The Association of Hippocampal Long-Term Potentiation-Induced Gene Expression with Genetic Risk for Psychosis
Abstract
1. Introduction
2. Results
2.1. Common Variant Association
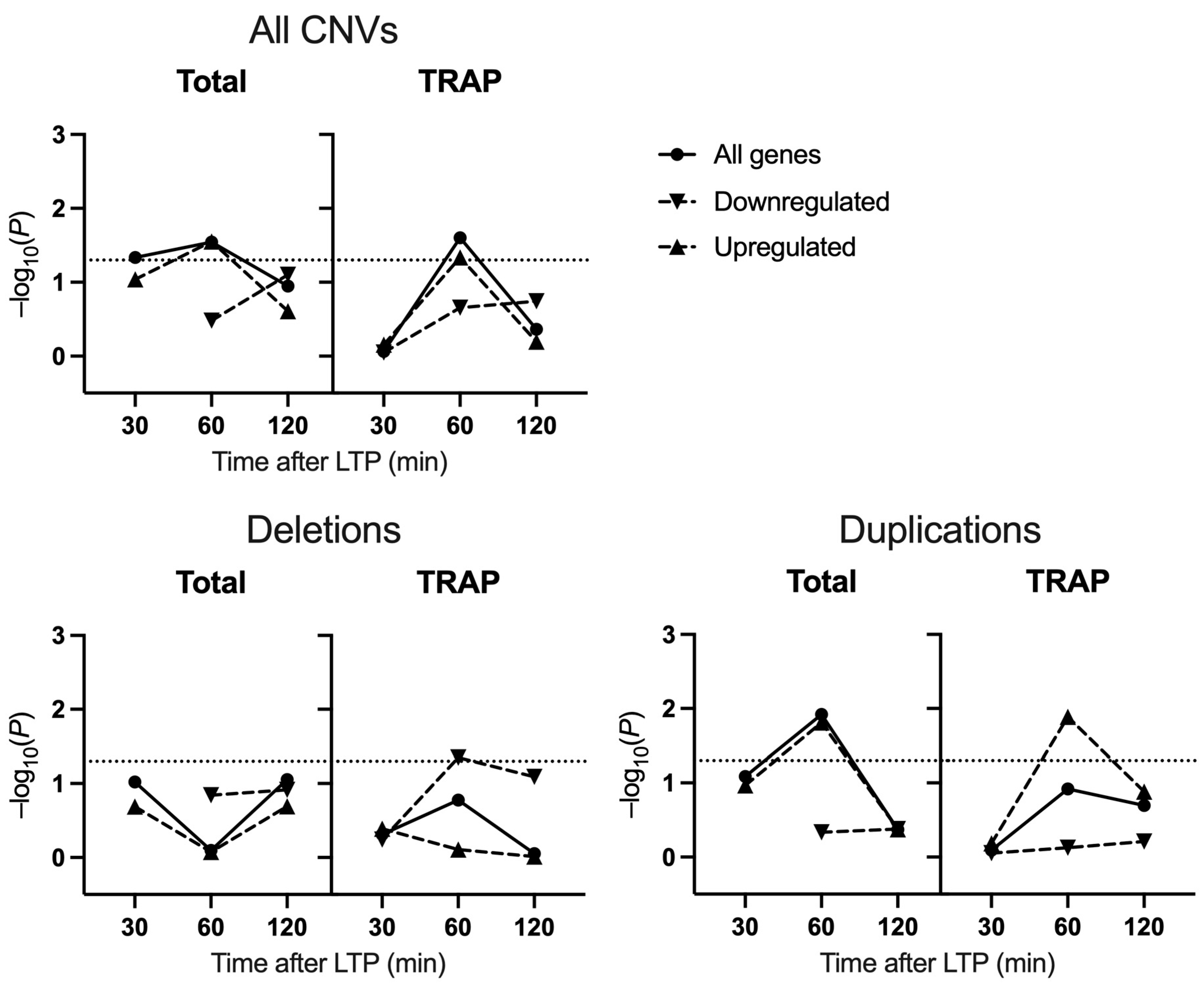
2.2. Schizophrenia CNV Enrichment
2.3. Schizophrenia De Novo Rare Coding Variant Enrichment
3. Discussion
4. Materials and Methods
4.1. LTP-Induced Gene Sets
4.2. Genotype Data
4.3. Gene Set Enrichment Analysis
probes + CNV study + number of gene set hits
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hall, J.; Bray, N.J. Schizophrenia genomics: Convergence on synaptic development, adult synaptic plasticity, or both? Biol. Psychiatry 2022, 91, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Mould, A.W.; Hall, N.A.; Milosevic, I.; Tunbridge, E.M. Targeting synaptic plasticity in schizophrenia: Insights from genomic studies. Trends Mol. Med. 2021, 27, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Trubetskoy, V.; Pardiñas, A.F.; Qi, T.; Panagiotaropoulou, G.; Awasthi, S.; Bigdeli, T.B.; Bryois, J.; Chen, C.-Y.; Dennison, C.A.; Hall, L.S.; et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 2022, 604, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Green, T.; Gothelf, D.; Glaser, B.; Debbane, M.; Frisch, A.; Kotler, M.; Weizman, A.; Eliez, S. Psychiatric Disorders and Intellectual Functioning Throughout Development in Velocardiofacial (22q11.2 Deletion) Syndrome. J. Am. Acad. Child Adolesc. Psychiatry 2009, 48, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.R.; Howrigan, D.P.; Merico, D.; Thiruvahindrapuram, B.; Wu, W.; Greer, D.S.; Antaki, D.; Shetty, A.; Holmans, P.A.; Pinto, D.; et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017, 49, 27–35. [Google Scholar] [CrossRef]
- Rees, E.; Walters, J.T.R.; Georgieva, L.; Isles, A.R.; Chambert, K.D.; Richards, A.L.; Mahoney-Davies, G.; Legge, S.E.; Moran, J.L.; McCarroll, S.A.; et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br. J. Psychiatry 2014, 204, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Stefansson, H.; Rujescu, D.; Cichon, S.; Pietiläinen, O.P.H.; Ingason, A.; Steinberg, S.; Fossdal, R.; Sigurdsson, E.; Sigmundsson, T.; Buiz er-Voskamp, J.E.; et al. Large recurrent microdeletions associated with schizophrenia. Nature 2008, 455, 232–236. [Google Scholar] [CrossRef]
- Rees, E.; Kendall, K.; Pardiñas, A.F.; Legge, S.E.; Pocklington, A.; Escott-Price, V.; MacCabe, J.H.; Collier, D.A.; Holmans, P.; O’donovan, M.C.; et al. Analysis of Intellectual Disability Copy Number Variants for Association With Schizophrenia. JAMA Psychiatry 2016, 73, 963–969. [Google Scholar] [CrossRef]
- Rees, E.; Carrera, N.; Morgan, J.; Hambridge, K.; Escott-Price, V.; Pocklington, A.J.; Richards, A.L.; Pardiñas, A.F.; McDonald, C.; Donohoe, G.; et al. Targeted Sequencing of 10,198 Samples Confirms Abnormalities in Neuronal Activity and Implicates Voltage-Gated Sodium Channels in Schizophrenia Pathogenesis. Biol. Psychiatry 2019, 85, 554–562. [Google Scholar] [CrossRef]
- Singh, T.; Poterba, T.; Curtis, D.; Akil, H.; Al Eissa, M.; Barchas, J.D.; Bass, N.; Bigdeli, T.B.; Breen, G.; Bromet, E.J.; et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 2022, 604, 509–516. [Google Scholar] [CrossRef]
- Rees, E.; Creeth, H.D.J.; Hwu, H.-G.; Chen, W.J.; Tsuang, M.; Glatt, S.J.; Rey, R.; Kirov, G.; Walters, J.T.R.; Holmans, P.; et al. Schizophrenia, autism spectrum disorders and developmental disorders share specific disruptive coding mutations. Nat. Commun. 2021, 12, 5353. [Google Scholar] [CrossRef]
- Kirov, G.; Pocklington, A.J.; Holmans, P.; Ivanov, D.; Ikeda, M.; Ruderfer, D.; Moran, J.; Chambert, K.; Toncheva, D.; Georgieva, L.; et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry 2012, 17, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Pocklington, A.J.; Rees, E.; Walters, J.T.R.; Han, J.; Kavanagh, D.H.; Chambert, K.D.; Holmans, P.; Moran, J.L.; McCarroll, S.A.; Kirov, G.; et al. Novel Findings from CNVs Implicate Inhibitory and Excitatory Signaling Complexes in Schizophrenia. Neuron 2015, 86, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Stahl, E.A.; Breen, G.; Forstner, A.J.; McQuillin, A.; Ripke, S.; Trubetskoy, V.; Mattheisen, M.; Wang, Y.; Coleman, J.R.I.; Gaspar, H.A.; et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019, 51, 793–803. [Google Scholar] [CrossRef]
- Mullins, N.; Forstner, A.J.; O’connell, K.S.; Coombes, B.; Coleman, J.R.I.; Qiao, Z.; Als, T.D.; Bigdeli, T.B.; Børte, S.; Bryois, J.; et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 2021, 53, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Pardiñas, A.F.; Holmans, P.; Pocklington, A.J.; Escott-Price, V.; Ripke, S.; Carrera, N.; Legge, S.E.; Bishop, S.; Cameron, D.; Hamshere, M.L.; et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018, 50, 381–389. [Google Scholar] [CrossRef]
- Bliss, T.V.P.; Lømo, T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 1973, 232, 331–356. [Google Scholar] [CrossRef]
- Whitlock, J.R.; Heynen, A.J.; Shuler, M.G.; Bear, M.F. Learning Induces Long-Term Potentiation in the Hippocampus. Science 2006, 313, 1093–1097. [Google Scholar] [CrossRef]
- Frey, U.; Krug, M.; Reymann, K.G.; Matthies, H. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res. 1988, 452, 57–65. [Google Scholar] [CrossRef]
- Bradshaw, K.D.; Emptage, N.J.; Bliss, T.V.P. A role for dendritic protein synthesis in hippocampal late LTP. Eur. J. Neurosci. 2003, 18, 3150–3152. [Google Scholar] [CrossRef]
- Redondo, R.L.; Morris, R.G.M. Making memories last: The synaptic tagging and capture hypothesis. Nat. Rev. Neurosci. 2011, 12, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Asok, A.; Leroy, F.; Rayman, J.B.; Kandel, E.R. Molecular Mechanisms of the Memory Trace. Trends Neurosci. 2019, 42, 14–22. [Google Scholar] [CrossRef]
- Chen, P.B.; Kawaguchi, R.; Blum, C.; Achiro, J.M.; Coppola, G.; O’Dell, T.J.; Martin, K.C. Mapping Gene Expression in Excitatory Neurons during Hippocampal Late-Phase Long-Term Potentiation. Front. Mol. Neurosci. 2017, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.J.; Orfila, J.E.; Achanta, P.; Martinez, J.L. Gene expression associated with in vivo induction of early phase-long-term potentiation (LTP) in the hippocampal mossy fiber-Cornus Ammonis (CA)3 pathway. Cell. Mol. Biol. 2003, 49, 1281–1287. [Google Scholar]
- Wibrand, K.; Messaoudi, E.; Håvik, B.; Steenslid, V.; Løvlie, R.; Steen, V.M.; Bramham, C.R. Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in adult rat dentate gyrus in vivo. Eur. J. Neurosci. 2006, 23, 1501–1511. [Google Scholar] [CrossRef]
- Valor, L.M.; Barco, A. Hippocampal gene profiling: Toward a systems biology of the hippocampus. Hippocampus 2012, 22, 929–941. [Google Scholar] [CrossRef]
- Ryan, M.M.; Ryan, B.; Kyrke-Smith, M.; Logan, B.; Tate, W.P.; Abraham, W.C.; Williams, J.M. Temporal Profiling of Gene Networks Associated with the Late Phase of Long-Term Potentiation In Vivo. PLoS ONE 2012, 7, e40538. [Google Scholar] [CrossRef] [PubMed]
- Park, C.S.; Gong, R.; Stuart, J.; Tang, S.-J. Molecular Network and Chromosomal Clustering of Genes Involved in Synaptic Plasticity in the Hippocampus. J. Biol. Chem. 2006, 281, 30195–30211. [Google Scholar] [CrossRef]
- Doyle, J.P.; Dougherty, J.D.; Heiman, M.; Schmidt, E.F.; Stevens, T.R.; Ma, G.; Bupp, S.; Shrestha, P.; Shah, R.D.; Doughty, M.L.; et al. Application of a Translational Profiling Approach for the Comparative Analysis of CNS Cell Types. Cell 2008, 135, 749–762. [Google Scholar] [CrossRef]
- Haimon, Z.; Volaski, A.; Orthgiess, J.; Boura-Halfon, S.; Varol, D.; Shemer, A.; Yona, S.; Zuckerman, B.; David, E.; Chappell-Maor, L.; et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 2018, 19, 636–644. [Google Scholar] [CrossRef]
- Sanz, E.; Bean, J.C.; Carey, D.P.; Quintana, A.; McKnight, G.S. RiboTag: Ribosomal Tagging Strategy to Analyze Cell-Type-Specific mRNA Expression In Vivo. Curr. Protoc. Neurosci. 2019, 88, e77. [Google Scholar] [CrossRef] [PubMed]
- Tsien, J.Z.; Chen, D.F.; Gerber, D.; Tom, C.; Mercer, E.H.; Anderson, D.J.; Mayford, M.; Kandel, E.R.; Tonegawa, S. Subregion- and Cell Type–Restricted Gene Knockout in Mouse Brain. Cell 1996, 87, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Skene, N.G.; Bryois, J.; Bakken, T.E.; Breen, G.; Crowley, J.J.; Gaspar, H.A.; Giusti-Rodriguez, P.; Hodge, R.D.; Miller, J.A.; Muñoz-Manchado, A.B.; et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 2018, 50, 825–833. [Google Scholar] [CrossRef] [PubMed]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef]
- Alberini, C.M.; Kandel, E.R. The Regulation of Transcription in Memory Consolidation. Cold Spring Harb. Perspect. Biol. 2014, 7, a021741. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, J.-J. Structural LTP: Signal transduction, actin cytoskeleton reorganization, and membrane remodeling of dendritic spines. Curr. Opin. Neurobiol. 2022, 74, 102534. [Google Scholar] [CrossRef]
- Lee, P.H.; Feng, Y.-C.A.; Smoller, J.W. Pleiotropy and Cross-Disorder Genetics Among Psychiatric Disorders. Biol. Psychiatry 2021, 89, 20–31. [Google Scholar] [CrossRef]
- Fromer, M.; Pocklington, A.J.; Kavanagh, D.H.; Williams, H.J.; Dwyer, S.; Gormley, P.; Georgieva, L.; Rees, E.; Palta, P.; Ruderfer, D.M.; et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014, 506, 179–184. [Google Scholar] [CrossRef]
- Pembroke, W.G.; Hartl, C.L.; Geschwind, D.H. Evolutionary conservation and divergence of the human brain transcriptome. Genome Biol. 2021, 22, 52. [Google Scholar] [CrossRef]
- Emes, R.D.; Grant, S.G.N. Evolution of Synapse Complexity and Diversity. Annu. Rev. Neurosci. 2012, 35, 111–131. [Google Scholar] [CrossRef] [PubMed]
- Gobert, D.; Topolnik, L.; Azzi, M.; Huang, L.; Badeaux, F.; Desgroseillers, L.; Sossin, W.S.; Lacaille, J.-C. Forskolin induction of late-LTP and up-regulation of 5′ TOP mRNAs translation via mTOR, ERK, and PI3K in hippocampal pyramidal cells. J. Neurochem. 2008, 106, 1160–1174. [Google Scholar] [CrossRef] [PubMed]
- Otmakhov, N.; Khibnik, L.; Otmakhova, N.; Carpenter, S.; Riahi, S.; Asrican, B.; Lisman, J. Forskolin-Induced LTP in the CA1 Hippocampal Region Is NMDA Receptor Dependent. J. Neurophysiol. 2004, 91, 1955–1962. [Google Scholar] [CrossRef]
- Bigdeli, T.B.; Genovese, G.; Georgakopoulos, P.; Meyers, J.L.; Peterson, R.E.; Iyegbe, C.O.; Medeiros, H.; Valderrama, J.; Achtyes, E.D.; Kotov, R.; et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol. Psychiatry 2020, 25, 2455–2467. [Google Scholar] [CrossRef]
- Wuttke, M.; Li, Y.; Li, M.; Sieber, K.B.; Feitosa, M.F.; Gorski, M.; Tin, A.; Wang, L.; Chu, A.Y.; Hoppmann, A.; et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 2020, 51, 957–972. [Google Scholar] [CrossRef]
- Rees, E.; Han, J.; Morgan, J.; Carrera, N.; Escott-Price, V.; Pocklington, A.J.; Duffield, M.; Hall, L.S.; Legge, S.E.; Pardiñas, F.; et al. De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nat. Neurosci. 2020, 23, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Ambalavanan, A.; Girard, S.L.; Ahn, K.; Zhou, S.; Dionne-Laporte, A.; Spiegelman, D.; Bourassa, C.V.; Gauthier, J.; Hamdan, F.F.; Xiong, L.; et al. De novo variants in sporadic cases of childhood onset schizophrenia. Eur. J. Hum. Genet. 2016, 24, 944–948. [Google Scholar] [CrossRef]
- Girard, S.L.; Gauthier, J.; Noreau, A.; Xiong, L.; Zhou, S.; Jouan, L.; Dionne-Laporte, A.; Spiegelman, D.; Henrion, E.; Diallo, O.; et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011, 43, 860–863. [Google Scholar] [CrossRef]
- Guipponi, M.; Santoni, F.; Setola, V.; Gehrig, C.; Rotharmel, M.; Cuenca, M.; Guillin, O.; Dikeos, D.; Georgantopoulos, G.; Papadimitriou, G.; et al. Exome Sequencing in 53 Sporadic Cases of Schizophrenia Identifies 18 Putative Candidate Genes. PLoS ONE 2014, 9, e112745. [Google Scholar] [CrossRef]
- Gulsuner, S.; Walsh, T.; Watts, A.C.; Lee, M.K.; Thornton, A.M.; Casadei, S.; Rippey, C.; Shahin, H.; Nimgaonkar, V.L.; Go, R.C.; et al. Spatial and Temporal Mapping of De Novo Mutations in Schizophrenia to a Fetal Prefrontal Cortical Network. Cell 2013, 154, 518–529. [Google Scholar] [CrossRef]
- E McCarthy, S.; Gillis, J.; Kramer, M.; Lihm, J.; Yoon, S.; Berstein, Y.; Mistry, M.; Pavlidis, P.; Solomon, R.; Ghiban, E.; et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatry 2014, 19, 652–658. [Google Scholar] [CrossRef]
- Takata, A.; Xu, B.; Ionita-Laza, I.; Roos, J.L.; Gogos, J.A.; Karayiorgou, M. Loss-of-Function Variants in Schizophrenia Risk and SETD1A as a Candidate Susceptibility Gene. Neuron 2014, 82, 773–780. [Google Scholar] [CrossRef]
- Wang, Q.; Li, M.; Yang, Z.; Hu, X.; Wu, H.-M.; Ni, P.; Ren, H.; Deng, W.; Li, M.; Ma, X.; et al. Increased co-expression of genes harboring the damaging de novo mutations in Chinese schizophrenic patients during prenatal development. Sci. Rep. 2015, 5, 18209. [Google Scholar] [CrossRef]
- Xu, B.; Ionita-Laza, I.; Roos, J.L.; Boone, B.; Woodrick, S.; Sun, Y.; Levy, S.; Gogos, J.A.; Karayiorgou, M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012, 44, 1365–1369. [Google Scholar] [CrossRef]
- Samocha, K.E.; Kosmicki, J.A.; Karczewski, K.J.; O’Donnell-Luria, A.H.; Pierce-Hoffman, E.; MacArthur, D.G.; Neale, B.M.; Daly, M.J. Regional missense constraint improves variant deleteriousness prediction. BioRxiv 2017. Preprint. [Google Scholar] [CrossRef]
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008, 455, 237–241. [Google Scholar] [CrossRef]
- Levinson, D.F.; Duan, J.; Oh, S.; Wang, K.; Sanders, A.R.; Shi, J.; Zhang, N.; Mowry, B.J.; Olincy, A.; Amin, F.; et al. Copy Number Variants in Schizophrenia: Confirmation of Five Previous Findings and New Evidence for 3q29 Microdeletions and VIPR2 Duplications. Am. J. Psychiatry 2011, 168, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Samocha, K.E.; Robinson, E.B.; Sanders, S.J.; Stevens, C.; Sabo, A.; McGrath, L.M.; Kosmicki, J.A.; Rehnström, K.; Mallick, S.; Kirby, A.; et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014, 46, 944–950. [Google Scholar] [CrossRef]

| Bipolar Disorder Subtype | β Value (SE) | p-Value | Adjusted p-Value | |
|---|---|---|---|---|
| 30 min TRAP-seq | Bipolar I | −0.090 (0.13) | 0.76 | 1.0 |
| Bipolar II | −0.090 (0.11) | 0.80 | 1.0 | |
| 30 min total RNA-seq | Bipolar I | −0.066 (0.22) | 0.62 | 1.0 |
| Bipolar II | −0.11 (0.18) | 0.72 | 1.0 | |
| 60 min TRAP-seq | Bipolar I | 0.13 (0.029) | 3.1 × 10−6 | 1.9 × 10−5 |
| Bipolar II | 0.0025 (0.025) | 0.46 | 1.0 | |
| 60 min total RNA-seq | Bipolar I | −0.027 (0.10) | 0.60 | 0.898 |
| Bipolar II | −0.096 (0.091) | 0.85 | 1.0 | |
| 120 min TRAP-seq | Bipolar I | 0.094 (0.034) | 0.0027 | 0.016 |
| Bipolar II | 0.0070 (0.029) | 0.40 | 1.0 | |
| 120 min total RNA-seq | Bipolar I | −0.028 (0.050) | 0.71 | 1.0 |
| Bipolar II | −0.028 (0.043) | 0.74 | 1.0 |
| LTP Gene Set | Schizophrenia Ultra-Rare Coding Variant Enrichment | |||
|---|---|---|---|---|
| Observed/Expected | p-Value | Adjusted p-Value | Rate Ratio (95% Confidence Intervals) | |
| 30-min total | 2/0.497 | 0.116 | 0.736 | 3.45 (0.416–12.5) |
| 30-min TRAP | 5/3.45 | 0.610 | 1.00 | 1.24 (0.401–2.91) |
| 60-min total | 7/3.27 | 0.114 | 0.642 | 1.84 (0.737–3.82) |
| 60-min TRAP | 80/57.4 | 0.101 | 0.931 | 1.23 (0.954–1.56) |
| 120-min total | 17/14.2 | 0.900 | 1.00 | 1.02 (0.592–1.66) |
| 120-min TRAP | 80/51.4 | 0.00892 | 0.181 | 1.39 (1.08–1.77) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wellard, N.L.; Clifton, N.E.; Rees, E.; Thomas, K.L.; Hall, J. The Association of Hippocampal Long-Term Potentiation-Induced Gene Expression with Genetic Risk for Psychosis. Int. J. Mol. Sci. 2024, 25, 946. https://doi.org/10.3390/ijms25020946
Wellard NL, Clifton NE, Rees E, Thomas KL, Hall J. The Association of Hippocampal Long-Term Potentiation-Induced Gene Expression with Genetic Risk for Psychosis. International Journal of Molecular Sciences. 2024; 25(2):946. https://doi.org/10.3390/ijms25020946
Chicago/Turabian StyleWellard, Natalie L., Nicholas E. Clifton, Elliott Rees, Kerrie L. Thomas, and Jeremy Hall. 2024. "The Association of Hippocampal Long-Term Potentiation-Induced Gene Expression with Genetic Risk for Psychosis" International Journal of Molecular Sciences 25, no. 2: 946. https://doi.org/10.3390/ijms25020946
APA StyleWellard, N. L., Clifton, N. E., Rees, E., Thomas, K. L., & Hall, J. (2024). The Association of Hippocampal Long-Term Potentiation-Induced Gene Expression with Genetic Risk for Psychosis. International Journal of Molecular Sciences, 25(2), 946. https://doi.org/10.3390/ijms25020946






