Impacts of Hyperglycemia on Epigenetic Modifications in Human Gingival Fibroblasts and Gingiva in Diabetic Rats
Abstract
1. Introduction
2. Results
2.1. Body Weight, Blood, and Alveolar Bone Evaluation
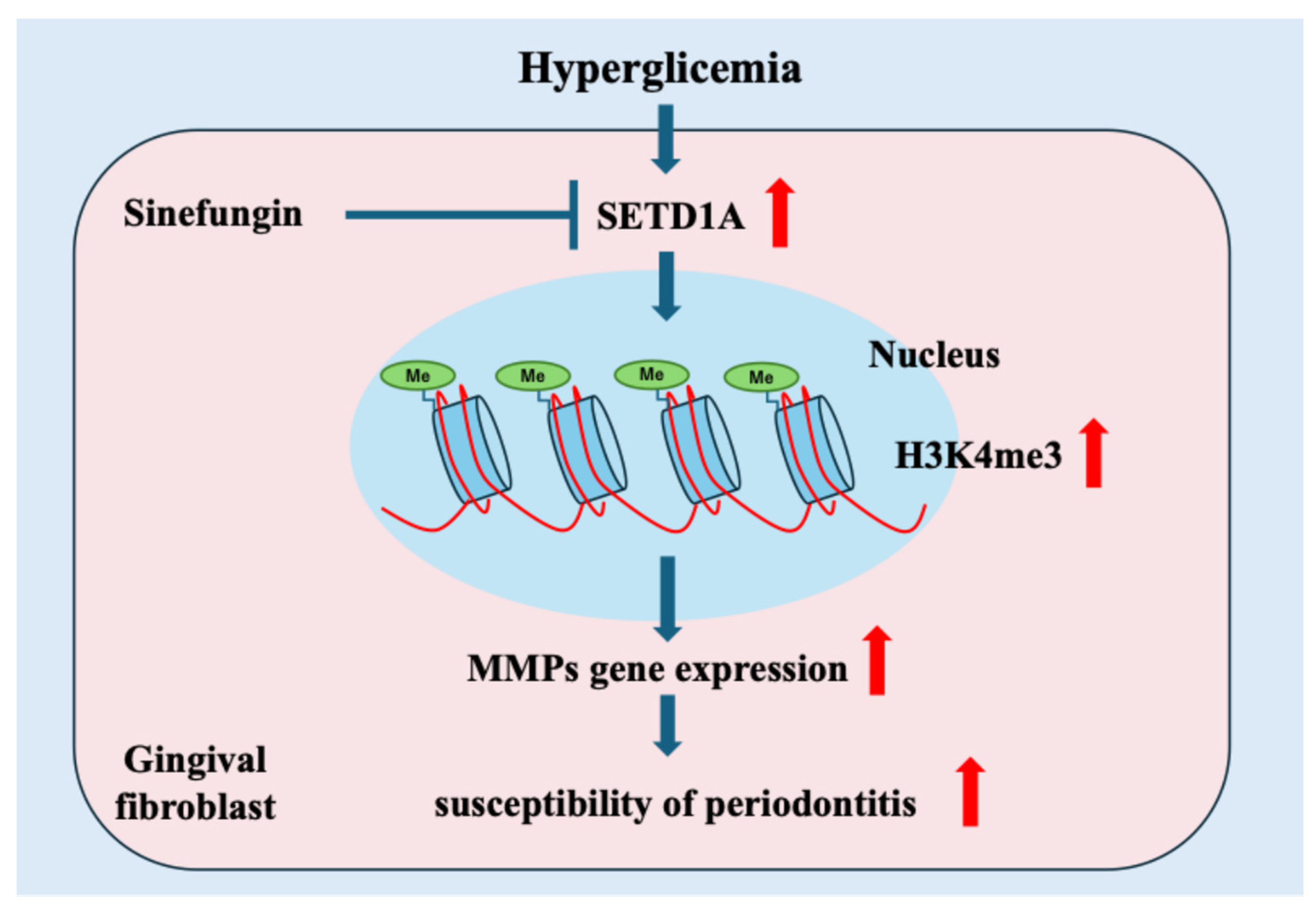
2.2. Diabetes Upregulates Histone Methylation and SETD1A in the Gingival Tissues
2.3. Expression of Histone-Methylated Proteins and SETD1A and the Effects of Histone Methyltransferase (HMT) Inhibitor in High-Glucose-Stimulated hGFs
2.4. Sinefungin Regulates Gene Expression of MMPs and Improves the MMP-TIMP Ratio (MMP/TIMP) in hGFs
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Induction of Diabetes
4.3. Tissue Collection
4.4. Microcomputed Tomography
4.5. Immunohistological Evaluation of Periodontal Tissue
4.6. Cell Culture
4.7. Immunohistochemical Staining of Gingival Fibroblasts
4.8. Gene Expression Analyses
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abe, T.; Hajishengallis, G. Optimization of the ligature-induced periodontitis model in mice. J. Immunol. Methods 2013, 394, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Slots, J. Periodontitis: Facts, fallacies and the future. Periodontol. 2000 2017, 75, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Grossi, S.G.; Genco, R.J. Periodontal disease and diabetes mellitus: A two-way relationship. Ann. Periodontol. 1998, 3, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Chapple, I.L.C.; Mealey, B.L.; Van Dyke, T.E.; Bartold, P.M.; Dommisch, H.; Eickholz, P.; Geisinger, M.L.; Genco, R.J.; Glogauer, M.; Goldstein, M.; et al. Periodontal health and gingival diseases and conditions on an intact and a reduced periodontium: Consensus report of workgroup 1 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. S1), S74–S84. [Google Scholar] [CrossRef]
- American Diabetes Association Professional Practice Committee. 2. Diagnosis and Classification of Diabetes: Standards of Care in Diabetes-2024. Diabetes Care 2024, 47 (Suppl. S1), S20–S42. [Google Scholar] [CrossRef]
- Graziani, F.; Gennai, S.; Solini, A.; Petrini, M. A systematic review and meta-analysis of epidemiologic observational evidence on the effect of periodontitis on diabetes An update of the EFP-AAP review. J. Clin. Periodontol. 2018, 45, 167–187. [Google Scholar] [CrossRef]
- Loe, H. Periodontal disease. The sixth complication of diabetes mellitus. Diabetes Care 1993, 16, 329–334. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Xiao, E.; Graves, D.T. Diabetes mellitus related bone metabolism and periodontal disease. Int. J. Oral. Sci. 2015, 7, 63–72. [Google Scholar] [CrossRef]
- Demmer, R.T.; Holtfreter, B.; Desvarieux, M.; Jacobs, D.R., Jr.; Kerner, W.; Nauck, M.; Volzke, H.; Kocher, T. The influence of type 1 and type 2 diabetes on periodontal disease progression: Prospective results from the Study of Health in Pomerania (SHIP). Diabetes Care 2012, 35, 2036–2042. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Clin. Periodontol. 2018, 45 (Suppl. S20), S149–S161. [Google Scholar] [CrossRef]
- Li, X.; Lu, L.; Hou, W.; Huang, T.; Chen, X.; Qi, J.; Zhao, Y.; Zhu, M. Epigenetics in the pathogenesis of diabetic nephropathy. Acta Biochim. Biophys. Sin. 2022, 54, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Yamada, S. Epigenetics in susceptibility, progression, and diagnosis of periodontitis. Jpn. Dent. Sci. Rev. 2022, 58, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Bhamidipati, T.; Kumar, M.; Verma, S.S.; Mohanty, S.K.; Kacar, S.; Reese, D.; Martinez, M.M.; Kamocka, M.M.; Dunn, K.W.; Sen, C.K.; et al. Epigenetic basis of diabetic vasculopathy. Front. Endocrinol. 2022, 13, 989844. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Lauberth, S.M.; Nakayama, T.; Wu, X.; Ferris, A.L.; Tang, Z.; Hughes, S.H.; Roeder, R.G. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 2013, 152, 1021–1036. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef]
- Benayoun, B.A.; Pollina, E.A.; Ucar, D.; Mahmoudi, S.; Karra, K.; Wong, E.D.; Devarajan, K.; Daugherty, A.C.; Kundaje, A.B.; Mancini, E.; et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 2014, 158, 673–688. [Google Scholar] [CrossRef]
- Howe, F.S.; Fischl, H.; Murray, S.C.; Mellor, J. Is H3K4me3 instructive for transcription activation? Bioessays 2017, 39, 1–12. [Google Scholar] [CrossRef]
- Okada, Y.; Feng, Q.; Lin, Y.; Jiang, Q.; Li, Y.; Coffield, V.M.; Su, L.; Xu, G.; Zhang, Y. hDOT1L links histone methylation to leukemogenesis. Cell 2005, 121, 167–178. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, F.; Gafken, P.R.; Gottschling, D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 2002, 109, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Marmorstein, R. Structure of SET domain proteins: A new twist on histone methylation. Trends Biochem. Sci. 2003, 28, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Qiu, T.; Wei, G.; Que, Y.; Wang, W.; Kong, Y.; Xie, T.; Chen, X. Role of Histone Post-Translational Modifications in Inflammatory Diseases. Front. Immunol. 2022, 13, 852272. [Google Scholar] [CrossRef]
- Yi, X.; Jiang, X.J.; Fang, Z.M. Histone methyltransferase SMYD2: Ubiquitous regulator of disease. Clin. Epigenetics 2019, 11, 112. [Google Scholar] [CrossRef]
- Amalinei, C.; Caruntu, I.D.; Giusca, S.E.; Balan, R.A. Matrix metalloproteinases involvement in pathologic conditions. Rom. J. Morphol. Embryol. 2010, 51, 215–228. [Google Scholar]
- Cabral-Pacheco, G.A.; Garza-Veloz, I.; Castruita-De la Rosa, C.; Ramirez-Acuna, J.M.; Perez-Romero, B.A.; Guerrero-Rodriguez, J.F.; Martinez-Avila, N.; Martinez-Fierro, M.L. The Roles of Matrix Metalloproteinases and Their Inhibitors in Human Diseases. Int. J. Mol. Sci. 2020, 21, 9739. [Google Scholar] [CrossRef]
- Yang, B.; Alimperti, S.; Gonzalez, M.V.; Dentchev, T.; Kim, M.; Suh, J.; Titchenell, P.M.; Ko, K.I.; Seykora, J.; Benakanakere, M.; et al. Reepithelialization of Diabetic Skin and Mucosal Wounds Is Rescued by Treatment With Epigenetic Inhibitors. Diabetes 2024, 73, 120–134. [Google Scholar] [CrossRef]
- Edgar, L.; Akbar, N.; Braithwaite, A.T.; Krausgruber, T.; Gallart-Ayala, H.; Bailey, J.; Corbin, A.L.; Khoyratty, T.E.; Chai, J.T.; Alkhalil, M.; et al. Hyperglycemia Induces Trained Immunity in Macrophages and Their Precursors and Promotes Atherosclerosis. Circulation 2021, 144, 961–982. [Google Scholar] [CrossRef]
- Sasaki, K.; Doi, S.; Nakashima, A.; Irifuku, T.; Yamada, K.; Kokoroishi, K.; Ueno, T.; Doi, T.; Hida, E.; Arihiro, K.; et al. Inhibition of SET Domain-Containing Lysine Methyltransferase 7/9 Ameliorates Renal Fibrosis. J. Am. Soc. Nephrol. 2016, 27, 203–215. [Google Scholar] [CrossRef]
- Kimball, A.S.; Joshi, A.; Carson, W.F., IV; Boniakowski, A.E.; Schaller, M.; Allen, R.; Bermick, J.; Davis, F.M.; Henke, P.K.; Burant, C.F.; et al. The Histone Methyltransferase MLL1 Directs Macrophage-Mediated Inflammation in Wound Healing and Is Altered in a Murine Model of Obesity and Type 2 Diabetes. Diabetes 2017, 66, 2459–2471. [Google Scholar] [CrossRef] [PubMed]
- Pandya Thakkar, N.; Pereira, B.M.V.; Katakia, Y.T.; Ramakrishnan, S.K.; Thakar, S.; Sakhuja, A.; Rajeev, G.; Soorya, S.; Thieme, K.; Majumder, S. Elevated H3K4me3 Through MLL2-WDR82 upon Hyperglycemia Causes Jagged Ligand Dependent Notch Activation to Interplay with Differentiation State of Endothelial Cells. Front. Cell Dev. Biol. 2022, 10, 839109. [Google Scholar] [CrossRef] [PubMed]
- Clark, I.M.; Swingler, T.E.; Sampieri, C.L.; Edwards, D.R. The regulation of matrix metalloproteinases and their inhibitors. Int. J. Biochem. Cell Biol. 2008, 40, 1362–1378. [Google Scholar] [CrossRef] [PubMed]
- Nitta, H.; Katagiri, S.; Nagasawa, T.; Izumi, Y.; Ishikawa, I.; Izumiyama, H.; Uchimura, I.; Kanazawa, M.; Chiba, H.; Matsuo, A.; et al. The number of microvascular complications is associated with an increased risk for severity of periodontitis in type 2 diabetes patients: Results of a multicenter hospital-based cross-sectional study. J. Diabetes Investig. 2017, 8, 677–686. [Google Scholar] [CrossRef]
- Waldron, A.L.; Schroder, P.A.; Bourgon, K.L.; Bolduc, J.K.; Miller, J.L.; Pellegrini, A.D.; Dubois, A.L.; Blaszkiewicz, M.; Townsend, K.L.; Rieger, S. Oxidative stress-dependent MMP-13 activity underlies glucose neurotoxicity. J. Diabetes Complicat. 2018, 32, 249–257. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, C.; Zhang, S.; Ke, Z.X.; Chen, D.X.; Li, Y.Q.; Li, Q. The Imbalance of MMP-2/TIMP-2 and MMP-9/TIMP-1 Contributes to Collagen Deposition Disorder in Diabetic Non-Injured Skin. Front. Endocrinol. 2021, 12, 734485. [Google Scholar] [CrossRef]
- Kubota, T.; Itagaki, M.; Hoshino, C.; Nagata, M.; Morozumi, T.; Kobayashi, T.; Takagi, R.; Yoshie, H. Altered gene expression levels of matrix metalloproteinases and their inhibitors in periodontitis-affected gingival tissue. J. Periodontol. 2008, 79, 166–173. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Bi, L.J. Alterations of collagen-I, MMP-1 and TIMP-1 in the periodontal ligament of diabetic rats under mechanical stress. J. Periodontal Res. 2011, 46, 448–455. [Google Scholar] [CrossRef]
- Guo, Q.; Li, X.; Han, H.; Li, C.; Liu, S.; Gao, W.; Sun, G. Histone Lysine Methylation in TGF-beta1 Mediated p21 Gene Expression in Rat Mesangial Cells. Biomed. Res. Int. 2016, 2016, 6927234. [Google Scholar] [CrossRef]
- Kera, Y.; Katoh, Y.; Ohta, M.; Matsumoto, M.; Takano-Yamamoto, T.; Igarashi, K. Methionine adenosyltransferase II-dependent histone H3K9 methylation at the COX-2 gene locus. J. Biol. Chem. 2013, 288, 13592–13601. [Google Scholar] [CrossRef]
- Villeneuve, L.M.; Reddy, M.A.; Lanting, L.L.; Wang, M.; Meng, L.; Natarajan, R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc. Natl. Acad. Sci. USA 2008, 105, 9047–9052. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Qian, X.; Shen, J.; Wang, Y.; Li, X.; Liu, R.; Xia, Y.; Chen, Q.; Peng, G.; Lin, S.Y.; et al. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat. Cell Biol. 2015, 17, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deng, P.; Liu, Y.; Wu, Y.; Chen, Y.; Guo, Y.; Zhang, S.; Zheng, X.; Zhou, L.; Liu, W.; et al. Alpha-ketoglutarate ameliorates age-related osteoporosis via regulating histone methylations. Nat. Commun. 2020, 11, 5596. [Google Scholar] [CrossRef] [PubMed]
- Montano, E.N.; Bose, M.; Huo, L.; Tumurkhuu, G.; De Los Santos, G.; Simental, B.; Stotland, A.B.; Wei, J.; Bairey Merz, C.N.; Suda, J.; et al. Alpha-Ketoglutarate-Dependent KDM6 Histone Demethylases and Interferon-Stimulated Gene Expression in Lupus. Arthritis Rheumatol. 2024, 76, 396–410. [Google Scholar] [CrossRef]
- Huang, F.; Luo, X.; Ou, Y.; Gao, Z.; Tang, Q.; Chu, Z.; Zhu, X.; He, Y. Control of histone demethylation by nuclear-localized alpha-ketoglutarate dehydrogenase. Science 2023, 381, eadf8822. [Google Scholar] [CrossRef]
- Adachi, K.; Miyajima, S.I.; Nakamura, N.; Miyabe, M.; Kobayashi, Y.; Nishikawa, T.; Suzuki, Y.; Kikuchi, T.; Kobayashi, S.; Saiki, T.; et al. Role of poly(ADP-ribose) polymerase activation in the pathogenesis of periodontitis in diabetes. J. Clin. Periodontol. 2017, 44, 971–980. [Google Scholar] [CrossRef]
- Miyajima, S.; Naruse, K.; Kobayashi, Y.; Nakamura, N.; Nishikawa, T.; Adachi, K.; Suzuki, Y.; Kikuchi, T.; Mitani, A.; Mizutani, M.; et al. Periodontitis-activated monocytes/macrophages cause aortic inflammation. Sci. Rep. 2014, 4, 5171. [Google Scholar] [CrossRef]
- Kondo, S.; Kojima, K.; Nakamura, N.; Miyabe, M.; Kikuchi, T.; Ohno, T.; Sawada, N.; Minato, T.; Saiki, T.; Ito, M.; et al. Increased expression of angiopoietin-like protein 4 regulates matrix metalloproteinase-13 expression in Porphyromonas gingivalis lipopolysaccharides-stimulated gingival fibroblasts and ligature-induced experimental periodontitis. J. Periodontal Res. 2023, 58, 43–52. [Google Scholar] [CrossRef]
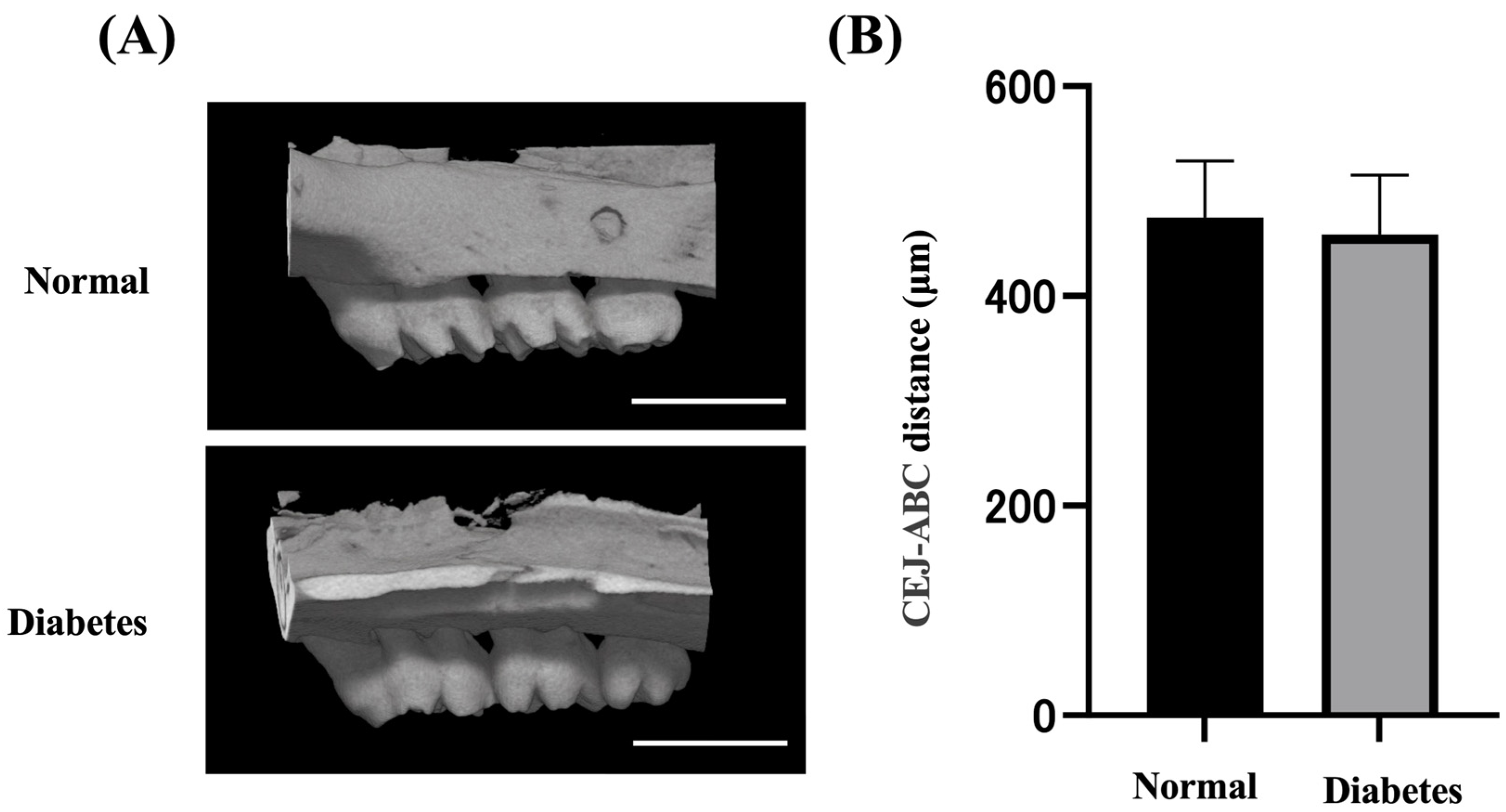
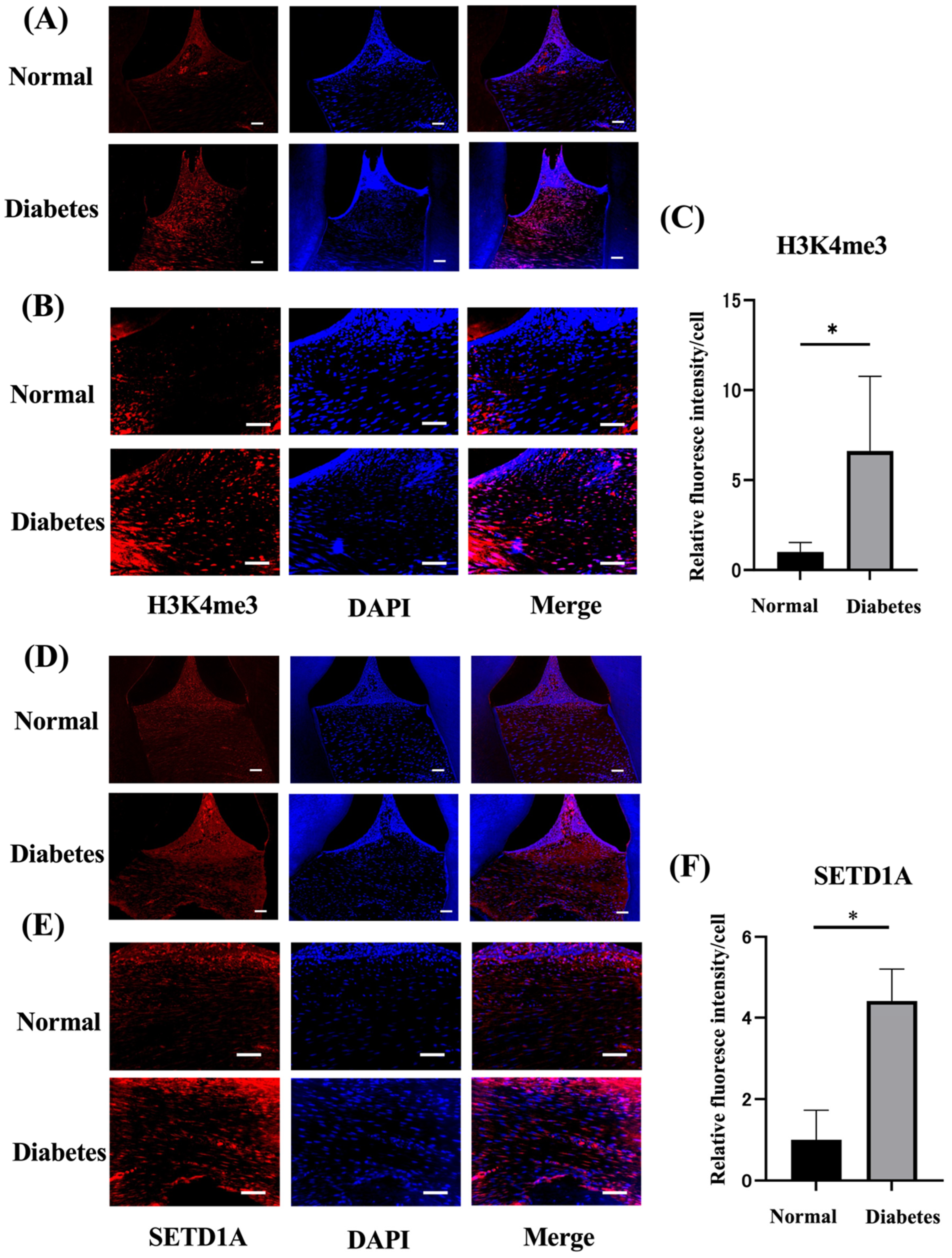
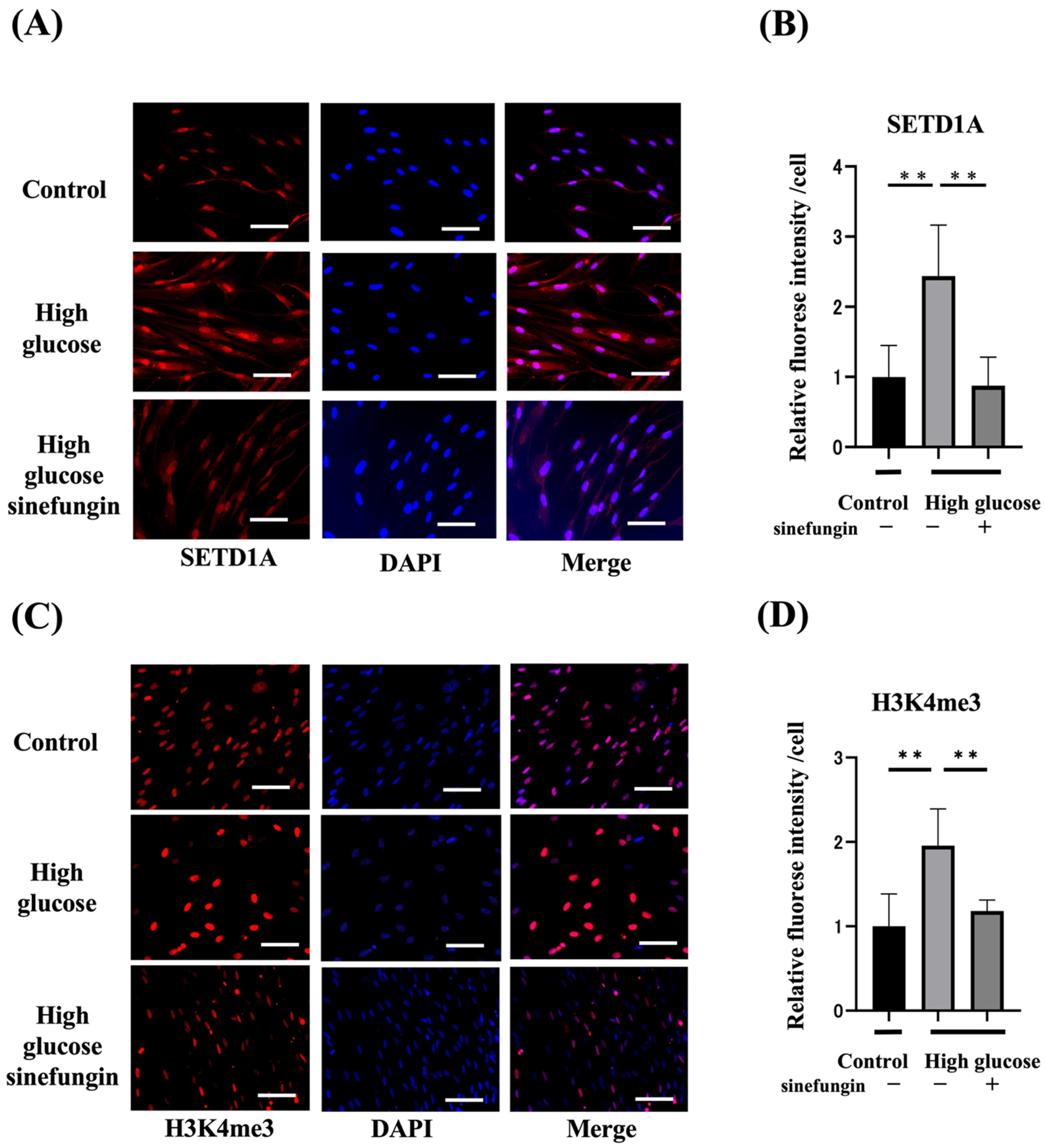
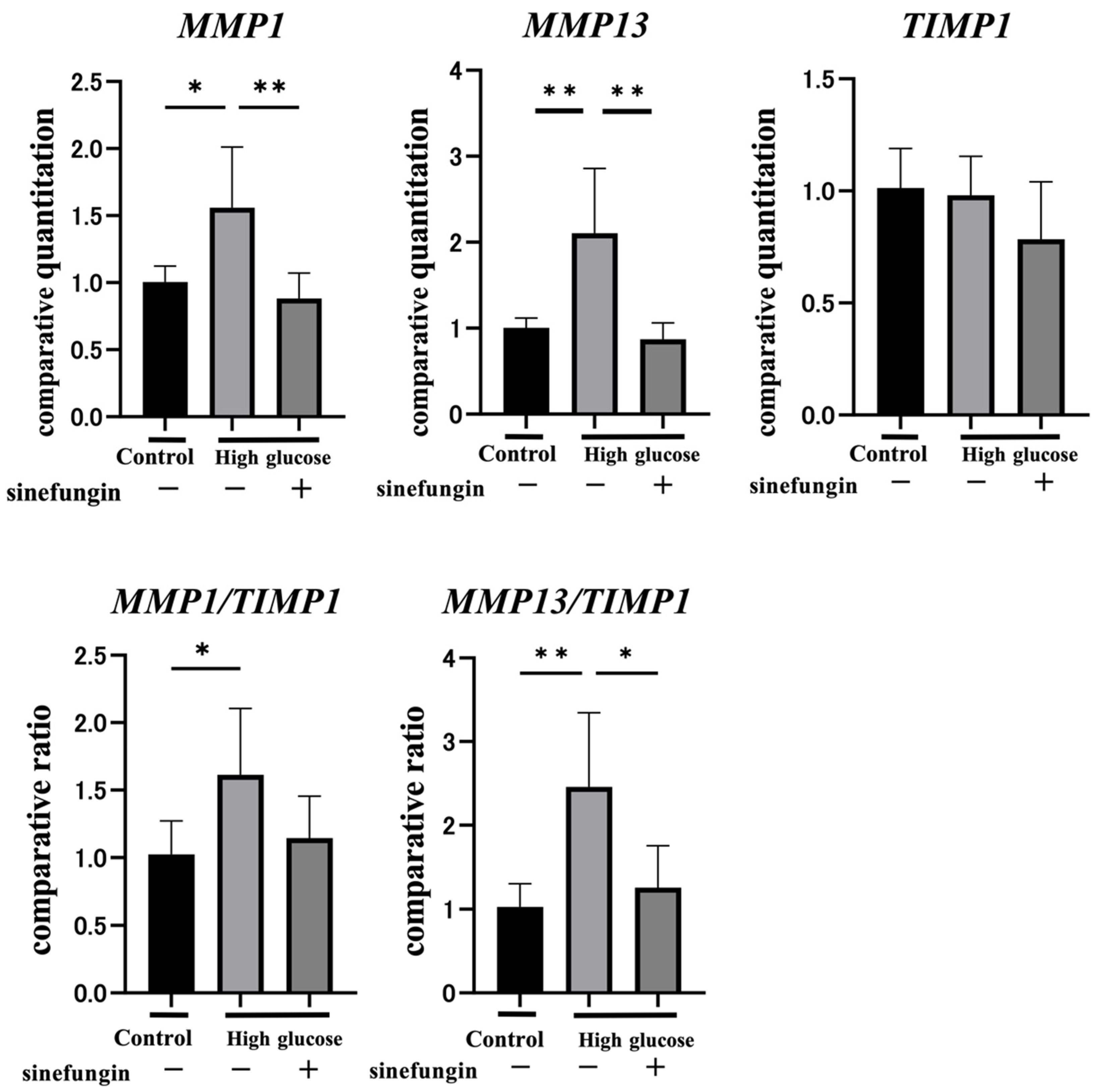
| Variable | Normal Rats | Diabetes Rats |
|---|---|---|
| Body weight (g) | 385.8 ± 22.1 | 276.7 ± 16.2 ** |
| Blood glucose (mmol/L) | 6.1 ± 0.9 | 26.4 ± 7.3 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kojima, K.; Nakamura, N.; Hayashi, A.; Kondo, S.; Miyabe, M.; Kikuchi, T.; Sawada, N.; Saiki, T.; Minato, T.; Ozaki, R.; et al. Impacts of Hyperglycemia on Epigenetic Modifications in Human Gingival Fibroblasts and Gingiva in Diabetic Rats. Int. J. Mol. Sci. 2024, 25, 10979. https://doi.org/10.3390/ijms252010979
Kojima K, Nakamura N, Hayashi A, Kondo S, Miyabe M, Kikuchi T, Sawada N, Saiki T, Minato T, Ozaki R, et al. Impacts of Hyperglycemia on Epigenetic Modifications in Human Gingival Fibroblasts and Gingiva in Diabetic Rats. International Journal of Molecular Sciences. 2024; 25(20):10979. https://doi.org/10.3390/ijms252010979
Chicago/Turabian StyleKojima, Kento, Nobuhisa Nakamura, Airi Hayashi, Shun Kondo, Megumi Miyabe, Takeshi Kikuchi, Noritaka Sawada, Tomokazu Saiki, Tomomi Minato, Reina Ozaki, and et al. 2024. "Impacts of Hyperglycemia on Epigenetic Modifications in Human Gingival Fibroblasts and Gingiva in Diabetic Rats" International Journal of Molecular Sciences 25, no. 20: 10979. https://doi.org/10.3390/ijms252010979
APA StyleKojima, K., Nakamura, N., Hayashi, A., Kondo, S., Miyabe, M., Kikuchi, T., Sawada, N., Saiki, T., Minato, T., Ozaki, R., Sasajima, S., Mitani, A., & Naruse, K. (2024). Impacts of Hyperglycemia on Epigenetic Modifications in Human Gingival Fibroblasts and Gingiva in Diabetic Rats. International Journal of Molecular Sciences, 25(20), 10979. https://doi.org/10.3390/ijms252010979






