Expression Pattern and Prognostic Significance of the Long Non-Coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 in Chronic Lymphocytic Leukemia
Abstract
1. Introduction
2. Results
2.1. Description of the Study Cohort
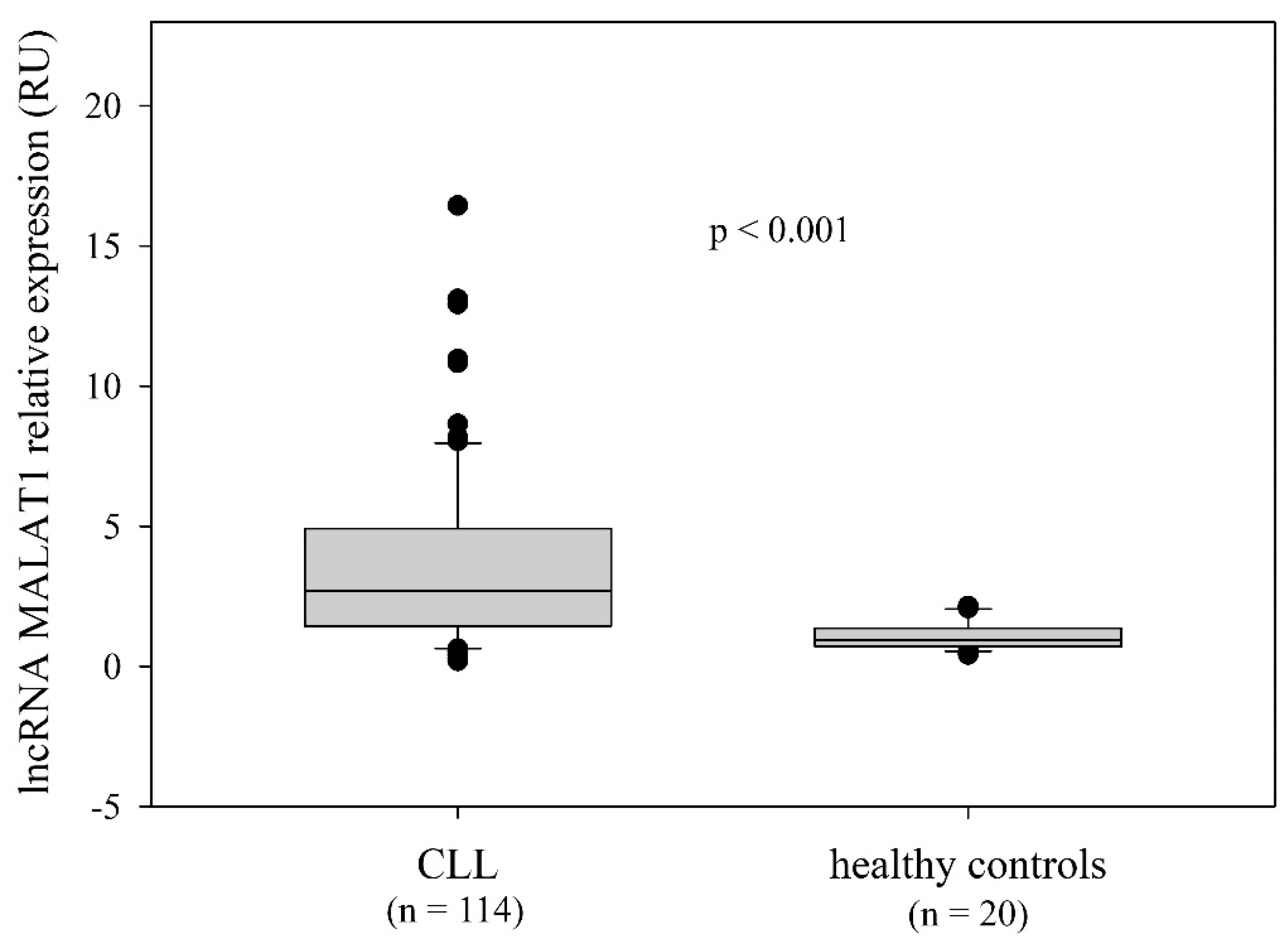
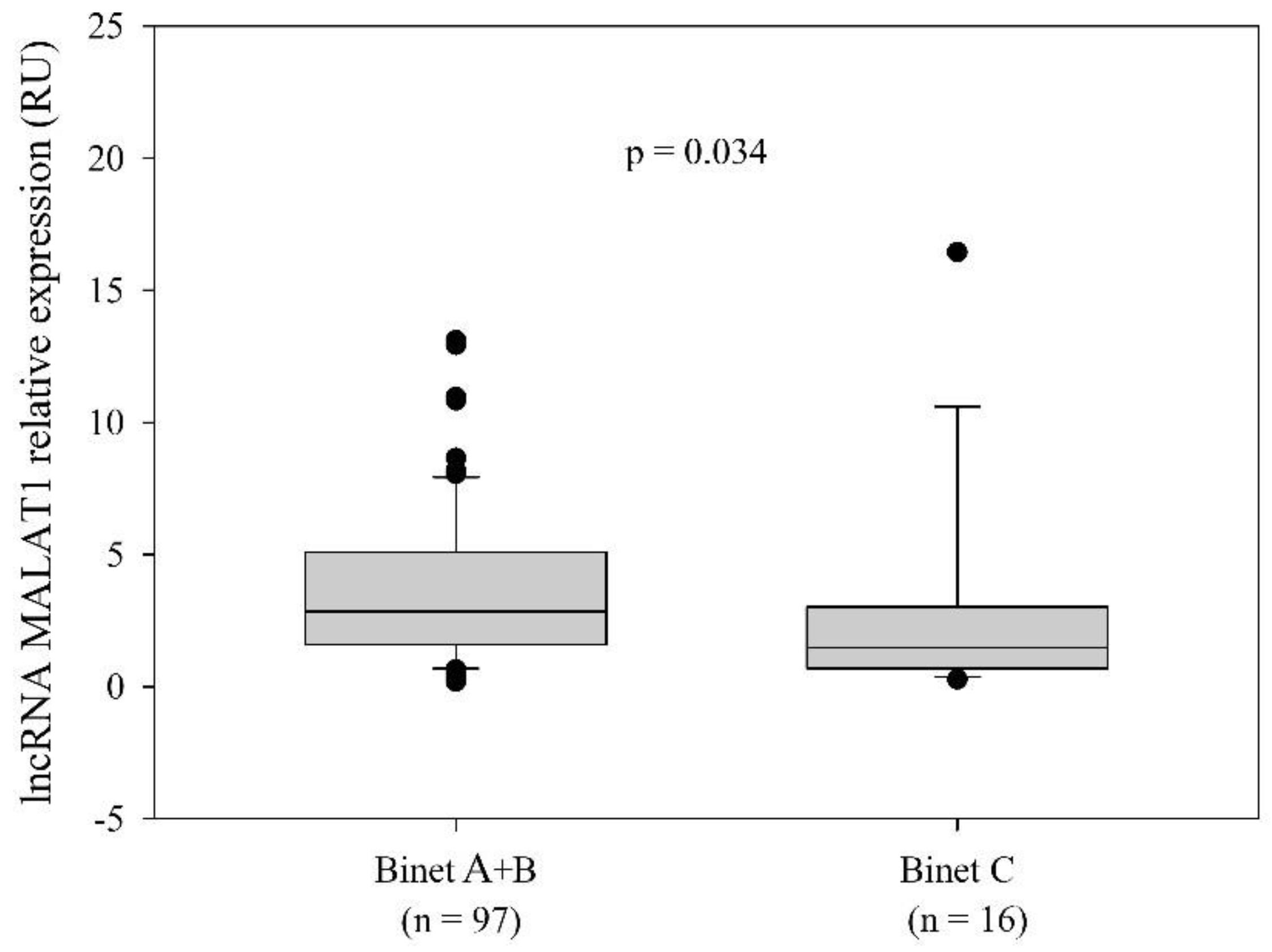
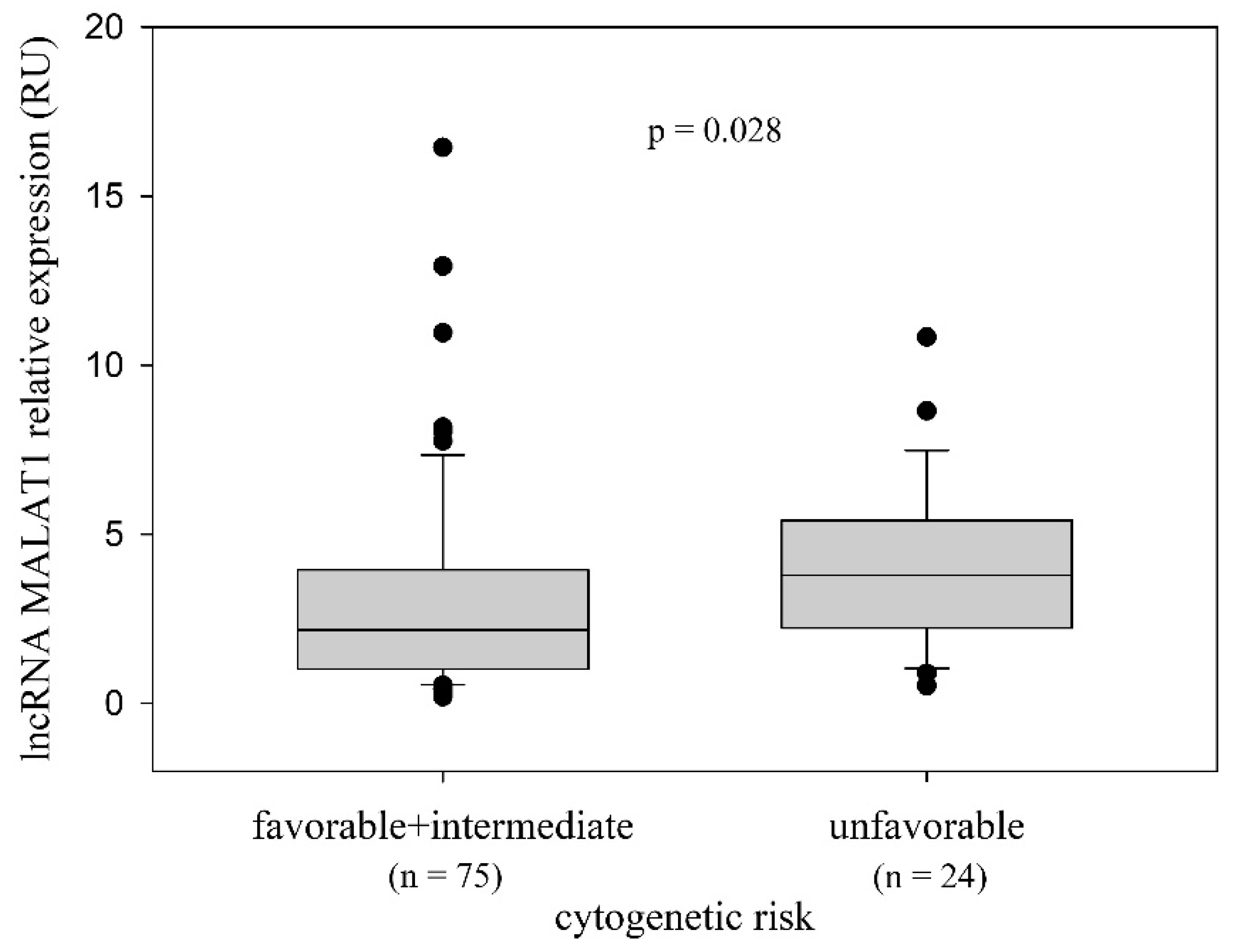
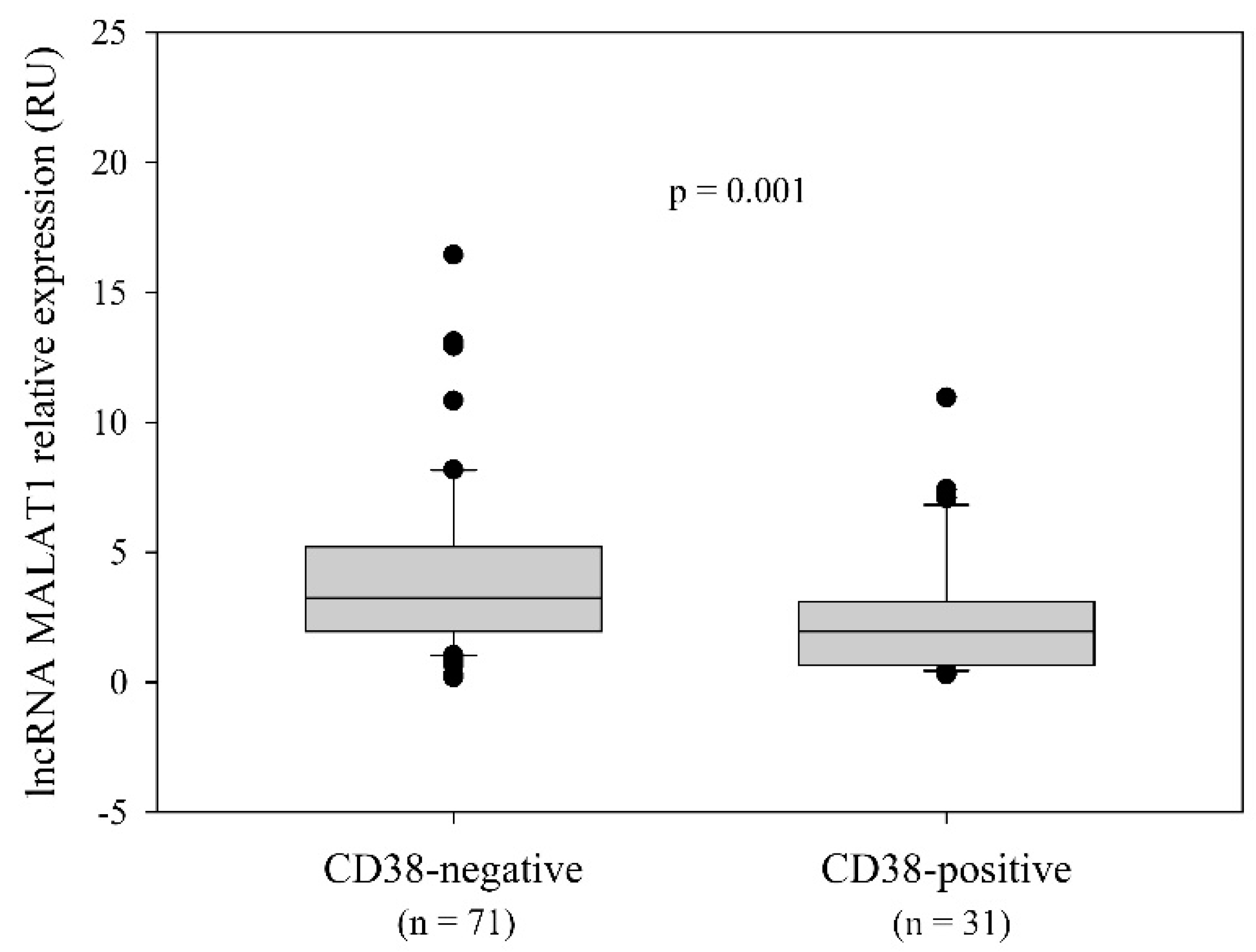
2.2. LncRNA MALAT1 Expression and the Relationship with the Disease-Related Variables
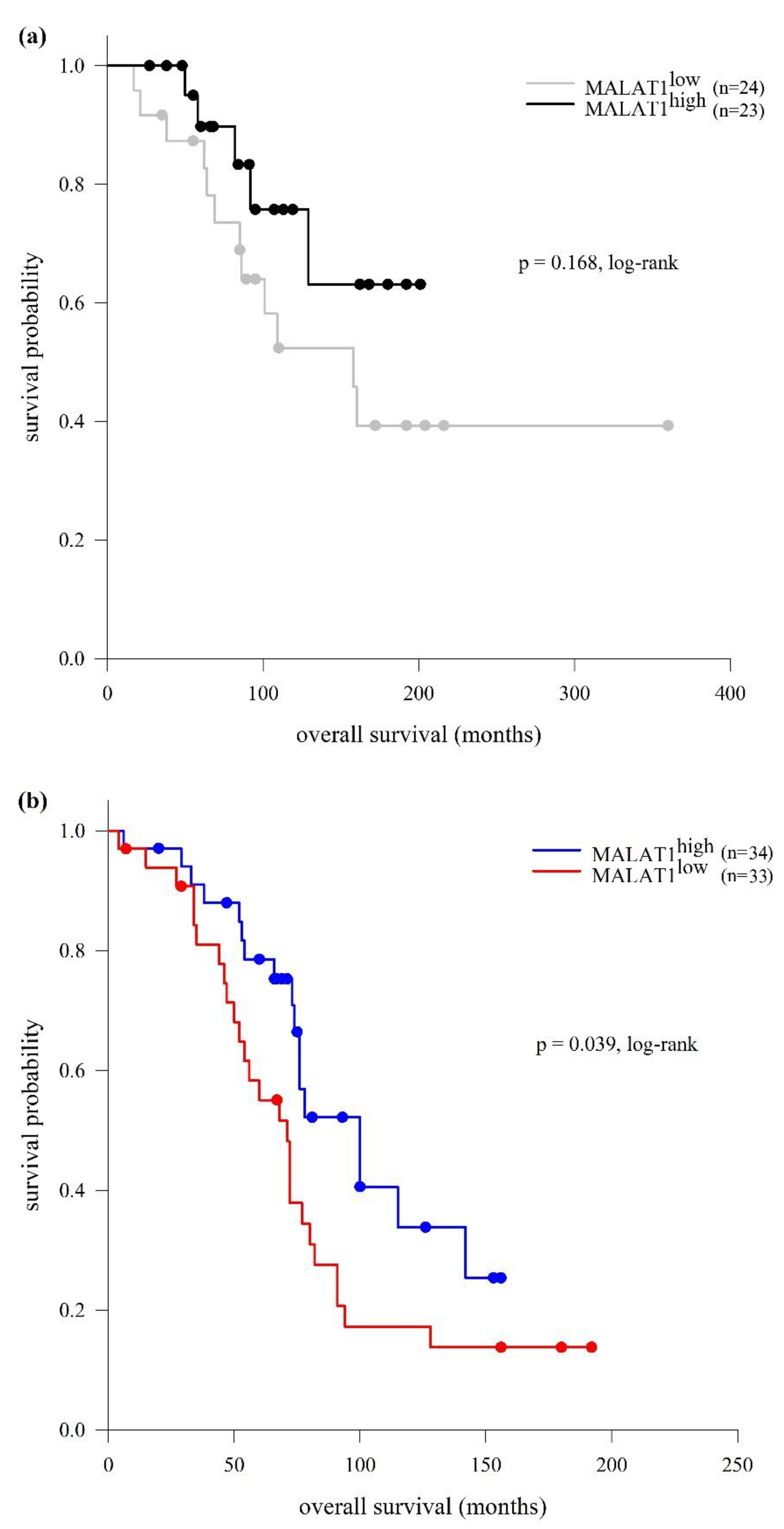
2.3. LncRNA MALAT1 Expression and Clinical Outcomes
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Quantification of LncRNA MALAT1
4.3. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed]
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 Long Non-Coding RNA: Functional Implications. Noncoding RNA 2020, 6, 22. [Google Scholar] [CrossRef]
- Johnsson, P.; Lipovich, L.; Grander, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef]
- Brown, J.A.; Bulkley, D.; Wang, J.; Valenstein, M.L.; Yario, T.A.; Steitz, T.A.; Steitz, J.A. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014, 21, 633–640. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef]
- Miyagawa, R.; Tano, K.; Mizuno, R.; Nakamura, Y.; Ijiri, K.; Rakwal, R.; Shibato, J.; Masuo, Y.; Mayeda, A.; Hirose, T.; et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 2012, 18, 738–751. [Google Scholar] [CrossRef]
- Ageeli, A.A.; McGovern-Gooch, K.R.; Kaminska, M.M.; Baird, N.J. Finely tuned conformational dynamics regulate the protective function of the lncRNA MALAT1 triple helix. Nucleic Acids Res. 2019, 47, 1468–1481. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Kabotyanski, E.B.; Reineke, L.C.; Shao, J.; Xiong, F.; Lee, J.H.; Dubrulle, J.; Johnson, H.; Stossi, F.; Tsoi, P.S.; et al. The SINEB1 element in the long non-coding RNA Malat1 is necessary for TDP-43 proteostasis. Nucleic Acids Res. 2020, 48, 2621–2642. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Sirokman, K.; McDonel, P.; Shishkin, A.A.; Surka, C.; Russell, P.; Grossman, S.R.; Chow, A.Y.; Guttman, M.; Lander, E.S. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 2014, 159, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, P.; Qu, J.; Shi, L.; Zhuang, W.; Fu, J.; Li, J.; Zhang, X.; Sun, Y.; Zhuang, W. Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J. Biol. Chem. 2014, 289, 29365–29375. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Xu, D.Y.; Sha, W.G.; Shen, L.; Lu, G.Y. Long non-coding RNA MALAT1 interacts with transcription factor Foxo1 to regulate SIRT1 transcription in high glucose-induced HK-2 cells injury. Biochem. Biophys. Res. Commun. 2018, 503, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Li, J.; Han, Z.; Chen, Z.; Zhang, Q. Silencing of lncRNA MALAT1 Prevents Inflammatory Injury after Lung Transplant Ischemia-Reperfusion by Downregulation of IL-8 via p300. Mol. Ther. Nucleic Acids 2019, 18, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lin, C.; Liu, W.; Zhang, J.; Ohgi, K.A.; Grinstein, J.D.; Dorrestein, P.C.; Rosenfeld, M.G. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011, 147, 773–788. [Google Scholar] [CrossRef]
- Stamato, M.A.; Juli, G.; Romeo, E.; Ronchetti, D.; Arbitrio, M.; Caracciolo, D.; Neri, A.; Tagliaferri, P.; Tassone, P.; Amodio, N. Inhibition of EZH2 triggers the tumor suppressive miR-29b network in multiple myeloma. Oncotarget 2017, 8, 106527–106537. [Google Scholar] [CrossRef]
- Wang, D.; Ding, L.; Wang, L.; Zhao, Y.; Sun, Z.; Karnes, R.J.; Zhang, J.; Huang, H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 2015, 6, 41045–41055. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, S.H.; Yang, W.I.; Kim, S.J.; Yoon, S.O. Association of the long non-coding RNA MALAT1 with the polycomb repressive complex pathway in T and NK cell lymphoma. Oncotarget 2017, 8, 31305–31317. [Google Scholar] [CrossRef]
- Wang, Q.M.; Lian, G.Y.; Song, Y.; Huang, Y.F.; Gong, Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019, 231, 116335. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.M.; Hu, W.W. Long non-coding RNA MALAT1 promotes oral squamous cell carcinoma development via microRNA-125b/STAT3 axis. J. Cell. Physiol. 2018, 233, 3384–3396. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; He, H.; Xiao, W.; Liu, Q.; Deng, Z.; Lu, Y.; Wang, Q.; Zheng, Q.; Li, Y. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget 2016, 7, 54733–54743. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Li, L.; Shi, Y.; Bu, X.; Xia, Y.; Wang, J.; Djangmah, H.S.; Liu, X.; You, Y.; Xu, B. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget 2016, 7, 63901–63912. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Chen, J.; Huang, J.; Lu, J.; Pei, S.; Ding, S.; Kang, L.; Xiao, R.; Zeng, Q. Functions and regulatory mechanisms of metastasis-associated lung adenocarcinoma transcript 1. J. Cell. Physiol. 2018, 234, 134–151. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, Y.; Li, H.; Chen, L.; Liu, Q. Regulatory Networks of LncRNA MALAT-1 in Cancer. Cancer Manag. Res. 2020, 12, 10181–10198. [Google Scholar] [CrossRef]
- Chen, Q.; Zhu, C.; Jin, Y. The Oncogenic and Tumor Suppressive Functions of the Long Noncoding RNA MALAT1: An Emerging Controversy. Front. Genet. 2020, 11, 93. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, D.; Li, Y.; Pan, X.; Zhang, X.; Kuang, B.; Zhou, M.; Li, X.; Xiong, W.; Li, G.; et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J. Cancer 2016, 7, 991–1001. [Google Scholar] [CrossRef]
- Hallek, M. Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am. J. Hematol. 2019, 94, 1266–1287. [Google Scholar] [CrossRef]
- Stevenson, F.K.; Forconi, F.; Packham, G. The meaning and relevance of B-cell receptor structure and function in chronic lymphocytic leukemia. Semin. Hematol. 2014, 51, 158–167. [Google Scholar] [CrossRef]
- Agathangelidis, A.; Chatzidimitriou, A.; Chatzikonstantinou, T.; Tresoldi, C.; Davis, Z.; Giudicelli, V.; Kossida, S.; Belessi, C.; Rosenquist, R.; Ghia, P.; et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: The 2022 update of the recommendations by ERIC, the European Research Initiative on CLL. Leukemia 2022, 36, 1961–1968. [Google Scholar] [CrossRef] [PubMed]
- Fabris, L.; Juracek, J.; Calin, G. Non-Coding RNAs as Cancer Hallmarks in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2020, 21, 6720. [Google Scholar] [CrossRef]
- Lingua, M.F.; Carra, G.; Maffeo, B.; Morotti, A. Non-Coding RNAs: The “Dark Side Matter” of the CLL Universe. Pharmaceuticals 2021, 14, 168. [Google Scholar] [CrossRef] [PubMed]
- Gasic, V.; Karan-Djurasevic, T.; Pavlovic, D.; Zukic, B.; Pavlovic, S.; Tosic, N. Diagnostic and Therapeutic Implications of Long Non-Coding RNAs in Leukemia. Life 2022, 12, 1770. [Google Scholar] [CrossRef] [PubMed]
- Subhash, S.; Andersson, P.-O.; Kosalai, S.T.; Kanduri, C.; Kanduri, M. Global DNA methylation profiling reveals new insights into epigenetically deregulated protein coding and long noncoding RNAs in CLL. Clin. Epigenetics 2016, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Manzoni, M.; Agnelli, L.; Vinci, C.; Fabris, S.; Cutrona, G.; Matis, S.; Colombo, M.; Galletti, S.; Taiana, E.; et al. lncRNA profiling in early-stage chronic lymphocytic leukemia identifies transcriptional fingerprints with relevance in clinical outcome. Blood Cancer J. 2016, 6, e468. [Google Scholar] [CrossRef]
- Ahmadi, A.; Kaviani, S.; Yaghmaie, M.; Pashaiefar, H.; Ahmadvand, M.; Jalili, M.; Alimoghaddam, K.; Eslamijouybari, M.; Ghavamzadeh, A. Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings. Blood Res. 2018, 53, 320–324. [Google Scholar] [CrossRef]
- Fernandez-Garnacho, E.M.; Nadeu, F.; Martin, S.; Mozas, P.; Rivero, A.; Delgado, J.; Gine, E.; Lopez-Guillermo, A.; Duran-Ferrer, M.; Salaverria, I.; et al. MALAT1 expression is associated with aggressive behavior in indolent B-cell neoplasms. Sci. Rep. 2023, 13, 16839. [Google Scholar] [CrossRef]
- Winkle, M.; Kluiver, J.L.; Diepstra, A.; van den Berg, A. Emerging roles for long noncoding RNAs in B-cell development and malignancy. Crit. Rev. Oncol. Hematol. 2017, 120, 77–85. [Google Scholar] [CrossRef]
- Dahl, M.; Kristensen, L.S.; Gronbaek, K. Long Non-Coding RNAs Guide the Fine-Tuning of Gene Regulation in B-Cell Development and Malignancy. Int. J. Mol. Sci. 2018, 19, 2475. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zou, J.; Wan, X.; Sun, C.; Peng, F.; Chu, Z.; Hu, Y. The Role of Noncoding RNAs in B-Cell Lymphoma. Front. Oncol. 2020, 10, 577890. [Google Scholar] [CrossRef]
- Li, L.J.; Chai, Y.; Guo, X.J.; Chu, S.L.; Zhang, L.S. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed. Pharmacother. 2017, 89, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sehgal, L.; Jain, N.; Khashab, T.; Mathur, R.; Samaniego, F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 2016, 14, 346. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.F.; Chang, Y.C.; Chang, C.S.; Lin, S.F.; Liu, Y.C.; Hsiao, H.H.; Chang, J.G.; Liu, T.C. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer 2014, 14, 809. [Google Scholar] [CrossRef] [PubMed]
- Handa, H.; Kuroda, Y.; Kimura, K.; Masuda, Y.; Hattori, H.; Alkebsi, L.; Matsumoto, M.; Kasamatsu, T.; Kobayashi, N.; Tahara, K.I.; et al. Long non-coding RNA MALAT1 is an inducible stress response gene associated with extramedullary spread and poor prognosis of multiple myeloma. Br. J. Haematol. 2017, 179, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Isin, M.; Ozgur, E.; Cetin, G.; Erten, N.; Aktan, M.; Gezer, U.; Dalay, N. Investigation of circulating lncRNAs in B-cell neoplasms. Clin. Chim. Acta 2014, 431, 255–259. [Google Scholar] [CrossRef]
- Ronchetti, D.; Agnelli, L.; Taiana, E.; Galletti, S.; Manzoni, M.; Todoerti, K.; Musto, P.; Strozzi, F.; Neri, A. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget 2016, 7, 14814–14830. [Google Scholar] [CrossRef]
- Hu, Y.; Lin, J.; Fang, H.; Fang, J.; Li, C.; Chen, W.; Liu, S.; Ondrejka, S.; Gong, Z.; Reu, F.; et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia 2018, 32, 2250–2262. [Google Scholar] [CrossRef]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, T.; Jia, Y.; Zou, J.; Wang, X.; Gu, W. LncRNA MALAT1/miR-181a-5p affects the proliferation and adhesion of myeloma cells via regulation of Hippo-YAP signaling pathway. Cell Cycle 2019, 18, 2509–2523. [Google Scholar] [CrossRef]
- Liu, N.; Feng, S.; Li, H.; Chen, X.; Bai, S.; Liu, Y. Long non-coding RNA MALAT1 facilitates the tumorigenesis, invasion and glycolysis of multiple myeloma via miR-1271-5p/SOX13 axis. J. Cancer Res. Clin. Oncol. 2020, 146, 367–379. [Google Scholar] [CrossRef]
- Jeffers, L.K.; Duan, K.; Ellies, L.G.; Seaman, W.T.; Burger-Calderon, R.A.; Diatchenko, L.B.; Webster-Cyriaque, J. Correlation of transcription of MALAT-1, a novel noncoding RNA, with deregulated expression of tumor suppressor p53 in small DNA tumor virus models. J. Cancer Ther. 2013, 4, 10–4236. [Google Scholar] [CrossRef]
- Ma, X.Y.; Wang, J.H.; Wang, J.L.; Ma, C.X.; Wang, X.C.; Liu, F.S. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genom. 2015, 16, 676. [Google Scholar] [CrossRef]
- Tripathi, V.; Shen, Z.; Chakraborty, A.; Giri, S.; Freier, S.M.; Wu, X.; Zhang, Y.; Gorospe, M.; Prasanth, S.G.; Lal, A.; et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013, 9, e1003368. [Google Scholar] [CrossRef]
- Chen, R.; Liu, Y.; Zhuang, H.; Yang, B.; Hei, K.; Xiao, M.; Hou, C.; Gao, H.; Zhang, X.; Jia, C.; et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017, 45, 9947–9959. [Google Scholar] [CrossRef]
- Tano, K.; Onoguchi-Mizutani, R.; Yeasmin, F.; Uchiumi, F.; Suzuki, Y.; Yada, T.; Akimitsu, N. Identification of Minimal p53 Promoter Region Regulated by MALAT1 in Human Lung Adenocarcinoma Cells. Front. Genet. 2017, 8, 208. [Google Scholar] [CrossRef]
- Molica, S.; Shanafelt, T.D.; Giannarelli, D.; Gentile, M.; Mirabelli, R.; Cutrona, G.; Levato, L.; Di Renzo, N.; Di Raimondo, F.; Musolino, C.; et al. The chronic lymphocytic leukemia international prognostic index predicts time to first treatment in early CLL: Independent validation in a prospective cohort of early stage patients. Am. J. Hematol. 2016, 91, 1090–1095. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Dohner, H.; Hillmen, P.; Keating, M.J.; Montserrat, E.; Rai, K.R.; et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 2008, 111, 5446–5456. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Dohner, H.; Hillmen, P.; Keating, M.; Montserrat, E.; Chiorazzi, N.; et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018, 131, 2745–2760. [Google Scholar] [CrossRef]
- Wood, B.L.; Arroz, M.; Barnett, D.; DiGiuseppe, J.; Greig, B.; Kussick, S.J.; Oldaker, T.; Shenkin, M.; Stone, E.; Wallace, P. 2006 Bethesda International Consensus recommendations on the immunophenotypic analysis of hematolymphoid neoplasia by flow cytometry: Optimal reagents and reporting for the flow cytometric diagnosis of hematopoietic neoplasia. Cytometry B Clin. Cytom. 2007, 72 (Suppl. S1), S14–S22. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.H.; Holden, J.T.; Bene, M.C.; Borowitz, M.J.; Braylan, R.C.; Cornfield, D.; Gorczyca, W.; Lee, R.; Maiese, R.; Orfao, A.; et al. 2006 Bethesda International Consensus recommendations on the flow cytometric immunophenotypic analysis of hematolymphoid neoplasia: Medical indications. Cytometry B Clin. Cytom. 2007, 72 (Suppl. S1), S5–S13. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, R.; Ghia, P.; Hadzidimitriou, A.; Sutton, L.A.; Agathangelidis, A.; Baliakas, P.; Darzentas, N.; Giudicelli, V.; Lefranc, M.P.; Langerak, A.W.; et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: Updated ERIC recommendations. Leukemia 2017, 31, 1477–1481. [Google Scholar] [CrossRef] [PubMed]
- Malcikova, J.; Tausch, E.; Rossi, D.; Sutton, L.A.; Soussi, T.; Zenz, T.; Kater, A.P.; Niemann, C.U.; Gonzalez, D.; Davi, F.; et al. ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-update on methodological approaches and results interpretation. Leukemia 2018, 32, 1070–1080. [Google Scholar] [CrossRef]
- International CLL-IPI working group. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): A meta-analysis of individual patient data. Lancet Oncol. 2016, 17, 779–790. [Google Scholar] [CrossRef]

| Variable | Global Cohort (n = 114) | MALAT1low Group (n = 57) | MALAT1high Group (n = 57) | p-Value |
|---|---|---|---|---|
| Sex (n = 114) a, male/female | 81/33 | 35/22 | 46/11 | 0.011 d |
| Age (years) | ||||
| overall (n = 113) a, median (range) | 59 (33−80) | 57 (33−80) | 61.5 (38−76) | 0.470 e |
| ˂65 (n = 76) a, median (range) | 56 (33−64) | 56 (33−64) | 55 (38−64) | 0.676 d |
| ≥65 (n = 37) a, median (range) | 68 (65−80) | 69 (66−80) | 66 (65−76) | 0.058 d |
| WBC [×109/L], (n = 99) a, median (range) | 37.6 (6.8−570) | 34.1 (6.8−476) | 44.4 (9.3−570) | 0.264 d |
| Ly [×109/L], (n = 90) a, median (range) | 28.2 (2.7−558.6) | 26.4 (2.7−447.4) | 33.7 (5.6–558.6) | 0.444 d |
| Platelets [×109/L], (n = 97) a, median (range) | 174 (1−432) | 172 (1−432) | 178 (3−361) | 0.710 e |
| Hb [g/L], (n = 98) a, median (range) | 139 (44−178) | 132.5 (57−166) | 143.5 (44−178) | 0.004 d |
| β2M [mg/L] (n = 59) a, median (range) | 3.2 (0.2−11.5) | 3.3 (0.2−11.5) | 3.2 (2−6.7) | 0.974 d |
| LDH level b, (n = 77) a, median (range) | 1.4 (0.3−4.2) | 1.5 (0.7−4.2) | 1.1 (0.3−3) | 0.004 d |
| Binet stage (n = 113) a, n (%) | 0.250 f | |||
| A | 53 (46.9) | 25 (44.6) | 28 (49.1) | |
| B | 44 (38.9) | 20 (35.7) | 24 (42.1) | |
| C | 16 (14.2) | 11 (19.6) | 5 (8.8) | |
| CD38 status (n = 102) a, n (%) | 0.001 d | |||
| positive (≥30%) | 31 (30.4) | 20 (40) | 11 (21.2) | |
| negative (˂30%) | 71 (69.6) | 30 (58.8) | 41 (78.8) | |
| Cytogenetic risk (n = 99) a, n (%) | 0.133 f | |||
| favorable (del13q14) c | 32 (32.3) | 18 (33.3) | 14 (31.1) | |
| intermediate (no aberrations, trisomy 12) | 43 (43.4) | 27 (50) | 16 (35.6) | |
| unfavorable (del11q22-23, del17p13) | 24 (24.2) | 9 (16.7) | 15 (33.3) | |
| IGHV SHM status (n = 114) a, n (%) | 1.000 g | |||
| mutated | 47 (41.2) | 24 (42.1) | 23 (40.4) | |
| unmutated | 67 (58.8) | 33 (57.9) | 34 (59.6) | |
| TP53 mutational status (n = 61) a, n (%) | 0.536 d | |||
| wt | 50 (82) | 31 (62) | 6 (54.5) | |
| mutated | 11 (18) | 19 (38) | 5 (45.5) | |
| CLL-IPI (n = 55) a, n (%) | 0.228 g | |||
| low risk (score 0−1) | 9 (16.4) | 8 (21.1) | 1 (5.9) | |
| intermediate risk (score 2−3) | 18 (32.7) | 14 (36.8) | 4 (23.5) | |
| high risk (score 4−6) | 23 (41.8) | 13 (34.2) | 10 (58.8) | |
| very high risk (score 7−10) | 5 (9.1) | 3 (7.9) | 2 (11.8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomic Vujovic, K.; Ugrin, M.; Tosic, N.; Vukovic, V.; Marjanovic, I.; Kostic, T.; Stankovic, S.; Otasevic, V.; Sarac, S.; Antic, D.; et al. Expression Pattern and Prognostic Significance of the Long Non-Coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2024, 25, 922. https://doi.org/10.3390/ijms25020922
Tomic Vujovic K, Ugrin M, Tosic N, Vukovic V, Marjanovic I, Kostic T, Stankovic S, Otasevic V, Sarac S, Antic D, et al. Expression Pattern and Prognostic Significance of the Long Non-Coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences. 2024; 25(2):922. https://doi.org/10.3390/ijms25020922
Chicago/Turabian StyleTomic Vujovic, Kristina, Milena Ugrin, Natasa Tosic, Vojin Vukovic, Irena Marjanovic, Tatjana Kostic, Sanja Stankovic, Vladimir Otasevic, Sofija Sarac, Darko Antic, and et al. 2024. "Expression Pattern and Prognostic Significance of the Long Non-Coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 in Chronic Lymphocytic Leukemia" International Journal of Molecular Sciences 25, no. 2: 922. https://doi.org/10.3390/ijms25020922
APA StyleTomic Vujovic, K., Ugrin, M., Tosic, N., Vukovic, V., Marjanovic, I., Kostic, T., Stankovic, S., Otasevic, V., Sarac, S., Antic, D., Pavlovic, S., & Karan-Djurasevic, T. (2024). Expression Pattern and Prognostic Significance of the Long Non-Coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences, 25(2), 922. https://doi.org/10.3390/ijms25020922







