactive acid cysteine-proteinase inhibitor in human lymphoid secondary follicles and in follicular-centre cell lymphomas. Int. J. Cancer 1985, 35, 319–325. [Google Scholar] [CrossRef]
- AlJohani, N.; Choi, S.J.; Day, A.G.; Alhejaily, A.; Virk, S.; Baetz, T.; LeBrun, D.P. Abundant expression of BMI1 in follicular lymphoma is associated with reduced overall survival. Leuk. Lymphoma 2018, 59, 2211–2219. [Google Scholar] [CrossRef]
- Sun, R.X.; Coste, J.; Segara, C.; Rousset, T.; Fabbro, M.; Rème, T.; Legouffe, E.; Klein, B.; Rossi, J.F. MBR rearrangement and P-glycoprotein expression are not independent prognostic factors like p53 protein in malignant lymphoma. Clin. Lab. Haematol. 1998, 20, 87–94. [Google Scholar] [CrossRef]
- Sandison, H.E.; Usher, S.; Karimiani, E.G.; Ashton, G.; Menasce, L.P.; Radford, J.A.; Linton, K.; Byers, R.J. PLK1 and YY1 interaction in follicular lymphoma is associated with unfavourable outcome. J. Clin. Pathol. 2013, 66, 764–767. [Google Scholar] [CrossRef]
- Banoei, M.M.; Mahé, E.; Mansoor, A.; Stewart, D.; Winston, B.W.; Habibi, H.R.; Shabani-Rad, M.T. NMR-based metabolomic profiling can differentiate follicular lymphoma from benign lymph node tissues and may be predictive of outcome. Sci. Rep. 2022, 12, 8294. [Google Scholar] [CrossRef]
- Nozaki, K.; Sugahara, H.; Ueda, S.; Ishikawa, J.; Karasuno, T.; Iida, M.; Kamae, T.; Moriyama, Y.; Kawakami, M.; Kosugi, S.; et al. Pretreatment levels of serum soluble interleukin-2 receptor are useful in selecting the treatment regimen for newly diagnosed advanced-stage follicular lymphoma with low tumor burden. Int. J. Hematol. 2021, 114, 217–221. [Google Scholar] [CrossRef]
- Yoshizato, T.; Nannya, Y.; Imai, Y.; Ichikawa, M.; Kurokawa, M. Clinical significance of serum-soluble interleukin-2 receptor in patients with follicular lymphoma. Clin. Lymphoma Myeloma Leuk. 2013, 13, 410–416. [Google Scholar] [CrossRef]
- Kusano, Y.; Yokoyama, M.; Terui, Y.; Inoue, N.; Takahashi, A.; Yamauchi, H.; Tsuyama, N.; Nishimura, N.; Mishima, Y.; Takeuchi, K.; et al. High pretreatment level of soluble interleukin-2 receptor is a robust prognostic factor in patients with follicular lymphoma treated with R-CHOP-like therapy. Blood Cancer J. 2017, 7, e614. [Google Scholar] [CrossRef][Green Version]
- Mozas, P.; Rivas-Delgado, A.; Rivero, A.; Dlouhy, I.; Nadeu, F.; Balagué, O.; González-Farré, B.; Baumann, T.; Giné, E.; Delgado, J.; et al. High serum levels of IL-2R, IL-6, and TNF-α are associated with higher tumor burden and poorer outcome of follicular lymphoma patients in the rituximab era. Leuk. Res. 2020, 94, 106371. [Google Scholar] [CrossRef]
- Nozaki, K.; Sugahara, H.; Ueda, S.; Ishikawa, J.; Fuji, S.; Masaie, H.; Tada, Y.; Karasuno, T.; Iida, M.; Mitsui, H.; et al. Pretreatment serum soluble interleukin-2 receptor level predicts survival in patients with newly diagnosed follicular lymphoma. Leuk. Lymphoma 2020, 61, 2113–2121. [Google Scholar] [CrossRef]
- Yamamoto, M.; Tanaka, K.; Umezawa, Y.; Nagao, T.; Toyota, S.; Miura, O. Soluble Interleukin-2 Receptor Level at Diagnosis Predicts Prognosis of Patients With Follicular Lymphoma Irrespective of Initial Management Strategy. Anticancer. Res. 2019, 39, 5115–5122. [Google Scholar] [CrossRef]
- Mir, M.A.; Maurer, M.J.; Ziesmer, S.C.; Slager, S.L.; Habermann, T.; Macon, W.R.; Link, B.K.; Syrbu, S.; Witzig, T.; Friedberg, J.W.; et al. Elevated serum levels of IL-2R, IL-1RA, and CXCL9 are associated with a poor prognosis in follicular lymphoma. Blood 2015, 125, 992–998. [Google Scholar] [CrossRef]
- Umino, K.; Fujiwara, S.I.; Ito, S.; Mashima, K.; Minakata, D.; Nakano, H.; Yamasaki, R.; Kawasaki, Y.; Sugimoto, M.; Ashizawa, M.; et al. Serum soluble interleukin-2 receptor level at diagnosis predicts transformation in patients with follicular lymphoma. Leuk. Lymphoma 2017, 58, 316–323. [Google Scholar] [CrossRef]
- Procházka, V.; Papajík, T.; Faber, E.; Raida, L.; Kapitáňová, Z.; Langová, K.; Prouzová, Z.; Jarošová, M.; Indrák, K. Soluble interleukin-2 receptor level predicts survival in patients with follicular lymphoma treated with cyclophosphamide, doxorubicin, vincristine and prednisone chemotherapy in the rituximab era. Leuk. Lymphoma 2014, 55, 1584–1590. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Grote, D.M.; Ziesmer, S.C.; Manske, M.K.; Witzig, T.E.; Novak, A.J.; Ansell, S.M. Soluble IL-2Rα facilitates IL-2-mediated immune responses and predicts reduced survival in follicular B-cell non-Hodgkin lymphoma. Blood 2011, 118, 2809–2820. [Google Scholar] [CrossRef]
- Catellani, S.; Poggi, A.; Bruzzone, A.; Dadati, P.; Ravetti, J.L.; Gobbi, M.; Zocchi, M.R. Expansion of Vdelta1 T lymphocytes producing IL-4 in low-grade non-Hodgkin lymphomas expressing UL-16-binding proteins. Blood 2007, 109, 2078–2085. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Grote, D.M.; Ziesmer, S.C.; Niki, T.; Hirashima, M.; Novak, A.J.; Witzig, T.E.; Ansell, S.M. IL-12 upregulates TIM-3 expression and induces T cell exhaustion in patients with follicular B cell non-Hodgkin lymphoma. J. Clin. Investig. 2012, 122, 1271–1282. [Google Scholar] [CrossRef]
- Li, Y.J.; Li, Z.M.; Xia, Z.J.; Li, S.; Xia, Y.; Huang, H.Q.; Huang, J.J.; Yi, P.Y.; Jiang, W.Q. High APRIL but not BAFF serum levels are associated with poor outcome in patients with follicular lymphoma. Ann. Hematol. 2015, 94, 79–88. [Google Scholar] [CrossRef]
- Rawal, S.; Chu, F.; Zhang, M.; Park, H.J.; Nattamai, D.; Kannan, S.; Sharma, R.; Delgado, D.; Chou, T.; Lin, H.Y.; et al. Cross talk between follicular Th cells and tumor cells in human follicular lymphoma promotes immune evasion in the tumor microenvironment. J. Immunol. 2013, 190, 6681–6693. [Google Scholar] [CrossRef]
- Rogers, L.M.; Mott, S.L.; Smith, B.J.; Link, B.K.; Sahin, D.; Weiner, G.J. Complement-Regulatory Proteins CFHR1 and CFHR3 and Patient Response to Anti-CD20 Monoclonal Antibody Therapy. Clin. Cancer Res. 2017, 23, 954–961. [Google Scholar] [CrossRef][Green Version]
- Procházka, V.; Faber, E.; Raida, L.; Kapitáňová, Z.; Langová, K.; Indrák, K.; Papajík, T. High serum carbohydrate antigen-125 (CA-125) level predicts poor outcome in patients with follicular lymphoma independently of the FLIPI score. Int. J. Hematol. 2012, 96, 58–64. [Google Scholar] [CrossRef]
- Witzig, T.E.; Maurer, M.J.; Habermann, T.M.; Link, B.K.; Micallef, I.N.; Nowakowski, G.S.; Ansell, S.M.; Colgan, J.P.; Inwards, D.J.; Porrata, L.F.; et al. Elevated monoclonal and polyclonal serum immunoglobulin free light chain as prognostic factors in B- and T-cell non-Hodgkin lymphoma. Am. J. Hematol. 2014, 89, 1116–1120. [Google Scholar] [CrossRef][Green Version]
- Rai, S.; Inoue, H.; Hanamoto, H.; Matsuda, M.; Maeda, Y.; Wada, Y.; Haeno, T.; Watatani, Y.; Kumode, T.; Hirase, C.; et al. Low absolute lymphocyte count is a poor prognostic factor for untreated advanced follicular lymphoma treated with rituximab plus bendamustine: Results of the prospective phase 2 CONVERT trial. Int. J. Hematol. 2021, 114, 205–216. [Google Scholar] [CrossRef]
- Yoshida, N.; Oda, M.; Kuroda, Y.; Katayama, Y.; Okikawa, Y.; Masunari, T.; Fujiwara, M.; Nishisaka, T.; Sasaki, N.; Sadahira, Y.; et al. Clinical significance of sIL-2R levels in B-cell lymphomas. PLoS ONE 2013, 8, e78730. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Ohwada, C.; Takeuchi, M.; Shimizu, N.; Sakaida, E.; Takeda, Y.; Sakai, S.; Tsukamoto, S.; Yamazaki, A.; Sugita, Y.; et al. LR11, a novel biomarker identified in follicular lymphoma. Br. J. Haematol. 2013, 163, 277–280. [Google Scholar] [CrossRef]
- Stevens, J.; Waters, R.; Sieniawska, C.; Kassam, S.; Montoto, S.; Fitzgibbon, J.; Rohatiner, A.; Lister, A.; Joel, S. Serum selenium concentration at diagnosis and outcome in patients with haematological malignancies. Br. J. Haematol. 2011, 154, 448–456. [Google Scholar] [CrossRef]
- Procházka, V.; Faber, E.; Raida, L.; Langová, K.; Indrák, K.; Papajík, T. High baseline serum thymidine kinase 1 level predicts unfavorable outcome in patients with follicular lymphoma. Leuk. Lymphoma 2012, 53, 1306–1310. [Google Scholar] [CrossRef]
- Xue, L.G.; Shen, H.R.; Gao, R.; Du, K.X.; Xing, T.Y.; Wang, W.T.; Wang, L.; Li, J.Y.; Liang, J.H.; Xu, W. Low T3 syndrome as a predictor of poor outcomes in patients with follicular lymphoma. Ann. Hematol. 2023, 102, 851–862. [Google Scholar] [CrossRef]
- Matsuo, T.; Tashiro, H.; Shirasaki, R.; Sumiyoshi, R.; Yamamoto, T.; Saito, S.; Matsumoto, K.; Ooi, J.; Shirafuji, N. Serum high-density lipoprotein cholesterol level has a significant prognostic impact on outcomes of follicular lymphoma patients. Medicine 2022, 101, e29541. [Google Scholar] [CrossRef]
- Jiménez-Ubieto, A.; Poza, M.; Martin-Muñoz, A.; Ruiz-Heredia, Y.; Dorado, S.; Figaredo, G.; Rosa-Rosa, J.M.; Rodriguez, A.; Barcena, C.; Navamuel, L.P.; et al. Real-life disease monitoring in follicular lymphoma patients using liquid biopsy ultra-deep sequencing, P.E.T./.C.T. Leukemia 2023, 37, 659–669. [Google Scholar] [CrossRef]
- Sarkozy, C.; Huet, S.; Carlton, V.E.; Fabiani, B.; Delmer, A.; Jardin, F.; Delfau-Larue, M.H.; Hacini, M.; Ribrag, V.; Guidez, S.; et al. The prognostic value of clonal heterogeneity and quantitative assessment of plasma circulating clonal IG-VDJ sequences at diagnosis in patients with follicular lymphoma. Oncotarget 2017, 8, 8765–8774. [Google Scholar] [CrossRef]
- Zhao, M.; Li, Q.; Yang, J.; Zhang, M.; Liu, X.; Zhang, H.; Huang, Y.; Li, J.; Bao, J.; Wang, J.; et al. Application of circulating tumour DNA in terms of prognosis prediction in Chinese follicular lymphoma patients. Front. Genet. 2023, 14, 1066808. [Google Scholar] [CrossRef]
- Shirouchi, Y.; Mishima, Y.; Takayama, T.; Minowa, S.; Ishihara, Y.; Tamba, M.; Hirano, M.; Onda, N.; Takeuchi, K.; Maruyama, D. Serum cell-free DNA concentration as a possible prognostic marker in newly diagnosed diffuse large B-cell lymphoma. Biomed. Res. 2022, 43, 99–106. [Google Scholar] [CrossRef]
- Casulo, C. Follicular lymphoma: Is there an optimal way to define risk? Hematology Am. Soc. Hematol. Educ. Program 2021, 2021, 313–319. [Google Scholar] [CrossRef]
- Rajamäki, A.; Sunela, K.; Prusila, R.E.I.; Kuusisto, M.E.L.; Mercadal, S.; Selander, T.; Kuitunen, H.; Pollari, M.; Jantunen, E.; Nystrand, I.; et al. Female patients with follicular lymphoma have a better prognosis if primary remission lasts over 24 months. Leuk. Lymphoma 2021, 62, 1639–1647. [Google Scholar] [CrossRef]
- Radkiewicz, C.; Bruchfeld, J.B.; Weibull, C.E.; Jeppesen, M.L.; Frederiksen, H.; Lambe, M.; Jakobsen, L.; El-Galaly, T.C.; Smedby, K.E.; Wästerlid, T. Sex differences in lymphoma incidence and mortality by subtype: A population-based study. Am. J. Hematol. 2023, 98, 23–30. [Google Scholar] [CrossRef]
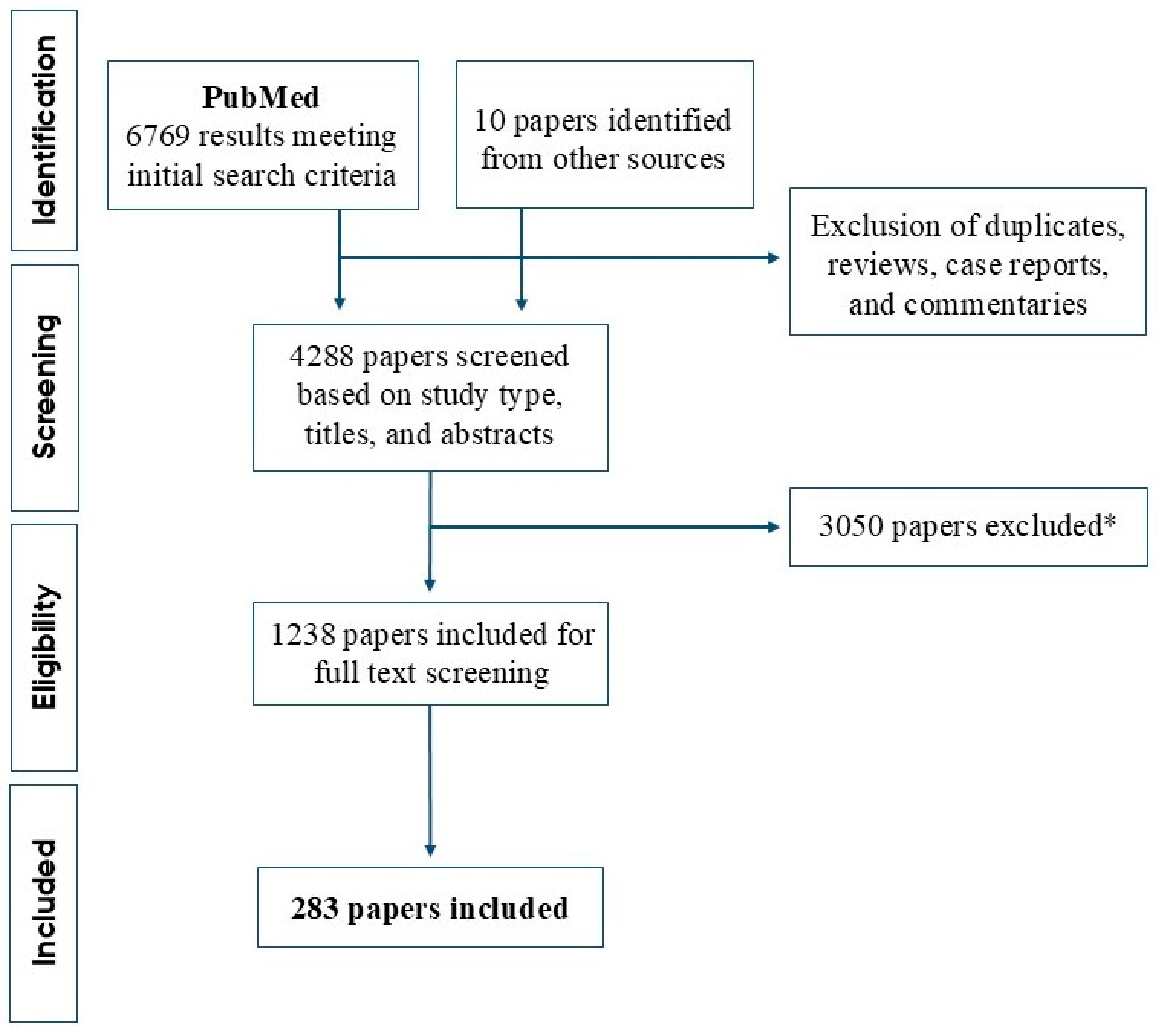
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enemark, M.H.; Hemmingsen, J.K.; Jensen, M.L.; Kridel, R.; Ludvigsen, M. Molecular Biomarkers in Prediction of High-Grade Transformation and Outcome in Patients with Follicular Lymphoma: A Comprehensive Systemic Review. Int. J. Mol. Sci. 2024, 25, 11179. https://doi.org/10.3390/ijms252011179
Enemark MH, Hemmingsen JK, Jensen ML, Kridel R, Ludvigsen M. Molecular Biomarkers in Prediction of High-Grade Transformation and Outcome in Patients with Follicular Lymphoma: A Comprehensive Systemic Review. International Journal of Molecular Sciences. 2024; 25(20):11179. https://doi.org/10.3390/ijms252011179
Chicago/Turabian StyleEnemark, Marie Hairing, Jonas Klejs Hemmingsen, Maja Lund Jensen, Robert Kridel, and Maja Ludvigsen. 2024. "Molecular Biomarkers in Prediction of High-Grade Transformation and Outcome in Patients with Follicular Lymphoma: A Comprehensive Systemic Review" International Journal of Molecular Sciences 25, no. 20: 11179. https://doi.org/10.3390/ijms252011179
APA StyleEnemark, M. H., Hemmingsen, J. K., Jensen, M. L., Kridel, R., & Ludvigsen, M. (2024). Molecular Biomarkers in Prediction of High-Grade Transformation and Outcome in Patients with Follicular Lymphoma: A Comprehensive Systemic Review. International Journal of Molecular Sciences, 25(20), 11179. https://doi.org/10.3390/ijms252011179






