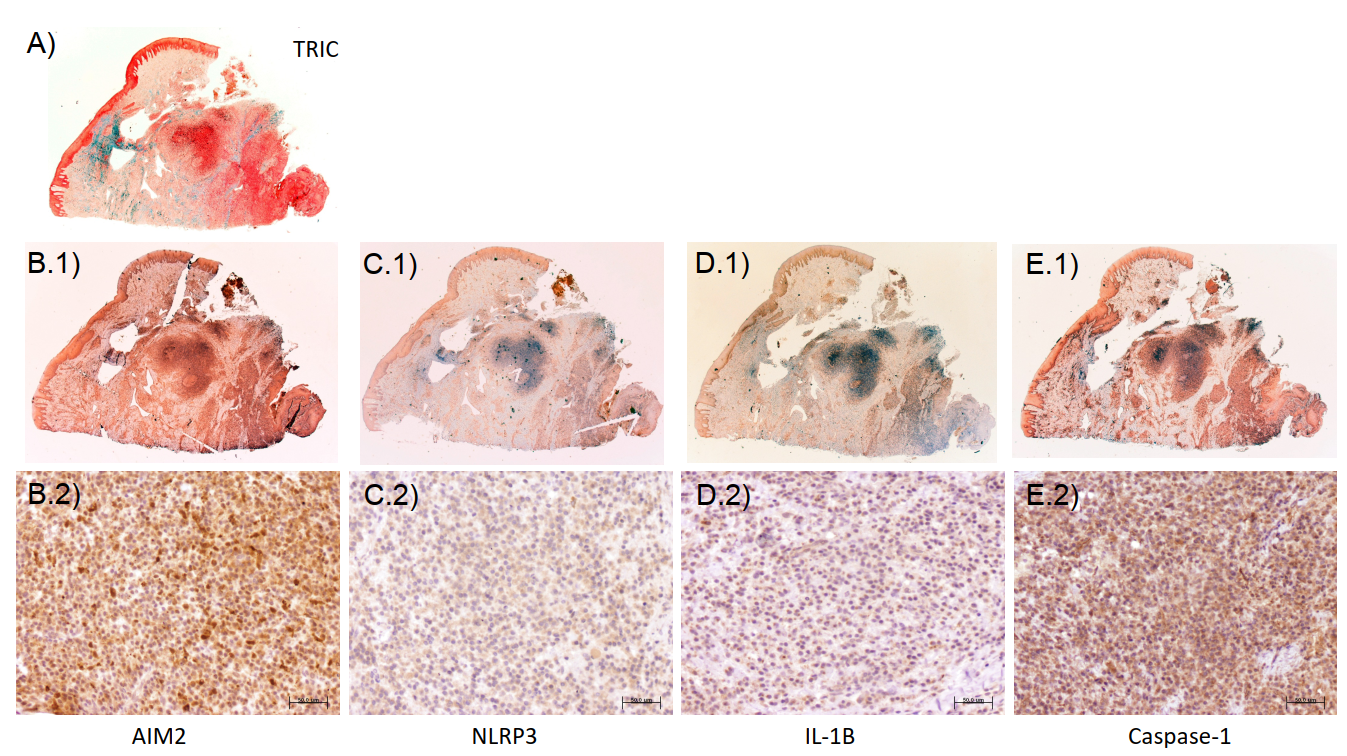
1. Introduction
Peri-implantitis, as defined, is “a plaque-associated pathological condition…characterized by inflammation in the peri-implant mucosa” [
1]. This inflammation in the soft tissue ends up affecting the surrounding bone, which leads to the destruction of the supportive peri-implant structures. To date, no successful therapy is available. In fact, the literature, comparatively, still indicates the same methods as 10 years ago both for cleaning and decontamination of the implant surface [
2,
3] and for reconstructing the tissues lost [
4,
5]. Thus, better understanding of the microbial–host interactions and additional factors must be achieved.
A fundamental step differentiating a reversible peri-implant disease (i.e., mucositis) from the state of bone destruction (i.e., peri-implantitis) is the increase in interleukin-1β (IL-1β) that disrupts the osteoclast–osteoblast balance [
6]. Within the context of periodontal and peri-implant diseases, that the production of IL-1β mediated by caspase-1 is a consequence of the activation of molecular complexes known as inflammasomes has been recently proposed [
7]. Inflammasomes can be activated in response to pathogen-associated molecular patterns (PAMPs) such as the interferon-inducible protein absent in melanoma 2 (AIM2). Other types of inflammasomes, such as the nucleotide-binding oligomerization domain-like receptors family pyrin domain containing 3 (NLRP3), are activated by both PAMPs and damage-associated molecular patterns (DAMPs) [
6]. Thus, the evaluation of the whole picture and of both pathways separately is important. Particularly, our group has recently published for the first time in the literature a characterization of the expression of both AIM2 and NLRP3 in peri-implantitis lesions from human patients. That study demonstrates that both inflammasomes are increased in peri-implantitis and correlate with other markers of inflammation, tissue destruction, and clinical variables such as probing pocket depth (PPD) [
8].
Non-bacterial-associated damage may originate from cement from the prosthesis. However, in the case of screw-retained implant connections, the only source of this type of DAMPs can be the release of debris from the friction between the two components. The role that implant debris, such as titanium particles and ions, may play in both osseointegration itself and in the initiation of peri-implant marginal bone resorption is being highly debated currently in dental implantology [
9,
10,
11,
12]. While debris from the implant itself may be released to the peri-implant bone, such debris may also be produced at the interface between the prosthesis and the implant or between the implant and the transmucosal abutments [
13]. In fact, numerous studies have found titanium particles released from the implants in the surrounding tissues [
14,
15,
16,
17,
18,
19,
20,
21]. Within this debate, the activation of NLRP3, specifically, may help in finding a biologically plausible explanation for the differences between periodontitis and peri-implantitis. Thus, it is being studied, particularly in vitro [
22]. Our group has also recently confirmed the activation in vitro of NLRP3 in response to titanium ions and of AIM2 in response to lipopolysaccharides (LPSs); interestingly, when both titanium and LPSs are combined, NLRP3 is predominant, while AIM2 decreases [
23]. In fact, the presence of titanium in the peri-implant sulcus has been associated with a modification of the structure and a reduction in the diversity of the microbiome associated with peri-implant health and disease [
24]. However, an analysis of the complex microbial communities present in clinical samples and how they correlate with the expression of inflammasomes is also required but, to our knowledge, has not been studied yet.
Thus, the current study aims to evaluate the microbial profiles in samples obtained from peri-implantitis lesions and correlate the findings with the characterization of the inflammatory process, particularly with respect to the expression of the inflammasomes AIM2 and NLRP3 as well as caspase-1 and interleukin-1β.
3. Discussion
The current study was aimed at analyzing the correlation between the abundance of specific microbes detectable in peri-implantitis sulci and the expression of the inflammasomes AIM2 and NLRP3 as well as caspase-1 and interleukin-1β. Our findings demonstrate significant negative and positive correlations between them.
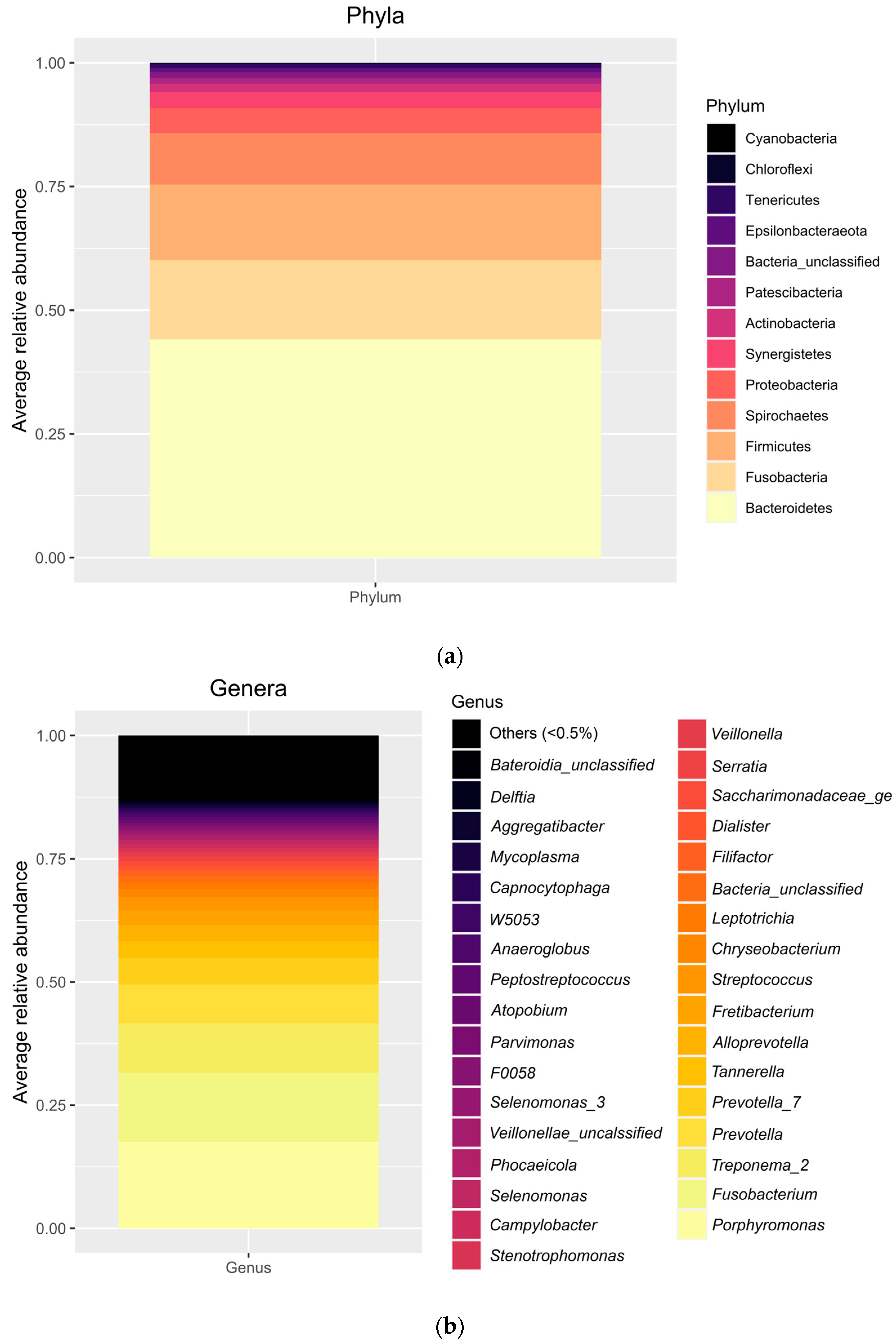
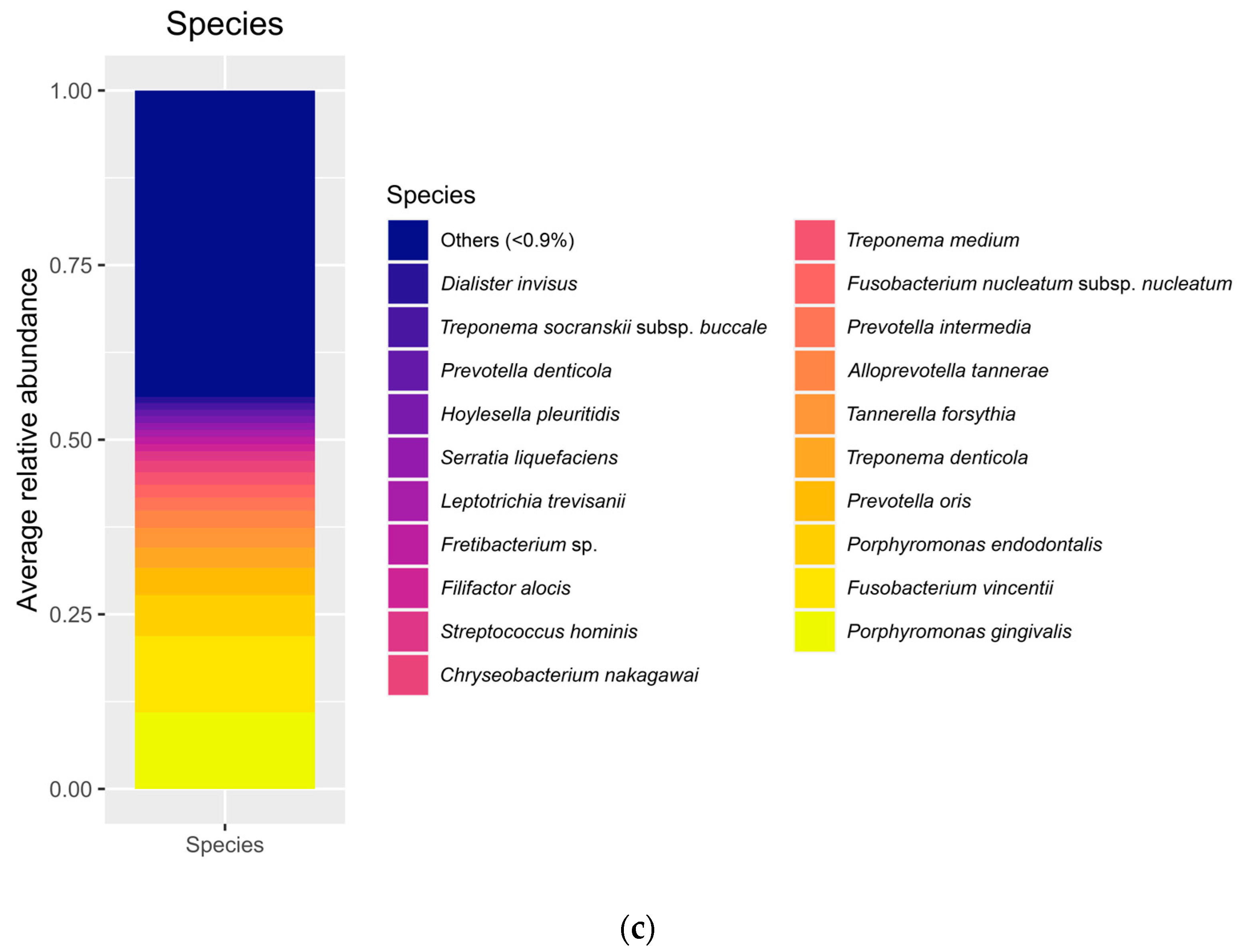
The relative abundance of several bacteria found in our samples represents the classical ecology of oral environments, and, particularly, of peri-implantitis lesions as described previously by a number of studies [
25,
26,
27,
28]. Specifically, the diversity indices can be considered high, in line with previous studies [
29,
30,
31]. Interestingly, the literature is consistent in reporting a decrease in diversity as the disease evolves [
32]. In contrast, we have found a negative correlation between several diversity indexes and the average probing depth, which would be more in line with recent theories about the understanding of peri-implantitis as a dysbiosis condition based on the loss of microbial diversity and the loss of keystone taxa that could favor the bloom of pathogens and changes in metabolic activities [
33].
Because of the absence of similar correlation studies between the variables of interest in the current analysis, we can only compare with those reporting on the presence or absence of microorganisms. According to this, it can be said that the phyla found in our study to be significantly correlated with inflammasomes in the lamina and the epithelium of peri-implantitis lesions (mainly Synergistetes and Tenericutes) had been previously associated with the severity of peri-implantitis lesions. In this sense, old studies such as that from 2008 by Shibli et al. using DNA-DNA checkerboard or that from 2010 by Koyanagi et al. using 16S rRNA gene analysis detected Synergistetes and Tenericutes only in peri-implantitis but not in periodontitis nor in healthy implants [
25,
34]. Most recent studies have found similar results [
35,
36,
37]. The positive correlation with the level of NLRP3 in the lamina propria we found in the current study may indicate a confirmation that these phyla play a role in the development of the disease.
Regarding the other significantly associated phylum, Epsilonbacteraeota, it must be said that very limited information has been published about it in the periodontal literature [
38]. Nothing has been found regarding its role in peri-implantitis. In fact, its role is mainly still unknown [
38]. Thus, our finding regarding its association with the levels of caspase-1 is speculative only.
Inflammasomes have been recently introduced in the peri-implant literature. As described earlier, both AIM2 and NLRP3 result in the activation of pro-caspase-1, which ultimately activates pro-IL-1β. Thus, the mechanisms by which this activation occurs might be of high interest, not only from a mechanistic point of view, but also as potential therapeutic targets [
39]. In fact, it has been described that NLRP3 specifically can play a determinant role in the impairment of fibroblasts’ migration capacities [
40] and alveolar bone loss. If suppressed, both age-related [
41] and ligature-induced [
42] periodontitis-like alveolar bone loss is decreased. Thus, the levels of NLRP3 in saliva have been proposed as markers of periodontal disease [
43].
If we take what is known in periodontitis and translate it to peri-implantitis, it is reasonable to think that similar effects may happen. However, although AIM2 would be activated by the same mechanisms in the periodontal and the peri-implant sulci, NLRP3 can be activated by bacteria, bacteria byproducts, and titanium [
22,
44]. Thus, there would be another activator in peri-implantitis that is not present in periodontitis. Even more, precisely because of the presence of the implant, the peri-implant niche must be considered to be very different compared with the periodontal niche; in fact, it has been described as a “unique microenvironment that force microbial adaptation and selection” [
45]. This is because, as recently proposed, titanium products, particularly ions, have the potential to change the microbiological composition of biofilms, specifically reducing their diversity [
24,
46]. The presence of titanium in the sulcus fluid [
14,
17,
18,
24] and the tissue of peri-implantitis lesions has been reported in several studies [
21]. Small particles (not visible in optical microscopy), even at the level of just titanium ions, are more frequently released as the consequence of wearing between the prosthesis and the implant; they have been identified in several studies as the most deleterious, as discussed in Wu et al. [
47]. Thus, the presence of titanium in the peri-implant sulcus would contribute to a microbial dysbiosis that, together with the titanium ions themselves, may ultimately end up in the development of peri-implantitis. Both inflammasome pathways would function: the pathway that responds to other biological stimuli only, such as AIM2, and the pathway that responds to both biological and physical products, such as NLRP3. This double path for pro-inflammatory inductors could explain the higher severity that is observed in peri-implantitis lesions compared with periodontitis.
However, very little is known so far about inflammasomes in peri-implantitis. Only a recent study by our group has demonstrated that both the inflammasomes AIM2 and NLRP3 are highly expressed in peri-implantitis lesions from human patients and correlate with other markers of inflammation, tissue destruction, and clinical variables such as PD [
8]. As expected, similar results regarding those correlations have also been found in the set of samples used in the current study. Moreover, in vitro, when both titanium and LPS are combined, NLRP3 is predominant while AIM2 decreases [
23].
The current study found several correlations between different species and clinical and immunohistochemical variables. First, like many other studies in the literature, we found a positive correlation between the average probing depth and the abundance of
Porphyromonas gingivalis. It was reported that
Porphyromonas gingivalis may reduce the expression of NLRP3 and IL-1β but not modify the expression of AIM2 in vitro, which would help dampen the host’s innate immune response [
48]. In contrast,
A. actinomycetemcomitans can increase the expression of NLRP3, while again keeping unaltered the expression of AIM2 [
49]. In a way, these results come to reinforce the findings of the current study, indicating that inflammasome activation may be increased or decreased depending on the bacteria in action. Our study, clinically and beyond the limitations of the single- or 10-species biofilm models like those executed in the referenced studies, correlates different species with both increased and decreased inflammasome activation. The specific mechanisms by which those bacteria influence the expression of inflammasomes need further investigation.
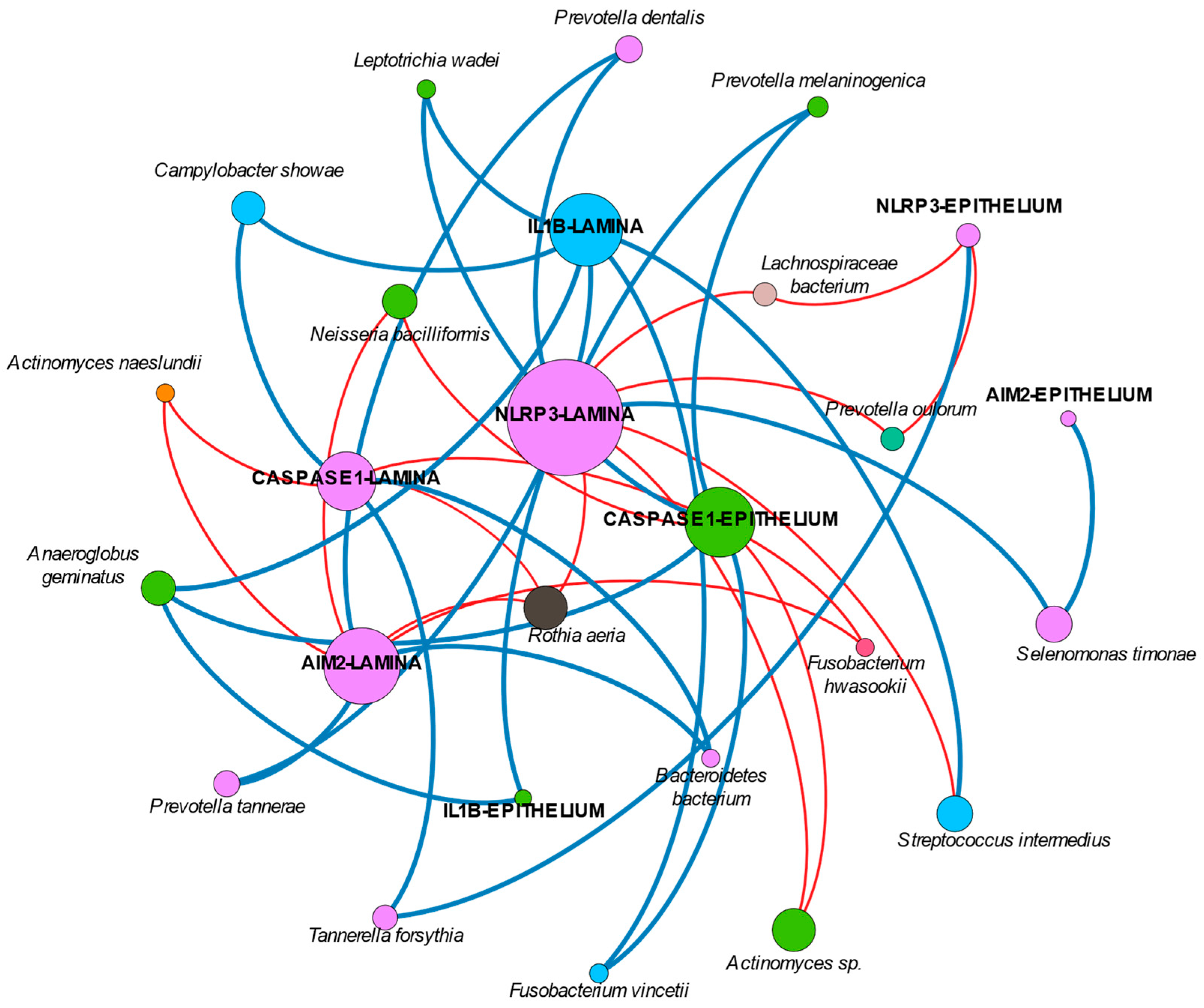
Interestingly, our network analysis found an isolated node of
Rothia aeria that is negatively correlated with the expression of NLRP3, AIM2, and caspase-1 in the lamina propria. Many studies have reported negative correlations between
Rothia and probing depth and disease [
28,
37,
50]. Our findings could be hypothesized as the explanation for those findings.
Finally, the available studies on IL-1β would have to be re-considered in the near future. As recently discussed, levels of IL-1β may suffer post-transcriptional modifications through inflammasomes that may not have been studied appropriately [
51]. Thus, differences in health and disease might be not properly represented, particularly if the expression of upstream regulatory molecules, predominantly NLRP3 in peri-implantitis as shown, are not looked at. The role of titanium might be a game-changer in this issue. In fact, if we consider the network analysis, it can be suggested that in peri-implantitis, NLRP3 plays a more relevant role compared with AIM2. In this sense, the association of NLRP3, IL-1β, and several genera has been found to be more dominant than those other clusters formed by AIM2 or caspase-1 and their respective bacteria. However, the current study did not evaluate the presence of titanium in the peri-implant sulcus nor in the lesion tissue, which would have added important and interesting information. In addition, we did not evaluate smoker patients separately nor exclude them from the current study, although tobacco consumption may influence both inflammation and biofilm composition. This is because the sample size was limited, as no previous data were available to propose a valid sample size calculation; so, a subgroups comparison would have reduced the number of patients per group even more. We did not exclude them from the study because smoking is common in patients with peri-implantitis; so, excluding them would have reduced the generalizability of the study. Furthermore, the evaluation of inflammasomes in peri-implantitis tissue has previously demonstrated no correlation with tobacco consumption [
8].
We must also acknowledge that this study evaluated samples from a clinical condition only and few markers of the inflammatory pathway, particularly related to the inflammasomes AIM2 and NLRP3. This information was obtained with immunohistochemical techniques. Furthermore, it was only a cross-sectional study, which misses the dynamics of a real clinical environment; a prospective study could provide interesting information on this topic, not only microbiologically but also from the inflammasome activation-deactivation dynamic. Even more, regardless of the remarkable association between bacteria and specific markers of the inflammasome complexes NLRP3 and AIM2 and their locations, we must acknowledge, as highlighted by Belibasakis et al., that current understanding of diseases initiated or modified by bacteria must be understood as community-driven interactions rather than a single type of bacteria orchestrating the whole complex [
32]. However, we truly believe that the information provided in this study is highly novel and would be helpful for future studies on the topic. Particularly, it would be interesting to keep the focus on the pathways evaluated here when evaluating more complex functional interactions between the host and the microbial community [
52]. Clinically, once more, patients and professionals must be aware of the complexity of the disease and the importance of attending regular maintenance visits to minimize any unbalance that may be initiated and result in peri-implant tissue destruction.