Abstract
Tight junction (TJ) proteins (Tjps), Tjp1 and Tjp2, are tight junction-associated scaffold proteins that bind to the transmembrane proteins of tight junctions and the underlying cytoskeleton. In this study, we first analyzed the tumorigenic characteristics of B16-F10 melanoma cells, including cell proliferation, migration, invasion, metastatic potential, and the expression patterns of related proteins, after the CRISPR–Cas9-mediated knockout (KO) of Tjp genes. The proliferation of Tjp1 and Tjp2 KO cells significantly increased in vitro. Other tumorigenic characteristics, including migration and invasion, were significantly enhanced in Tjp1 and Tjp2 KO cells. Zonula occludens (ZO)-associated protein Claudin-1 (CLDN-1), which is a major component of tight junctions and functions in controlling cell-to-cell adhesion, was decreased in Tjp KO cells. Additionally, Tjp KO significantly stimulated tumor growth and metastasis in an in vivo mouse model. We performed a transcriptome analysis using next-generation sequencing (NGS) to elucidate the key genes involved in the mechanisms of action of Tjp1 and Tjp2. Among the various genes affected by Tjp KO-, cell cycle-, cell migration-, angiogenesis-, and cell–cell adhesion-related genes were significantly altered. In particular, we found that the Ninjurin-1 (Ninj1) and Catenin alpha-1 (Ctnna1) genes, which are known to play fundamental roles in Tjps, were significantly downregulated in Tjp KO cells. In summary, tumorigenic characteristics, including cell proliferation, migration, invasion, tumor growth, and metastatic potential, were significantly increased in Tjp1 and Tjp2 KO cells, and the knockout of Tjp genes significantly affected the expression of related proteins.
1. Introduction
Tight junction proteins (Tjps), also known as zonula occludens (ZO), are scaffold proteins that are involved in the formation and maintenance of tight junctions and are critical for the integrity and barrier function of the epithelial and endothelial cell layers. Tjp1, Tjp2, and Tjp3 are peripheral membrane proteins that play crucial roles in linking the actomyosin cytoskeleton to other tight junction transmembrane proteins, such as occludins and claudins [1]. They also control apical–junctional organization and promote the formation of a functional barrier [2,3,4,5,6]. Previous studies on Tjp1 and Tjp2 have mainly focused on the regulation of epithelial barrier function. Tight junction protein compositions are involved in the maintenance of tight junction integrity, regulation of paracellular transport, and control of epithelial permeability. ZO proteins contain three N-terminal PDZ domains, followed by an SH3 domain, a GuK domain, and a carboxy-terminal end that includes both an acidic domain and a proline-rich region. PDZ domains are abundant protein interaction modules that frequently recognize short amino acid motifs at the C-termini of target proteins. They play a regulatory role in various biological processes, including transport, ion channel signaling, and other signal transduction systems [7,8].
Tjp1 and Tjp2 also interact with cytoplasmic proteins to form higher-order molecular structures at junctions, where various proteins involved in signaling and transcriptional modulation are recruited [9,10,11]. These proteins also include those involved in epithelial proliferation and differentiation, and tight junction protein compositions are known to function beyond regulating cell permeability [12]. Moreover, Tjp1 and Tjp2 have been reported to be associated with signaling pathways and cytoskeletal organization. Tjp1 interacts with various signaling molecules and cytoskeletal components, thereby participating in cell signaling and cytoskeletal organization. A number of functional studies have investigated the role of Tjp1 in cell polarity, migration, and cytoskeletal dynamics. Previous studies have reported the involvement of Tjp1 in cadherin-based cell adhesion via direct binding to catenin and actin filaments in exogenous E-cadherin (EL cells) [13]. Depletion of Tjp1 results in a decrease in non-muscle myosin-2B (NM2B) integration at the junction [14,15,16], an increase in conical stiffness [17], a loss of the twisting of the tight junction membrane [14,18], and an increase in joint contractility, combined with tissue changes in the conical actin filament [19,20]. Tjp1 partly influences cell proliferation by binding transcription factors that can localize to both adhesion complexes and the nucleus, where they play a role in regulating gene expression [21,22,23]. In a recent study on the regulation of the mechanical properties of apical and junctional membranes, the organization of apical actomyosin filaments and the junctional assembly of Tjp1 were reported to undergo changes and interactions in barrier structures in CRISPR-knockout (KO) epithelial cell lines [24]. Furthermore, Tjp1 and Tjp2 are reported to be associated with several diseases, including cancer, autoimmune disorders, and gastrointestinal disorders. Celiac disease (CD) is a representative autoimmune and inflammatory disease in which tight junctions open in the early stages, leading to severe intestinal damage [25,26,27,28]. Recently, the association between inflammation and TJ proteins has been studied. In particular, it has been reported that ZO-1 expression is reduced in inflammation-related human lung disease [8]. Type 1 diabetes is also associated with tight junctions. Observations in animal models have suggested that increased infiltration occurs prior to the appearance of histological or obvious signs of diabetes. Previous studies support these findings by reporting that zone-dependent intestinal permeability increases in the same rat model two to three weeks before the onset of type 1 diabetes [29].
The effect of Tjp on the tumorigenic characteristics of cancer cells varies depending on the specific context and type of cancer. In some studies, the loss or downregulation of Tjp1 has been associated with increased tumorigenic characteristics and cancer progression. A reduced expression of Tjp1 has been observed in various cancer types, including breast, ovarian, and colorectal cancers, and the loss of Tjp1 may contribute to the disruption of cell–cell adhesion and increase cell migration, invasion, and metastasis, all of which are hallmarks of aggressive cancer behavior [30,31,32]. However, there are also studies suggesting that Tjp1 has tumor suppressive properties. Tjp1 expression is capable of inhibiting cell proliferation, inducing cell cycle arrest, and promoting apoptosis in cancer cells. In some instances, Tjp1 has been found to regulate key signaling pathways involved in tumor growth and survival, such as the Akt and ERK pathways [33,34]. Therefore, although it is difficult to make a generalized statement, in certain cancer types, the knockout of Tjp1 may indeed lead to an increase in tumorigenic characteristics and cancer progression. However, in other contexts, Tjp1 loss may contribute to a decrease in tumorigenic characteristics. In humans, Tjp1 and Tjp2 are known to play a role in the invasion and metastasis of cancer by regulating the expression of Tjp1 and Tjp2 and causing changes in barrier function due to structural alterations in tight junctions [35]. Early studies found a correlation between reduced tight junctions and tumor differentiation [36,37,38], and research on the functional roles of tight junctions in tumor development has increased, although most studies have been conducted in vitro [30,39]. In breast cancer, Tjp1 is reduced in poorly differentiated tumors and is associated with an increase in tumor grade and TNM (tumor-nodal) status [32]. Studies exploring the effects of tight junction molecules on tumor progression are currently accumulating [35,40]. Although TJ proteins are closely related to cancer and other diseases, studies on their mechanisms of action are scarce. A recent study revealed that the expression of Tjp1 is decreased in human lung diseases associated with inflammation [8] and in pancreatic cancer cell lines, and the knockdown (KD) of Tjp1 and claudin-1 contributes to tumor migration and invasion in xenograft tumors [41].
A tight junction protein is a protein that plays a role in cell-to-cell binding. However, the investigation into how this binding influences the characteristics of cancer growth, metastasis, migration, invasion in cancer cells has not yet been conclusively conducted.
In this study, we first knocked out Tjp1 and Tjp2 in B16-F10 mouse melanoma cells using the CRISPR–Cas9 system and analyzed the tumorigenic characteristics, such as invasion and migration, in vitro. Additionally, we analyzed the effects of Tjp1 and Tjp2 KO on tumor growth and experimental metastasis in an in vivo model. We confirmed TJ assembly by re-expressing Tjp in KO cells. An RNA-seq analysis was used to identify genes that are modulated by TJ proteins and are associated with tumorigenic characteristics and adhesion.
2. Results
2.1. Knockout of Tjp1 and Tjp2 in Mouse Melanoma Cell Lines
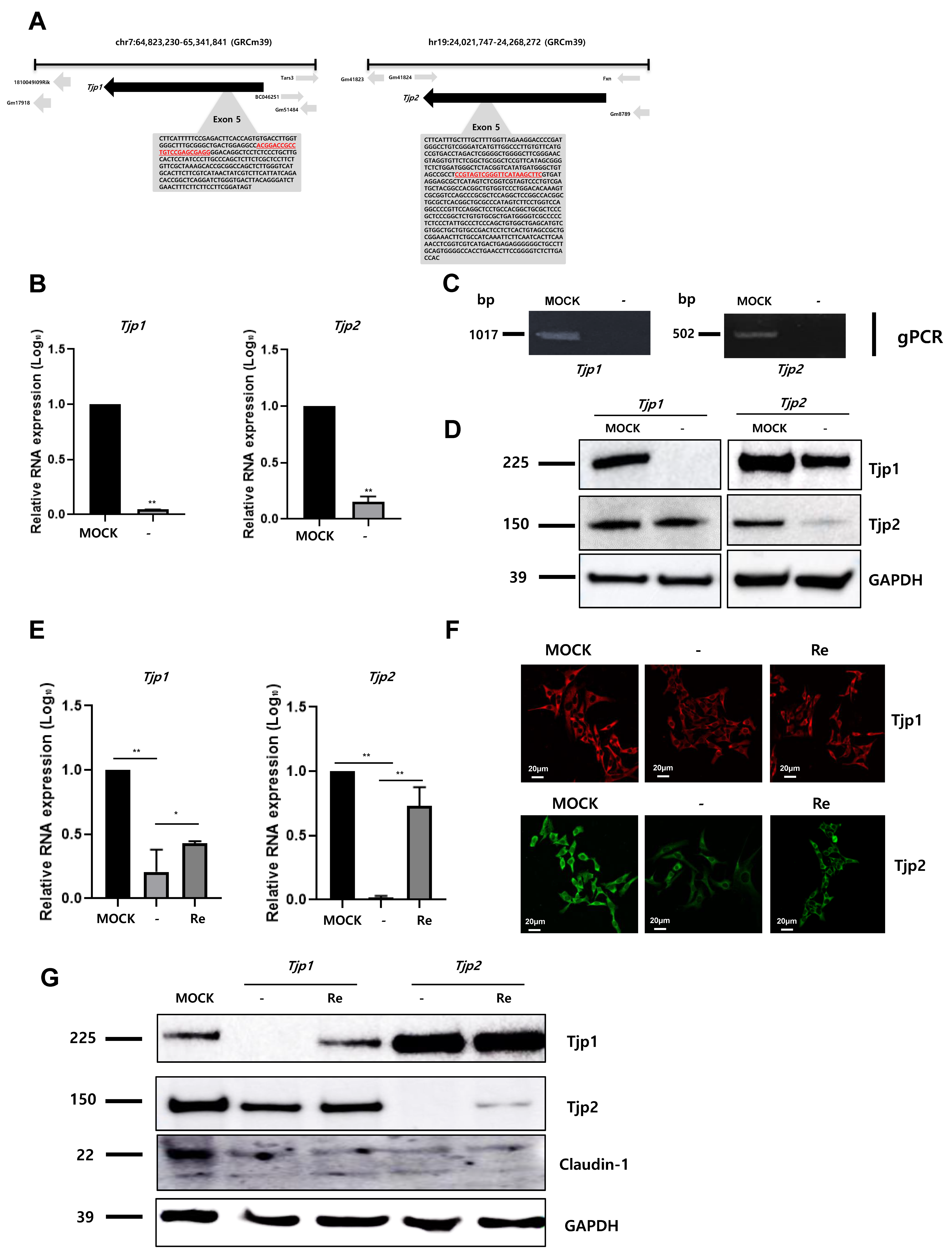
To identify the role of Tjp1 and Tjp2 in the tumorigenic characteristics of mouse melanoma, Tjp1 and Tjp2 KO B16-F10 melanoma cell lines were generated using the CRISPR/Cas9 gene editing system. gRNA was designed with the Tjp1 and Tjp2 exon 5 region, which encodes the most functional TJ protein, and Tjp1 and Tjp2 KO stable clones were selected through hygromycin selection. The locations and sequences of Tjp1 and Tjp2 gRNAs are displayed in Figure 1A and Supplementary Figure S1. After the selection of CRISPR/Cas9 system-transfected cells, Tjp1 and Tjp2 KO clones were confirmed with RT-PCR (Supplementary Figure S2). The RNA levels of Tjp1 and Tjp2 were confirmed using real-time PCR, and Tjp1 and Tjp2 expression was significantly reduced in each KO clone (Figure 1B). Genomic PCR (Figure 1C) containing the gRNA-selected region with specific primers (Supplementary Figure S2) confirmed the complete removal of the TJ protein genes, while other derivatives at different loci remained unaffected. The protein levels of Tjp1 and Tjp2 were also significantly reduced in KO cells (Figure 1D). To further investigate the impact and mechanism of Tjp gene KO, we generated re-expressing cells by overexpressing Tjp1 and Tjp2 in each KO cell line. The expression of Tjp1 and Tjp2 at the RNA level was partially recovered in each re-expressed cell line (Figure 1E), and IF and Western blot analyses confirmed that the protein levels were also recovered in each re-expressed cell line (Figure 1F,G). The expression of claudin-1, which interacts with Tjps to form tight junctions, was significantly reduced in both Tjp1 and Tjp2 KO cells; however, the reduced expression level was not recovered after re-expression in both Tjp1 and Tjp2 KO cells (Figure 1G).
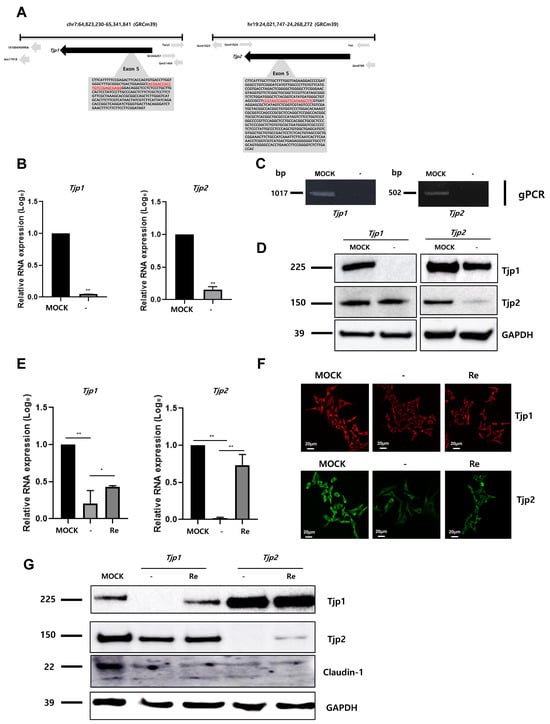
Figure 1.
Knockout of tight junction (TJ) protein expression in B16-F10 melanoma cells. (A) Selection of guide RNAs (gRNAs, in red) for the targeting of Tjp1 and Tjp2 in knockout system. gRNA was designed with the Tjp1 and Tjp2 exon 5 region, which encodes the most functional TJ proteins. (B) Expressions of Tjp1 and Tjp2 RNA in Tjp1 and Tjp2 knockout (KO) cells, respectively. Real-time polymerase chain reaction (RT-qPCR) performed for general region of Tjp genes. Real-time PCR reactions were coupled to melting-curve analysis to confirm the amplification specificity. Non-template controls were included for each primer pair to check for any significant level of contaminants. Real-time PCR was performed in three independent experiments with three different samples per group. The mRNA expression levels were calculated by normalizing to GAPDH and presented relative to the control with calculated mean values and 95% confidence intervals. The statistical significance of differences between groups was determined using a two-tailed Student’s t-test. ** p < 0.01. (C) Genomic PCR was performed for specific regions of Tjp derivatives. (D) Protein expression of Tjp1 and Tjp2 in Tjp1 KO and Tjp2 KO B16-F10 cells. Western blotting was performed to analyze the protein levels of TJ proteins. (E) Relative mRNA expression levels of Tjp1 KO and Tjp2 KO and re-expressed B16-F10 melanoma cells. The mRNA expression levels were determined with real-time qPCR and calculated by normalizing to GAPDH. * p < 0.05, ** p < 0.01. (F) Immunofluorescence (IF) staining of the tight junction protein expressions in Tjp1 and Tjp2 KO cells and re-expressed cells. (G) Protein expressions of Tjp1, Tjp2, and Claudin-1 in MOCK, Tjp1KO, Tjp2 KO, and re-expressed B16-F10 melanoma cells. Protein expressions were analyzed with Western blot.
2.2. Decreased Expression of Tight Junction Proteins Stimulates Tumorigenic Characteristics, Including Invasion, Migration, Cell Proliferation, Tumor Growth, and Experimental Metastasis
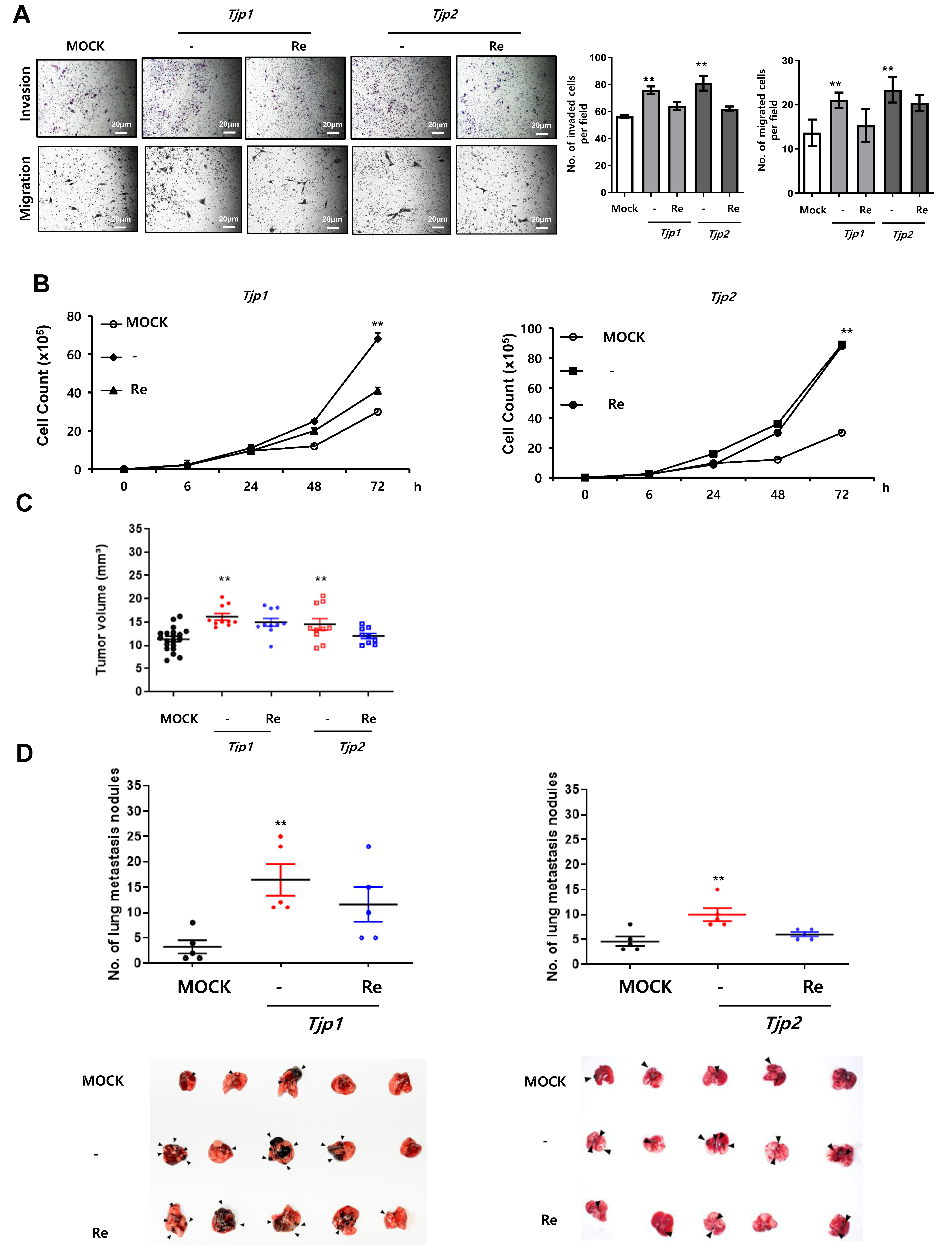
We conducted in vitro migration and invasion assays to examine the effects of Tjp KO on the migration and invasion of B16-F10 melanoma cells. As shown in Figure 2A, the migration and invasion of Tjp KO cells was significantly increased compared to those of the MOCK cells. These results suggest that the depletion of Tjp1 and Tjp2 stimulates the invasion and migration of B16-F10 melanoma cells. A significant increase in cell proliferation was the most prominent characteristic of Tjp KO cells. As shown in Figure 2B, cell growth was significantly higher in Tjp1 and Tjp2 KO cells, whereas cell growth was recovered in Tjp1 and Tjp2 re-expressed cells compared with MOCK cells.
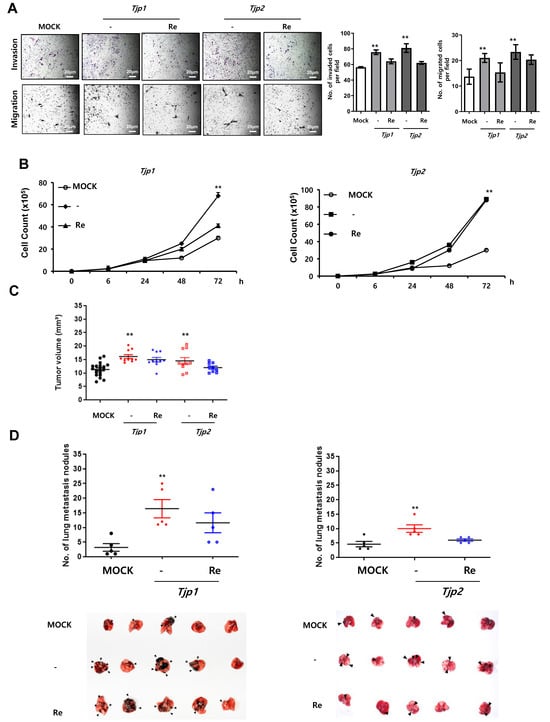
Figure 2.
Tjp1 and Tjp2 KO increased tumorigenic characteristics, including invasion, migration, cell proliferation, tumor growth, and experimental metastasis, in B16-F10 cells. (A) Invasion and migration of MOCK, Tjp1 KO, Tjp2 KO, and re-expressed cells. Invasion and migration were significantly increased in Tjp1 KO and Tjp2 KO B16-F10 melanoma cells and were recovered in re-expressed cells of each KO group. (B) Cell proliferation of MOCK, Tjp1 KO, Tjp2 KO, and re-expressed cells. Cell proliferation was significantly increased in Tjp1 KO and Tjp2 KO cells and was recovered in re-expressed cells of each KO group. (C) Subcutaneous tumor growth assay. Dot plot of xenograft tumor volume of MOCK (black), Tjp1 KO, Tjp2 KO (red), and re-expressed (blue) cells at 14 days after subcutaneous injection of cells. Tumor size was significantly increased in Tjp KO cells and was recovered in re-expressed cells. (D) Experimental metastasis assay. MOCK (black), Tjp KO (red), and re-expressed (blue) cells were injected into the tail veins of mice (10 mice per group), and the mice were sacrificed 3 weeks after injection. The number of metastatic lung nodules was determined by directly counting the nodules on the lung. ** p < 0.01.
We examined the effects of Tjp1 and Tjp2 KO expression on tumor growth and metastasis by subcutaneously and intravenously injecting separate groups of mice (10 per test group) with MOCK, Tjp1 and Tjp2 KO, or Tjp1 and Tjp2 re-expressed B16-F10 melanoma cells. Fourteen days after the subcutaneous injection, the mean tumor size of MOCK B16-F10 cells was 11.32 mm (95% CI_6.72 to 15.57). The mean tumor size was 16.1 mm (95% CI_ 13.84 to 20.33 mm) for mice injected with Tjp1 KO B16-F10 cells and 14.9 mm (95% CI_ 9.71 to 18.56 mm) for mice injected with Tjp1 re-expressed B16-F10 cells. Furthermore, the mean tumor size was 14.5 mm (95% CI_ 9.37 to 20.6 mm) for mice injected with Tjp2 KO B16-F10 cells and 12.8 mm (95% CI_ 9.94 to 20.23 mm) for mice injected with Tjp2 re-expressed B16-F10 cells. After comparing the mean tumor sizes of all treatment groups, we discovered that tumor growth was significantly increased in the Tjp1 and Tjp2 KO injected groups and growth rates recovered in the Tjp1 and Tjp2 re-expressing groups. These results confirmed that tumor growth was significantly promoted in Tjp1 and Tjp2 KO mice in vivo (Figure 2C). We further investigated whether Tjp1 and Tjp2 KO stimulated the metastatic potential of B16-F10 melanoma cells. Tumor cells were injected into the tail vein of the mice to evaluate their differential colonization abilities in the lungs. Three weeks after the mice were intravenously injected with the cells, the mean number of metastatic lung nodules was visually monitored (Figure 2D). Compared with the MOCK mice, significant increases in the size and number of lung nodules were observed in Tjp1 and Tjp2 KO-injected mice, but metastasis rates recovered in re-expressing cells. These results suggest that Tjp1 and Tjp2 KO not only promotes tumor growth but also enhances the metastatic potential of B16-F10 melanoma cells.
2.3. RNA Sequencing (RNA-Seq) for Analysis of Gene Expression Profile
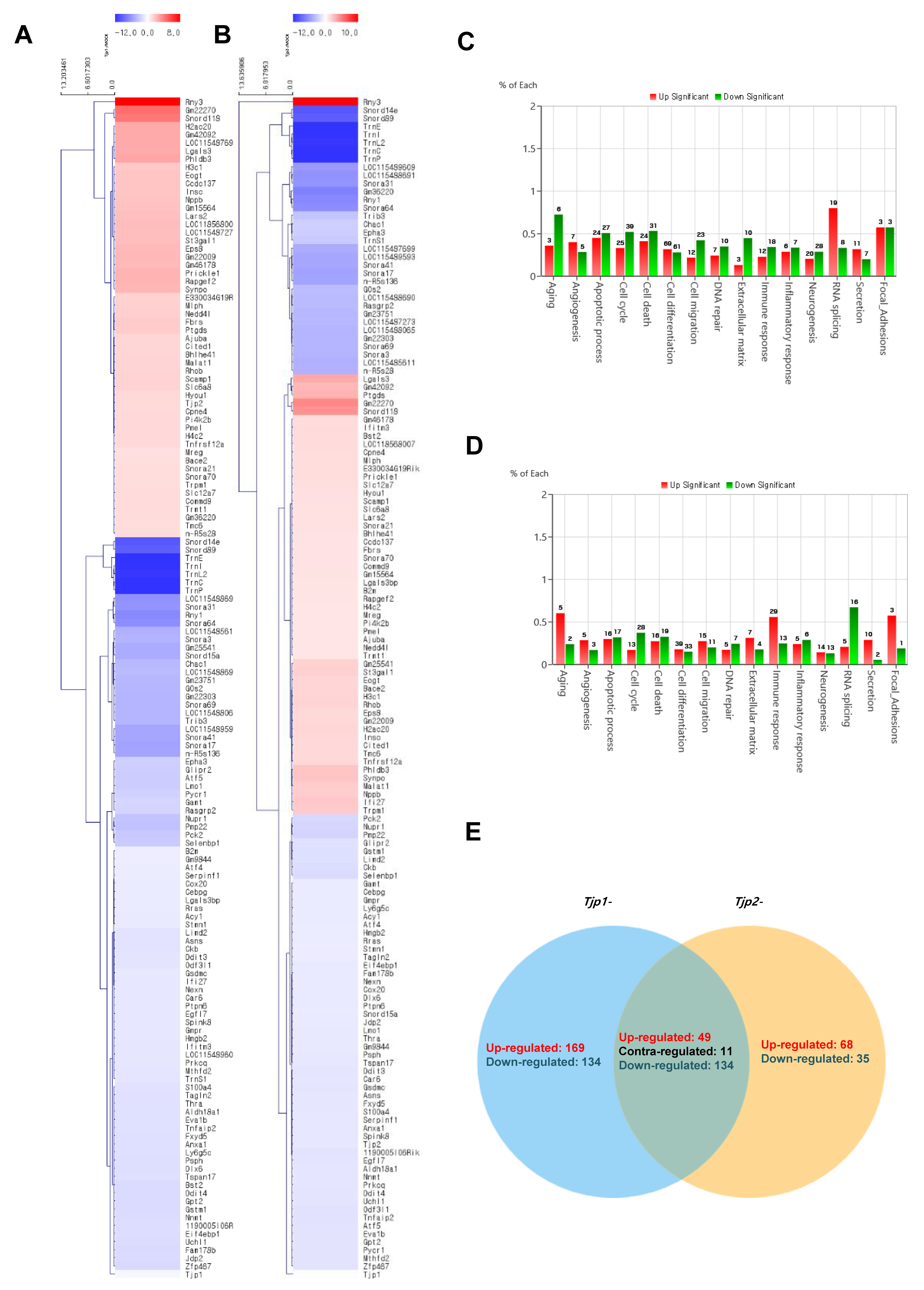
The cluster analysis of mRNAs related to Tjp KO is displayed in Figure 3. To identify the functional mechanism of TJ proteins in tumorigenic characteristics, the transcriptomes of Tjp1 KO and Tjp2 KO B16-F10 melanoma cells were analyzed and compared with those of MOCK cells using RNA sequencing. Heat maps of gene expression changes are displayed in Figure 3A for the Tjp1 KO group (89 downregulated and 55 upregulated; top panel) and in Figure 3B for the Tjp2 KO group (90 downregulated and 54 upregulated; top panel). The expression fold changes (FC) and various category-specific analyses of Tjp1 and Tjp2 KO cells were compared with those of MOCK cells using the ExDEGA program (Figure 3C,D). Among these categories, we analyzed the genes associated with tumor characteristics and the cell barrier. To identify the meaningful targets of tight junction proteins, Venn diagrams were analyzed in Tjp1 and Tjp2 KO B16-F10 melanoma cells. Approximately 49,196 transcripts were identified, and 144 of these differentially expressed genes (DEGs) were significantly different (Figure 3E).
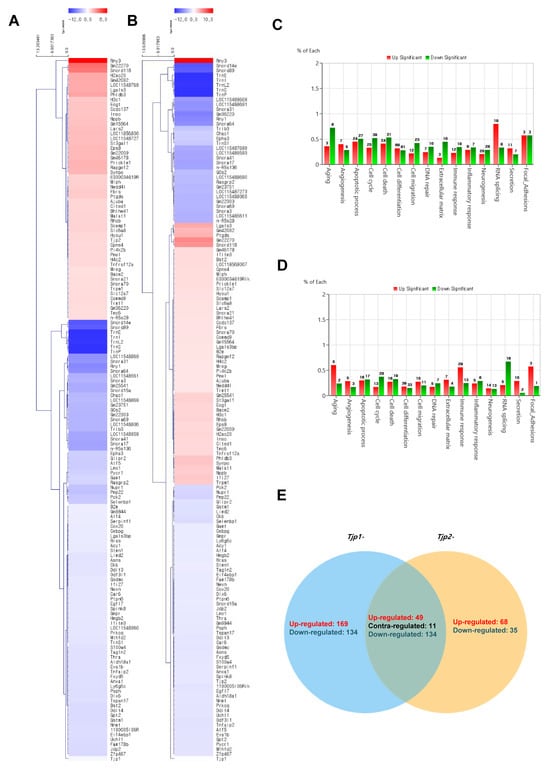
Figure 3.
Transcriptome analysis of Tjp1 KO and Tjp2 KO B16-F10 melanoma cells. (A) Clustering analysis of differentially expressed mRNAs related to Tjp1 KO. (B) Clustering analysis of differentially expressed mRNAs related to Tjp2 KO. The fold change 2, log2-normalized read counts of four were selected in (A,B). (C) Gene ontology analysis of differentially expressed genes (DEGs) in Tjp1 KO cells. (D) Gene ontology analysis of DEGs in Tjp2 KO cells. (E) Comparison of the DEG expression patterns observed in the Tjp1 KO and Tjp2 KO cells.
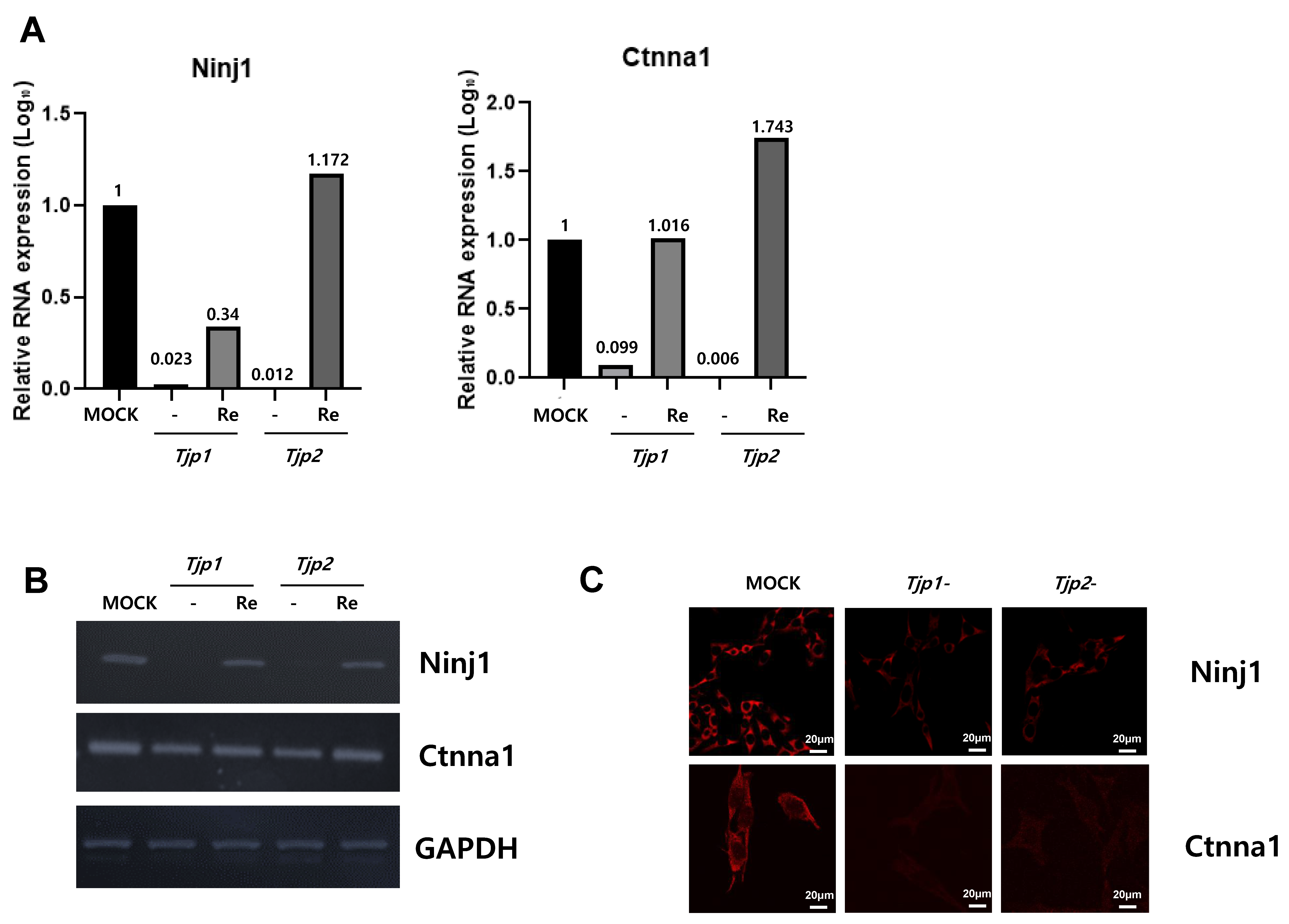
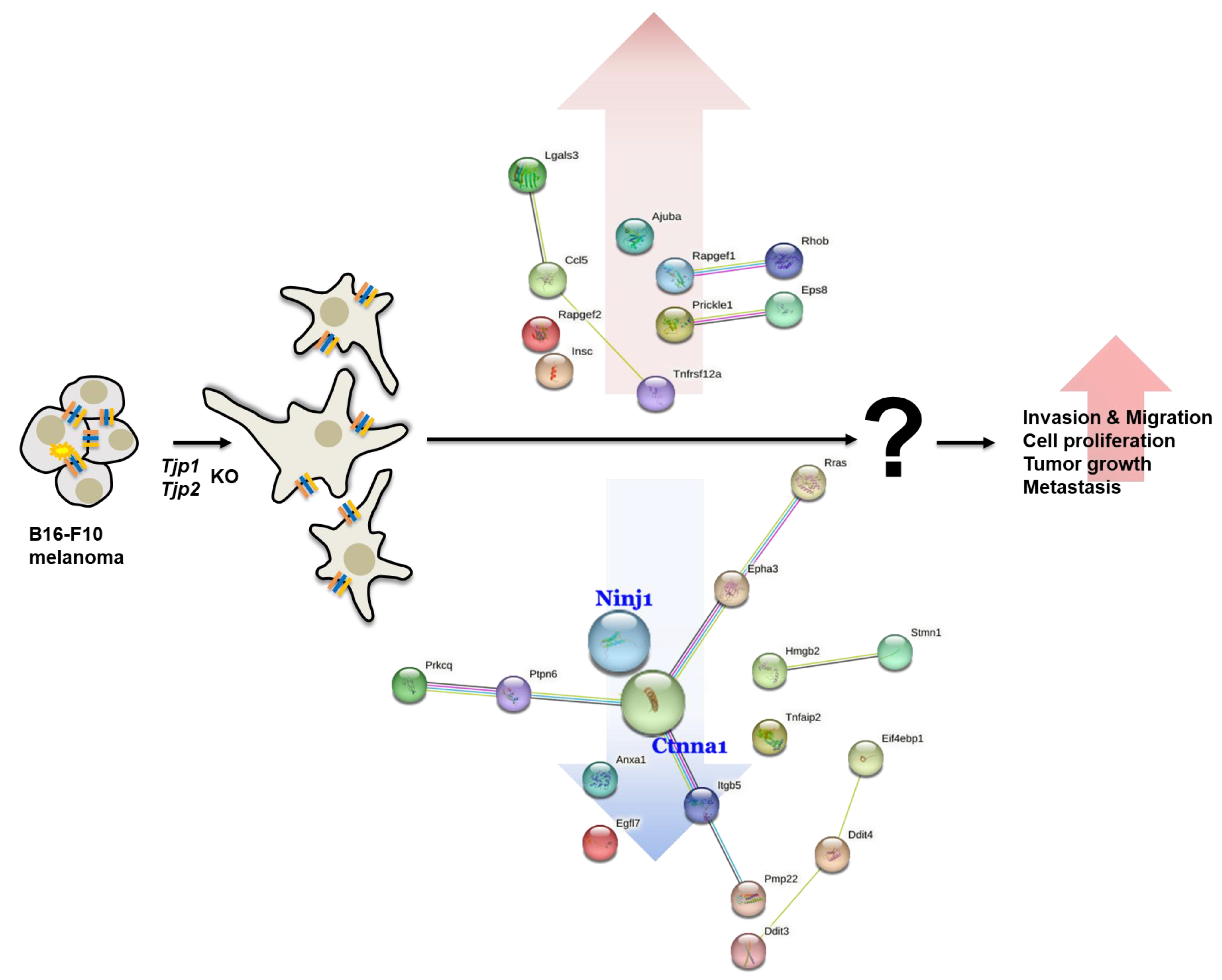
Among the 144 DEGs, genes related to cell cycle, cell migration, angiogenesis, and cell–cell adhesion appeared to be related to the tumorigenic characteristics of tight junction proteins. From the multitude of genes expressed in the context of Tjp1 and Tjp2 KO, we have chosen genes that commonly exhibit increases and decreases in tumorigenesis-related characteristics, including cell cycle, cell migration, angiogenesis, and cell–cell adhesion. This selection is based on consideration of cancer-related phenotypes that manifest (Table 1 and Table 2). The genes related to Tjp1 and Tjp2 KO, Ninj1, and Ctnna1, which are important cell–cell adherence factors, were significantly downregulated in both Tjp KO groups. As shown in Figure 4A,B, Ninj1 and Ctnna1 were downregulated at the mRNA level. IF confirmed that Ninj1 and Ctnna1 were downregulated at the protein level (Figure 4C).

Table 1.
Differential gene expression in Tjp1 knockout cells.

Table 2.
Differential gene expression in Tjp2 knockout cells.
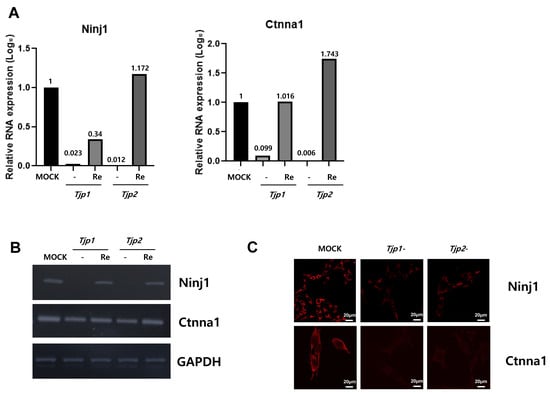
Figure 4.
Expression of Ninj1 and Ctnna1 in Tjp1 KO, Tjp2 KO, and re-expressed B16-F10 melanoma cells. (A) Real-time PCR of Ninj1 and Ctnna1 in Tjp1 KO, Tjp2 KO, and re-expressed B16-F10 melanoma cells. The mRNA expression levels were calculated by normalizing to GAPDH and presented relative to the control with calculated mean values and 95% confidence intervals. (B) RT-PCR of Ninj1 and Ctnna1 in Tjp1 KO, Tjp2 KO, and re-expressed B16-F10 melanoma cells. (C) Immunofluorescence staining of Ninj1 and Ctnna1 in Tjp1 and Tjp2 KO cells.
3. Discussion
The role of tight junction protein in cancer has been extensively studied; however, its significance remains controversial. Tight junction proteins contribute to cell proliferation via several mechanisms. In contrast to our observations, the TJP-1 protein was found to be highly expressed in adenocarcinoma samples compared to healthy tissues [42]. Importantly, high claudin expression in human cancer tissues is inconsistent with boundary localization and barrier regulation [43]. In addition, previous studies distinguish between claudin expression level and tight junction formation and conclude that deleting ZO proteins results in no tight junctions despite normal claudin levels in mouse mammary gland tumor Eph4 cells [44]. In our results, the expression of claudin was significantly reduced upon Tjp KO cells. However, the re-expression of Tjp did not show a significant expression of claudin difference compared to that of Tjp KO cells. It is worth noting that B16F10 cells, being highly aggressive melanoma cells, exhibit variations that may be attributed to their cell type.
Overall, previous studies have suggested that ZO proteins contribute to the control of the contact regulation of cell proliferation. Decreased TJP-1 expression is correlated with increased invasiveness in breast cancer [45], colorectal cancer [46], and cancers of the human digestive tract [47]. Additionally, TJP-1 has been reported to be involved in epithelial–mesenchymal transition (EMT) processes associated with tumor invasion [48]. Therefore, TJP-1 is believed to play a significant role in the processes underlying tumor growth, and its expression is closely associated with patient prognosis. TJP-2 is also a tumor suppressor protein. Previous studies have reported that TJP-2 exhibits a loss of localization at the cell borders and displays cytoplasmic staining in testicular carcinoma in situ and in bronchopulmonary cancers. This abnormal localization of TJP-2 is associated with the disruption of the blood–testis barrier [49] and the invasive characteristics of lung cancer cells in vitro [50]. The expression of TJP-2 has also been found to be downregulated in various carcinomas, such as breast [51,52] and pancreatic [53] as well as in hypoxia-resistant cancer cell lines derived from scirrhous gastric carcinoma [54]. Notably, patients with incompletely enhanced glioblastoma multiforme (GBM), who exhibit an increased survival rate, display higher levels of TJP-2 expression than those with completely enhanced GBM, which is associated with shortened survival [55]. In the context of tumorigenesis, the altered expression or dysfunction of tight junction proteins has been implicated in increased tumor invasiveness and metastasis. The loss of tight junction integrity promotes the migration and invasion of cancer cells through tissues. Furthermore, the disruption of tight junctions can lead to changes in cellular polarity, aberrant signaling pathways, and enhanced angiogenesis, all of which are hallmark features associated with tumor progression [12]. However, in Madin–Darby Canine Kidney (MDCK) cells, TJP-1 KO or double knockdown (dKD) decreased the migration of individual cells within a confluent monolayer. In addition, the velocity of single TJP-1 and TJP-2 dKD cells in the absence of cell–cell contact was even higher than that of single WT cells [56]. These results suggest that the newly discovered function of TJP1 and TJP2 also plays a role in efficient population cell migration by maintaining tissue fluidity and regulating proliferation. In this study, we successfully generated Tjp1 and Tjp2 KO melanoma cells for the first time using the CRISPR/Cas9 system and demonstrated that TJ proteins are not only important for barrier function but also have a significant effect on cancer progression. We observed significant increases in cell migration, invasion, and proliferation in vitro. In vivo experiments revealed a significant increase in both the size and metastasis of cancer cells compared to the MOCK groups. Our results confirmed that Tjp KO in B16-F10 cells increased cancer progression and improved tumor characteristics (Figure 5). These results are equivalent to those of previous reports that suggested that the tight junction protein, zonula occludens (ZO)-1, regulates cell proliferation and gene expression [57]. Moreover, studies on metastasis and tight junction proteins have demonstrated that the expression of TJP1 mRNA is correlated with lymph node metastasis in patients with human bladder cancer [58]. Tjp2 may represent a novel target for molecular therapies aimed at preventing the invasion and metastasis of hamster pancreatic cancer [59]. Tight junction proteins could have a fundamental role in preventing the metastasis of breast cancer cells [35]. In this study, we explored the post-extravasation process through experimental metastasis. Even in the step where the binding force is weakened, allowing for free cells to penetrate blood vessels, they can easily exit the host cells without entrapment, in contrast to general cancer cells. There is a high likelihood of avoiding the binding of immune cells, and metastasis may be promoted by the tight junction proteins’ knockout even during growth in secondary tumors.
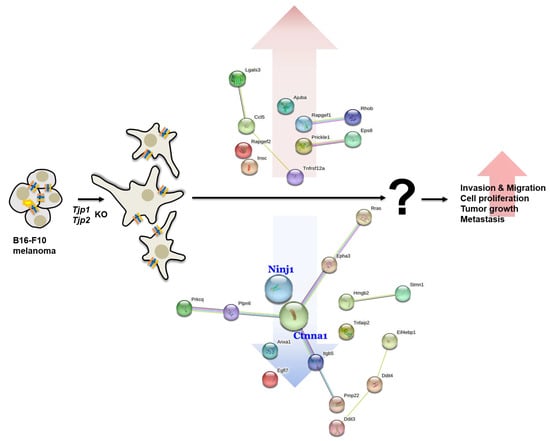
Figure 5.
Schematic diagram of the effect of Tjp1 and Tjp2 KO on tumorigenic characteristics.
We also found that Ninj1 and Ctnna1, which are important factors for cell–cell adhesion in a transcriptome analysis, were significantly reduced in the Tjp KO group. Ninj1 is known to play an important role, not only in cell–cell adhesion but also in immune function. Ninj1 also plays a crucial role in pulmonary fibrosis by promoting the interactions between macrophages and alveolar epithelial cells [60]. Ninj1 expression leads to macrophage activation in intestinal inflammatory conditions [61] and its upregulation in macrophages enhances cell–cell and cell–matrix adhesion in the hyaloid vascular system [62]. Its role in immunity and inflammatory reactions potentially affects the expression of TJ proteins. Ctnna1 (catenin) is primarily associated with adherens junctions, which are cell–cell junctions that link neighboring cells. It is part of a complex of proteins that helps to anchor and connect cells to one another. This complex, known as the adherens junction, plays fundamental roles in cell adhesion, signaling, and tissue integrity. It forms a complex with other proteins, including cadherins and catenin, and connects its cytoplasmic domain to the actin cytoskeleton. This linkage is essential for the stability and integrity of adherens junctions [63,64,65]. In particular, TJP-1 is an actin filament (F-actin)-binding protein located at a close junction that connects claudin to the actin cytoskeleton of epithelial cells. In non-cortical cells without tight junctions, TJP-1 is localized to adhesive junctions (AJ) and can indirectly link cytosines to the actin cytoskeleton via beta- and alpha-catenin [66]. Although a direct relationship between Tjp2 and Ctnna1 has not yet been reported, our study revealed a reduction in Ctnna1 expression in both Tjp1 KO and Tjp2 KO cells. Ctnna1 and tight junction proteins are distinct but complementary components of cell junctions that contribute to the overall stability, integrity, and functionality of tissues and cell layers in the body. However, the relationships between Ninj1, Ctnna1, and Tjp remain unclear. Further research is needed to determine how Tjp1 and Tjp2 KO regulates the protein expression of Ninj1 and Ctnna1 and how the functions of Tjp and these proteins are correlated.
Our study identifies genes modified by KO and highlights the genetic characteristics of tumors. Specifically, Tjp1 and Tjp2 play crucial roles in accurately regulating these processes. Previous studies have reported that the inhibition of ZO-1 restrains the proliferation and invasion of oral squamous carcinoma cells [67] while enhancing the cell proliferation and invasion capacity of endometrial cancer [68], pancreatic cancer [41], and liver cancer [69]. Recently, a known regulatory mechanism has emerged in pancreatic cancer. Mechanistically, the zinc transporter protein 4 (ZIP4) exhibits high expression, suppressing ZO-1 and claudin-1 expressions by modulating the mesenchymal cell marker zinc finger E-box binding homeobox 1 (ZEB1). ZEB1 directly binds to the promoters of ZO-1 and claudin-1, repressing their transcription. Inhibition of ZIP4 elevates the expression levels of ZO-1 and claudin-1, as well as the phosphorylation levels of focal adhesion kinase (FAK) and Paxillin—two molecules related to cell adhesion and motility. The subsequent silencing of ZO-1 or claudin-1 rescues the phosphorylation levels and mitigates the phenomena of attenuated cell proliferation and invasion. The controversies surrounding the regulation of cancer cell biological processes by ZO proteins may be explained through the complicated and heterogeneous signal-regulating backgrounds of different cancer cells [70].
In addition, various studies have revealed binding genes in the context of previous melanoma treatments. Tight junction proteins regulate the paracellular transport of molecules, but the staining of a tissue microarray revealed that claudin-1 was overexpressed in melanoma and aberrantly expressed in the cytoplasm of malignant cells, suggesting a role other than transport [71]. The normal phenotype and controlled proliferation of melanocytes is strictly regulated by keratinocytes via E-cadherin. The extracellular domains of separate E-cadherin molecules are tethered together, and the intracellular domain is anchored to the actin fibers of the cytoskeleton via a complex of catenins [72]. The barrier molecule junctions plakoglobin, filaggrin, and dystonin play roles in melanoma growth and angiogenesis [73]. Further exploration is necessary to assess the potential translational implications of these findings in clinical melanoma treatment.
These other regulatory genes are involved; we identified expression changes in several genes involved in the cell cycle, cell migration, angiogenesis, and cell–cell adhesion in Tjp1 and Tjp2 KO cells. Based on these results, a further correlation analysis with genes whose functions have not been extensively studied is also needed. In addition, another characteristic feature indicated by our results is the promotion of cell division and metastasis in Tjp1 and Tjp2 KO regarding the association between tight junction proteins and cell division; these studies remain controversial, lacking clear evidence. Therefore, further follow-up studies are warranted.
4. Materials and Methods
4.1. Generation of Knockout Cell Line with CRISPR–Cas9
Guide RNA (gRNA) sequences for the CRISPR/Cas9 system were designed using the CRISPR design provided by Bioneer (Daejeon, Republic of Korea). The insert oligonucleotides for tight junction protein-1 gRNA #1 were 5′-ACGGACCGCCTGTCCGAGCGAGG-3′ and 5′-GAAGCTTATGAACCCGACTACGG-3’ for tight junction protein-2 gRNA #2. The complementary oligonucleotides for gRNAs were annealed and cloned into the CRISPR/Cas9-Puro and pRGEN-Cas9-CMV/T7-Hygro-EGFP vectors (Toolgen, Seoul, Republic of Korea). Mouse melanoma B16-F10 cells were transfected with CRISPR/Cas9+gRNA #1 and #2. Approximately 18 h after transfection, the cells were treated with 100 μg/mL hygromycin for 2 days. After 2 weeks, colonies were isolated using cloning cylinders, RT-PCR, qRT-PCR, and Western blotting.
4.2. Cell Culture
Mouse melanoma B16-F10 cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). B16-F10 cells were cultured in RPMI1620 containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA), 1% penicillin, and L-glutamine (Gibco, Brooklyn, NY, USA). All cell lines were maintained at 37 °C in a humidified atmosphere containing 5% CO2 and 90% humidity.
4.3. Plasmid Construct for Over-Expression
The plasmid construct used in the present study was the pcDNA3.1+/C-(K)-DYK expression vector. Tjp1_OMu20010 (catalogue number U9042HE240-1) and Tjp2_OMu15570 (Cat. NO NM_001198985.2) were purchased from Genescript (Piscataway, NJ, USA). Plasmid transfection was carried out using the Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) and according to the Lipofectamine 2000 protocol.
4.4. RT-PCR, qRT-PCR, and Genomic PCR (Polymerase Chain Reaction)
RNA was extracted from the cells using TRIzol Reagent (Invitrogen), cDNA was synthesized using a cDNA synthesis kit (Bioneer, Daejeon, Republic of Korea), qRT-PCR was performed using TB Green Premix Taq (Takara, Japan), and the results were recorded using QuantStudio 3 (Thermo Fisher Scientific). The relative gene expression levels were quantified based on the 2−ΔΔCt method and normalized to the reference gene, GAPDH. The primer sequences for Tjp genes are as follows: Tjp1 sense, 5′-GCC TCT GCA GTT AAG CAT-3′; antisense, 5′-AAG AGC TGG CTG TTT TA-3′; Tjp2 sense, 5′-ATG GGA GCA GTA CAC CGT GA-3′; antisense, 5′-TGA CCA CCC TGT CAT TTT CTT G-3. mGAPDH was used as a control (sense primer, 5′-CTCATGACCACAGTCCAT-3′; antisense primer, 5′-CACATTGGGGGTAGGAAC-3′). The Tjp1 RT-PCR cycling conditions were as follows: 94 °C for 2 min to activate DNA polymerase; 38 cycles at 94 °C for 1 min; 58 °C for 1 min; 72 °C for 1 min; and 72 °C for 10 min for post-elongation. The Tjp2 RT-PCR cycling conditions were as follows: 94 °C for 2 min to activate DNA polymerase; 38 cycles of 94 °C for 1 min; 60 °C for 1 min; 72 °C for 1 min; and 72°C for 10 min for post-elongation. Genomic PCR was performed using chromosomal DNA from the cells and PCR primers. Tjp1 sense, 5′-TGT GTT GGG GAA ATG TGC TG-3′, antisense 5′-CTG GCC CAA CAT TTC TTG CT-3′; Tjp2 sense, 5′-ACG ACC GAG GTT TTG AAG TG-3′, antisense 5′-TGT TGG CCC TTG TGT TCA TG-3′. The Tjp1 and Tjp2 genomic PCR cycling conditions were as follows: 94 °C for 10 min to activate DNA polymerase; 30 cycles of 94 °C for 1 min; 60 °C for 1 min; 72 °C for 1 min; and 72 °C for 5 min for post-elongation. The products were analyzed on a 1.8% agarose gel and photographed under LED light.
4.5. Western Blot Analysis
Western blotting was performed as previously described [74]. The following antibodies were used: Tjp1 (1:1000, Invitrogen, USA); ZO-2 (1:1000, Cell Signaling, Denver, MA, USA); Claudin-1 (1:1000, Invitrogen, USA); and GAPDH (1:5000, R&D Systems, Minneapolis, MN, USA). Secondary antibodies linked to HRP (RSA1122, RSA1221, BioActs, Incheon, Republic of Korea) were also used.
4.6. In Vitro Migration and Invasion Assays
In vitro migration and invasion assays were performed as previously described [75]. Briefly, transwell chambers containing membranes with an 8 μm pore size (Invitrogen) were used for both assays. For the migration assay, 600 μL of conditioned medium, which was obtained by culturing B16-F10 cells for 18 h in serum-free RPMI, was placed in the lower chambers of each well. B16-F10 cells (1 × 104 cells) were resuspended in 100 μL of serum-free RPMI and placed in the upper chambers of each well. The chambers were incubated for 18 h at 37 °C, and the cells in the lower chambers were fixed and stained with Diff Quit (Sysmex, Tokyo, Japan) following the Diff Quit protocol. The invasion assay was performed in a similar fashion, except that the upper surface of the transwell filter was coated with 20 µL of 0.5 mg/mL Matrigel (BD Biosciences, Bedford, MA, USA) before the cells were added to the upper chambers. All experiments were repeated at least three times, and each data point was measured in triplicate. Mean values and 95% confidence intervals (CIs) were calculated.
4.7. Cell Proliferation Assay
The in vitro cell proliferation assay was performed as described previously [76]. Briefly, cells (1 × 104 cells per well) were plated in complete medium in a 6-well plate and incubated for 72 h. The cells were then harvested, and cell proliferation rates were measured by counting viable cells using the trypan blue dye exclusion method.
4.8. Immunofluorescence (IF)
Immunofluorescence analysis was performed as described previously [77]. The chamber slides (1 × 104 cells) were incubated for 48 h at 37 °C, and cells were fixed with methanol. Slides were then permeabilized by incubation in 10% normal serum in phosphate buffered saline (PBS) for 1 h to block nonspecific antibodies. Slides were stained with antibodies to ZO-1 (1:500 dilution, Invitrogen, USA), ZO-2 (1:500 dilution, Cell Signaling, USA), Ninj1 (1:100 dilution, ABclonal, Woburn, MA, USA) [78], and Ctnna1 (1:500 dilution, Cell Signaling, USA) overnight at 4 °C. After washing thrice with PBS for 5 min, the slides were incubated with secondary antibodies, including Alexa Fluor 546 anti-mouse and Alexa Fluor 488 anti-rabbit antibodies (Invitrogen, Carlsbad, CA, USA). Specimen epifluorescence was determined using a confocal laser scanning microscope (LSM510 META; ZEISS, Jena, Germany). Confocal images were analyzed using AlphaEase FC image analysis IS-2200 software (Alpga Innotecgh, Randburg, Gauteng, South Africa). Fluorescence was measured using a Leica DMi8 microscope (Leica, Wetzlar, Germany).
4.9. Subcutaneous Tumor Growth and Experimental Metastasis Assays
Six-week-old female C57BL/6 mice (NCI, Frederick, MD, USA) were used for the tumor xenograft assay. Cells (2 × 105 cells/200 μL; 10 mice per group) were injected subcutaneously, and tumor diameters were measured every 2 days for 2 weeks post-injection using digital calipers. The metastatic potential of Tjp1, Tjp2 KO, and re-expressing cells was also examined using a lung colonization assay as previously described [75]. Briefly, C57BL/6 mice (10 mice per group) were infected with either the control or KO and re-expressing cells (1 × 105 cells/200 μL) via the tail veins. Three weeks later, the mice were sacrificed via asphyxiation with CO2, and their lungs were removed. The metastatic nodules on the lung surfaces were counted. Animal experiments were approved and conducted under the guidance of Kosin University College of Medicine Institutional Animal Care and Use Committee (KUCMIACUC; KMAP-22-12) [79].
4.10. Library Preparation and Sequencing
Libraries were prepared from total RNA using the NEBNext Ultra II Directional RNA-Seq Kit (NEW ENGLAND BioLabs, Inc., Hitchin, UK). mRNA was isolated using a Poly(A) RNA Selection Kit (LEXOGEN, Inc., Wien, Austria). The isolated mRNAs were used for cDNA synthesis and shearing following the manufacturer’s instructions. Indexing was performed using Illumina indexes 1–12. Enrichment was performed using PCR. Subsequently, libraries were examined using a TapeStation HS D1000 Screen Tape (Agilent Technologies, Amstelveen, The Netherlands) to evaluate the mean fragment size. Quantification was performed using a library quantification kit and StepOne Real-Time PCR System (Life Technologies, Inc., Carlsbad, CA, USA). High-throughput sequencing was performed via paired-end 100 sequencing using a NovaSeq 6000 (Illumina, Inc., San Diego, CA, USA).
4.11. Data Analysis
Transcriptome analysis was performed using Ebiogen (Ebiogen Inc., Seoul, Republic of Korea). Quality control of the raw sequencing data was performed using Fast QC. Adapter and low-quality reads (<Q20) were removed using FASTX_Trimmer and BBMap. Trimmed reads were mapped to the reference genome using TopHat. The Read Count (RC) data were processed based on the fragments per kilobase per million (FPKM+G) + geometric normalization method using EdgeR within R. Fragments per kilobase per million (FPKM) reads were estimated using Cufflinks. Data mining and graphic visualization were performed using ExDEGA v4.1 (Ebiogen Inc., Seoul, Republic of Korea).
4.12. Statistical Analysis
Descriptive statistics were calculated for the patient characteristics. Statistical significance was defined as a two-tailed p-value < 0.05. The fluorescence intensity was read using image analysis software, and the intensity was measured to calculate the mean values and 95% CIs. The statistical significance of the differences among groups was determined using a two-tailed Student’s t-test.
5. Conclusions
In conclusion, Tjp1 and Tjp2 play a significant role in cell proliferation, migration, cancer growth, and metastasis, involving various genes.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25020833/s1.
Author Contributions
Conceptualization, E.-J.K. and H.-J.C.; data curation, E.-J.K., D.-Y.K., M.-H.K. and H.A.; formal analysis, E.-J.K.; funding acquisition, K.S.S. and H.-J.C.; investigation, E.-J.K., D.-Y.K., M.-H.K. and H.A.; project administration, E.-J.K., K.S.S. and H.-J.C.; supervision, J.K., J.-Y.J., K.S.S. and H.-J.C.; writing—original draft, E.-J.K. and H.-J.C.; writing—review and editing, J.K., J.-Y.J., K.S.S. and H.-J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korean government (NRF-2021R1A4A1031380).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional r Ethics Committee of Kosin University College of Medicine Institutional Animal Care and Use Committee (KUCMIACUC; KMAP-22-12).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE240070.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviation
Tjp, Tight junction protein; ZO, zonula occludens; KO, Knockout; EMT, epithelial-mesenchymal transition; DEGs, differentially expressed genes (DEGs); dKD, double knockdown; Ninj1, Ninjurin-1; Ctnna1, Catenin alpha-1; FPKM: Fragments per kilobase per million; FC: fold change; zinc transporter protein 4 (ZIP4); zinc finger E-box binding homeobox 1 (ZEB1); focal adhesion kinase (FAK)
References
- Stevenson, B.R.; Siliciano, J.D.; Mooseker, M.S.; Goodenough, D.A. Identification of ZO-1: A high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 1986, 103, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Fanning, A.S.; Jameson, B.J.; Jesaitis, L.A.; Anderson, J.M. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 1998, 273, 29745–29753. [Google Scholar] [CrossRef] [PubMed]
- Van Itallie, C.M.; Tietgens, A.J.; Anderson, J.M. Visualizing the dynamic coupling of claudin strands to the actin cytoskeleton through ZO-1. Mol. Biol. Cell 2017, 28, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Fanning, A.S.; Van Itallie, C.M.; Anderson, J.M. Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell 2012, 23, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Odenwald, M.A.; Choi, W.; Kuo, W.T.; Singh, G.; Sailer, A.; Wang, Y.; Shen, L.; Fanning, A.S.; Turner, J.R. The scaffolding protein ZO-1 coordinates actomyosin and epithelial apical specializations in vitro and in vivo. J. Biol. Chem. 2018, 293, 17317–17335. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Acharya, B.R.; Peyret, G.; Fardin, M.A.; Mege, R.M.; Ladoux, B.; Yap, A.S.; Fanning, A.S.; Peifer, M. Remodeling the zonula adherens in response to tension and the role of afadin in this response. J. Cell Biol. 2016, 213, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Meerschaert, K.; Tun, M.P.; Remue, E.; De Ganck, A.; Boucherie, C.; Vanloo, B.; Degeest, G.; Vandekerckhove, J.; Zimmermann, P.; Bhardwaj, N.; et al. The PDZ2 domain of zonula occludens-1 and -2 is a phosphoinositide binding domain. Cell. Mol. Life Sci. 2009, 66, 3951–3966. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.J.; Choi, Y.H.; Song, K.S. The PDZ motif peptide of ZO-1 attenuates Pseudomonas aeruginosa LPS-induced airway inflammation. Sci. Rep. 2020, 10, 19644. [Google Scholar] [CrossRef]
- Balda, M.S.; Matter, K. Epithelial cell adhesion and the regulation of gene expression. Trends Cell Biol. 2003, 13, 310–318. [Google Scholar] [CrossRef]
- Matter, K.; Balda, M.S. Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 2003, 4, 225–236. [Google Scholar] [CrossRef]
- Gonzalez-Mariscal, L.; Tapia, R.; Chamorro, D. Crosstalk of tight junction components with signaling pathways. Biochim. Biophys. Acta 2008, 1778, 729–756. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Roth, I.; Feraille, E.; Hasler, U. Different effects of ZO-1, ZO-2 and ZO-3 silencing on kidney collecting duct principal cell proliferation and adhesion. Cell Cycle 2014, 13, 3059–3075. [Google Scholar] [CrossRef] [PubMed]
- Itoh, M.; Nagafuchi, A.; Moroi, S.; Tsukita, S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. J. Cell Biol. 1997, 138, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Savino, B.; Locati, M.; Zammataro, L.; Allavena, P.; Bonecchi, R. The chemokine system in cancer biology and therapy. Cytokine Growth Factor Rev. 2010, 21, 27–39. [Google Scholar] [CrossRef]
- Waugh, D.J.; Wilson, C. The interleukin-8 pathway in cancer. Clin. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef]
- Mehrad, B.; Keane, M.P.; Strieter, R.M. Chemokines as mediators of angiogenesis. Thromb. Haemost. 2007, 97, 755–762. [Google Scholar] [CrossRef]
- Salcedo, R.; Resau, J.H.; Halverson, D.; Hudson, E.A.; Dambach, M.; Powell, D.; Wasserman, K.; Oppenheim, J.J. Differential expression and responsiveness of chemokine receptors (CXCR1-3) by human microvascular endothelial cells and umbilical vein endothelial cells. FASEB J. 2000, 14, 2055–2064. [Google Scholar] [CrossRef]
- Singh, S.; Sadanandam, A.; Singh, R.K. Chemokines in tumor angiogenesis and metastasis. Cancer Metastasis Rev. 2007, 26, 453–467. [Google Scholar] [CrossRef]
- Polette, M.; Gilles, C.; Nawrocki-Raby, B.; Lohi, J.; Hunziker, W.; Foidart, J.M.; Birembaut, P. Membrane-type 1 matrix metalloproteinase expression is regulated by zonula occludens-1 in human breast cancer cells. Cancer Res. 2005, 65, 7691–7698. [Google Scholar] [CrossRef]
- Lazennec, G.; Richmond, A. Chemokines and chemokine receptors: New insights into cancer-related inflammation. Trends Mol. Med. 2010, 16, 133–144. [Google Scholar] [CrossRef]
- Bauer, H.; Zweimueller-Mayer, J.; Steinbacher, P.; Lametschwandtner, A.; Bauer, H.C. The dual role of zonula occludens (ZO) proteins. J. Biomed. Biotechnol. 2010, 2010, 402593. [Google Scholar] [CrossRef]
- Balda, M.S.; Matter, K. Tight junctions and the regulation of gene expression. Biochim. Biophys. Acta 2009, 1788, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Matter, K.; Balda, M.S. Epithelial tight junctions, gene expression and nucleo-junctional interplay. J. Cell Sci. 2007, 120 Pt 9, 1505–1511. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, E.; Spadaro, D.; Rouaud, F.; King, J.M.; Flinois, A.; Shah, J.; Sluysmans, S.; Mean, I.; Jond, L.; Turner, J.R.; et al. Cingulin binds to the ZU5 domain of scaffolding protein ZO-1 to promote its extended conformation, stabilization, and tight junction accumulation. J. Biol. Chem. 2022, 298, 101797. [Google Scholar] [CrossRef] [PubMed]
- Madara, J.L.; Trier, J.S. Structural abnormalities of jejunal epithelial cell membranes in celiac sprue. Lab. Investig. 1980, 43, 254–261. [Google Scholar] [PubMed]
- Wolters, V.M.; Alizadeh, B.Z.; Weijerman, M.E.; Zhernakova, A.; van Hoogstraten, I.M.; Mearin, M.L.; Wapenaar, M.C.; Wijmenga, C.; Schreurs, M.W. Intestinal barrier gene variants may not explain the increased levels of antigliadin antibodies, suggesting other mechanisms than altered permeability. Hum. Immunol. 2010, 71, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Szakal, D.N.; Gyorffy, H.; Arato, A.; Cseh, A.; Molnar, K.; Papp, M.; Dezsofi, A.; Veres, G. Mucosal expression of claudins 2, 3 and 4 in proximal and distal part of duodenum in children with coeliac disease. Virchows Arch. 2010, 456, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Schumann, M.; Gunzel, D.; Buergel, N.; Richter, J.F.; Troeger, H.; May, C.; Fromm, A.; Sorgenfrei, D.; Daum, S.; Bojarski, C.; et al. Cell polarity-determining proteins Par-3 and PP-1 are involved in epithelial tight junction defects in coeliac disease. Gut 2012, 61, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Watts, T.; Berti, I.; Sapone, A.; Gerarduzzi, T.; Not, T.; Zielke, R.; Fasano, A. Role of the intestinal tight junction modulator zonulin in the pathogenesis of type I diabetes in BB diabetic-prone rats. Proc. Natl. Acad. Sci. USA 2005, 102, 2916–2921. [Google Scholar] [CrossRef]
- Kaihara, T.; Kawamata, H.; Imura, J.; Fujii, S.; Kitajima, K.; Omotehara, F.; Maeda, N.; Nakamura, T.; Fujimori, T. Redifferentiation and ZO-1 reexpression in liver-metastasized colorectal cancer: Possible association with epidermal growth factor receptor-induced tyrosine phosphorylation of ZO-1. Cancer Sci. 2003, 94, 166–172. [Google Scholar] [CrossRef]
- Smalley, K.S.; Brafford, P.; Haass, N.K.; Brandner, J.M.; Brown, E.; Herlyn, M. Up-regulated expression of zonula occludens protein-1 in human melanoma associates with N-cadherin and contributes to invasion and adhesion. Am. J. Pathol. 2005, 166, 1541–1554. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Watkins, G.; Mansel, R.E.; Jiang, W.G. Loss of tight junction plaque molecules in breast cancer tissues is associated with a poor prognosis in patients with breast cancer. Eur. J. Cancer 2004, 40, 2717–2725. [Google Scholar] [CrossRef] [PubMed]
- Mitsiades, N.; Mitsiades, C.S.; Poulaki, V.; Chauhan, D.; Fanourakis, G.; Gu, X.; Bailey, C.; Joseph, M.; Libermann, T.A.; Treon, S.P.; et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. USA 2002, 99, 14374–14379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Baladandayuthapani, V.; Lin, H.; Mulligan, G.; Li, B.; Esseltine, D.W.; Qi, L.; Xu, J.; Hunziker, W.; Barlogie, B.; et al. Tight Junction Protein 1 Modulates Proteasome Capacity and Proteasome Inhibitor Sensitivity in Multiple Myeloma via EGFR/JAK1/STAT3 Signaling. Cancer Cell 2016, 29, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Jiang, W.G. Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta 2009, 1788, 872–891. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulos, D. Mortality and cardiovascular and bleeding outcomes in patients with CKD receiving antiplatelet therapy. Am. J. Kidney Dis. 2013, 61, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Zhong, Y.; Isomura, H.; Saitoh, M.; Enomoto, K.; Sawada, N.; Mori, M. Localization of 7H6 tight junction-associated antigen along the cell border of vascular endothelial cells correlates with paracellular barrier function against ions, large molecules, and cancer cells. Exp. Cell Res. 1996, 222, 269–274. [Google Scholar] [CrossRef]
- Hoevel, T.; Macek, R.; Mundigl, O.; Swisshelm, K.; Kubbies, M. Expression and targeting of the tight junction protein CLDN1 in CLDN1-negative human breast tumor cells. J. Cell. Physiol. 2002, 191, 60–68. [Google Scholar] [CrossRef]
- Borka, K.; Kaliszky, P.; Szabo, E.; Lotz, G.; Kupcsulik, P.; Schaff, Z.; Kiss, A. Claudin expression in pancreatic endocrine tumors as compared with ductal adenocarcinomas. Virchows Arch. 2007, 450, 549–557. [Google Scholar] [CrossRef]
- Gonzalez-Mariscal, L.; Lechuga, S.; Garay, E. Role of tight junctions in cell proliferation and cancer. Prog. Histochem. Cytochem. 2007, 42, 1–57. [Google Scholar] [CrossRef]
- Liu, M.; Yang, J.; Zhang, Y.; Zhou, Z.; Cui, X.; Zhang, L.; Fung, K.M.; Zheng, W.; Allard, F.D.; Yee, E.U.; et al. ZIP4 Promotes Pancreatic Cancer Progression by Repressing ZO-1 and Claudin-1 through a ZEB1-Dependent Transcriptional Mechanism. Clin. Cancer Res. 2018, 24, 3186–3196. [Google Scholar] [CrossRef]
- Takai, E.; Tan, X.; Tamori, Y.; Hirota, M.; Egami, H.; Ogawa, M. Correlation of translocation of tight junction protein Zonula occludens-1 and activation of epidermal growth factor receptor in the regulation of invasion of pancreatic cancer cells. Int. J. Oncol. 2005, 27, 645–651. [Google Scholar]
- Diaz-Coranguez, M.; Liu, X.; Antonetti, D.A. Tight Junctions in Cell Proliferation. Int. J. Mol. Sci. 2019, 20, 5972. [Google Scholar] [CrossRef]
- Umeda, K.; Ikenouchi, J.; Katahira-Tayama, S.; Furuse, K.; Sasaki, H.; Nakayama, M.; Matsui, T.; Tsukita, S.; Furuse, M.; Tsukita, S. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 2006, 126, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Hoover, K.B.; Liao, S.Y.; Bryant, P.J. Loss of the tight junction MAGUK ZO-1 in breast cancer: Relationship to glandular differentiation and loss of heterozygosity. Am. J. Pathol. 1998, 153, 1767–1773. [Google Scholar] [CrossRef] [PubMed]
- Kaihara, T.; Kusaka, T.; Nishi, M.; Kawamata, H.; Imura, J.; Kitajima, K.; Itoh-Minami, R.; Aoyama, N.; Kasuga, M.; Oda, Y.; et al. Dedifferentiation and decreased expression of adhesion molecules, E-cadherin and ZO-1, in colorectal cancer are closely related to liver metastasis. J. Exp. Clin. Cancer Res. 2003, 22, 117–123. [Google Scholar] [PubMed]
- Kimura, Y.; Shiozaki, H.; Hirao, M.; Maeno, Y.; Doki, Y.; Inoue, M.; Monden, T.; Ando-Akatsuka, Y.; Furuse, M.; Tsukita, S.; et al. Expression of occludin, tight-junction-associated protein, in human digestive tract. Am. J. Pathol. 1997, 151, 45–54. [Google Scholar] [PubMed]
- Polette, M.; Mestdagt, M.; Bindels, S.; Nawrocki-Raby, B.; Hunziker, W.; Foidart, J.M.; Birembaut, P.; Gilles, C. Beta-catenin and ZO-1: Shuttle molecules involved in tumor invasion-associated epithelial-mesenchymal transition processes. Cells Tissues Organs 2007, 185, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Fink, C.; Weigel, R.; Hembes, T.; Lauke-Wettwer, H.; Kliesch, S.; Bergmann, M.; Brehm, R.H. Altered expression of ZO-1 and ZO-2 in Sertoli cells and loss of blood-testis barrier integrity in testicular carcinoma in situ. Neoplasia 2006, 8, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Luczka, E.; Syne, L.; Nawrocki-Raby, B.; Kileztky, C.; Hunziker, W.; Birembaut, P.; Gilles, C.; Polette, M. Regulation of membrane-type 1 matrix metalloproteinase expression by zonula occludens-2 in human lung cancer cells. Clin. Exp. Metastasis 2013, 30, 833–843. [Google Scholar] [CrossRef]
- Chlenski, A.; Ketels, K.V.; Korovaitseva, G.I.; Talamonti, M.S.; Oyasu, R.; Scarpelli, D.G. Organization and expression of the human zo-2 gene (tjp-2) in normal and neoplastic tissues. Biochim. Biophys. Acta 2000, 1493, 319–324. [Google Scholar] [CrossRef]
- Tokes, A.M.; Szasz, A.M.; Juhasz, E.; Schaff, Z.; Harsanyi, L.; Molnar, I.A.; Baranyai, Z.; Besznyak, I., Jr.; Zarand, A.; Salamon, F.; et al. Expression of tight junction molecules in breast carcinomas analysed by array PCR and immunohistochemistry. Pathol. Oncol. Res. 2012, 18, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Chlenski, A.; Ketels, K.V.; Tsao, M.S.; Talamonti, M.S.; Anderson, M.R.; Oyasu, R.; Scarpelli, D.G. Tight junction protein ZO-2 is differentially expressed in normal pancreatic ducts compared to human pancreatic adenocarcinoma. Int. J. Cancer 1999, 82, 137–144. [Google Scholar] [CrossRef]
- Kato, Y.; Yashiro, M.; Noda, S.; Tendo, M.; Kashiwagi, S.; Doi, Y.; Nishii, T.; Matsuoka, J.; Fuyuhiro, Y.; Shinto, O.; et al. Establishment and characterization of a new hypoxia-resistant cancer cell line, OCUM-12/Hypo, derived from a scirrhous gastric carcinoma. Br. J. Cancer 2010, 102, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Pope, W.B.; Chen, J.H.; Dong, J.; Carlson, M.R.; Perlina, A.; Cloughesy, T.F.; Liau, L.M.; Mischel, P.S.; Nghiemphu, P.; Lai, A.; et al. Relationship between gene expression and enhancement in glioblastoma multiforme: Exploratory DNA microarray analysis. Radiology 2008, 249, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Skamrahl, M.; Pang, H.; Ferle, M.; Gottwald, J.; Rubeling, A.; Maraspini, R.; Honigmann, A.; Oswald, T.A.; Janshoff, A. Tight Junction ZO Proteins Maintain Tissue Fluidity, Ensuring Efficient Collective Cell Migration. Adv. Sci. 2021, 8, e2100478. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, A.; Tschernutter, M.; Bainbridge, J.W.; Balaggan, K.S.; Mowat, F.; West, E.L.; Munro, P.M.; Thrasher, A.J.; Matter, K.; Balda, M.S.; et al. The tight junction associated signalling proteins ZO-1 and ZONAB regulate retinal pigment epithelium homeostasis in mice. PLoS ONE 2010, 5, e15730. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C.; Tsai, K.W.; Liao, J.B.; Kuo, W.T.; Chang, Y.C.; Yang, Y.F. High expression of tight junction protein 1 as a predictive biomarker for bladder cancer grade and staging. Sci. Rep. 2022, 12, 1496. [Google Scholar] [CrossRef]
- Zhou, L.; Tan, X.; Wang, W.; Wang, B.; Dai, X.; Liu, J. Analysis of invasion-metastasis in pancreatic cancer: Correlation between the expression and arrangement of tight junction protein-2 and cell dissociation in pancreatic cancer cells. Mol. Med. Rep. 2010, 3, 149–153. [Google Scholar]
- Choi, S.; Woo, J.K.; Jang, Y.S.; Kang, J.H.; Hwang, J.I.; Seong, J.K.; Yoon, Y.S.; Oh, S.H. Ninjurin1 Plays a Crucial Role in Pulmonary Fibrosis by Promoting Interaction between Macrophages and Alveolar Epithelial Cells. Sci. Rep. 2018, 8, 17542. [Google Scholar] [CrossRef]
- Jung, H.J.; Kang, J.H.; Pak, S.; Lee, K.; Seong, J.K.; Oh, S.H. Detrimental Role of Nerve Injury-Induced Protein 1 in Myeloid Cells under Intestinal Inflammatory Conditions. Int. J. Mol. Sci. 2020, 21, 614. [Google Scholar] [CrossRef]
- Lee, H.J.; Ahn, B.J.; Shin, M.W.; Jeong, J.W.; Kim, J.H.; Kim, K.W. Ninjurin1 mediates macrophage-induced programmed cell death during early ocular development. Cell Death Differ. 2009, 16, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Beavon, I.R. The E-cadherin-catenin complex in tumour metastasis: Structure, function and regulation. Eur. J. Cancer 2000, 36, 1607–1620. [Google Scholar] [CrossRef] [PubMed]
- Giannini, A.L.; Vivanco, M.; Kypta, R.M. alpha-catenin inhibits beta-catenin signaling by preventing formation of a beta-catenin*T-cell factor*DNA complex. J. Biol. Chem. 2000, 275, 21883–21888. [Google Scholar] [CrossRef]
- Forster, S.; Hehlgans, S.; Rodel, F.; Otto, B.; Cordes, N. Differential effects of alpha-catenin on the invasion and radiochemosensitivity of human colorectal cancer cells. Int. J. Oncol. 2018, 52, 1117–1128. [Google Scholar] [PubMed]
- Yokoyama, S.; Tachibana, K.; Nakanishi, H.; Yamamoto, Y.; Irie, K.; Mandai, K.; Nagafuchi, A.; Monden, M.; Takai, Y. alpha-catenin-independent recruitment of ZO-1 to nectin-based cell-cell adhesion sites through afadin. Mol. Biol. Cell 2001, 12, 1595–1609. [Google Scholar] [CrossRef] [PubMed]
- Babkair, H.; Yamazaki, M.; Uddin, M.S.; Maruyama, S.; Abe, T.; Essa, A.; Sumita, Y.; Ahsan, M.S.; Swelam, W.; Cheng, J.; et al. Aberrant expression of the tight junction molecules claudin-1 and zonula occludens-1 mediates cell growth and invasion in oral squamous cell carcinoma. Hum. Pathol. 2016, 57, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, F.; Zhang, L.; Jia, Y.; Chen, H. MicroRNA-103 regulates the progression in endometrial carcinoma through ZO-1. Int. J. Immunopathol. Pharmacol. 2019, 33, 2058738419872621. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Zhang, H.; Tu, F.; Qiang, Y.; Nie, C. Decreased expression of ZO-1 is associated with tumor metastases in liver cancer. Oncol. Lett. 2019, 17, 1859–1864. [Google Scholar] [CrossRef]
- Yu, S.; He, J.; Xie, K. Zonula Occludens Proteins Signaling in Inflammation and Tumorigenesis. Int. J. Biol. Sci. 2023, 19, 3804–3815. [Google Scholar] [CrossRef]
- Leotlela, P.D.; Wade, M.S.; Duray, P.H.; Rhode, M.J.; Brown, H.F.; Rosenthal, D.T.; Dissanayake, S.K.; Earley, R.; Indig, F.E.; Nickoloff, B.J.; et al. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene 2007, 26, 3846–3856. [Google Scholar] [CrossRef] [PubMed]
- Kuphal, S.; Bosserhoff, A.K. E-cadherin cell-cell communication in melanogenesis and during development of malignant melanoma. Arch. Biochem. Biophys. 2012, 524, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Leick, K.M.; Rodriguez, A.B.; Melssen, M.M.; Benamar, M.; Lindsay, R.S.; Eki, R.; Du, K.P.; Parlak, M.; Abbas, T.; Engelhard, V.H.; et al. The Barrier Molecules Junction Plakoglobin, Filaggrin, and Dystonin Play Roles in Melanoma Growth and Angiogenesis. Ann. Surg. 2019, 270, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Ko, E.J.; Ock, M.S.; Choi, Y.H.; Iovanna, J.L.; Mun, S.; Han, K.; Kim, H.S.; Cha, H.J. Human Endogenous Retrovirus (HERV)-K env Gene Knockout Affects Tumorigenic Characteristics of nupr1 Gene in DLD-1 Colorectal Cancer Cells. Int. J. Mol. Sci. 2021, 22, 3941. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.J.; Jeong, M.J.; Kleinman, H.K. Role of thymosin beta4 in tumor metastasis and angiogenesis. J. Natl. Cancer Inst. 2003, 95, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Mizoguchi, J.; Ohtaki, T. Effect of oestrogen on Pasteurella pneumotropica in rat vagina. Lab. Anim. 1986, 20, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Jeon, K.Y.; Ko, E.J.; Chang, H.K.; Lee, S.H.; Ahn, B.K.; Ock, M.S.; Cha, H.J. Correlation of long interspersed element-1 open reading frame 1 and c-Met proto-oncogene protein expression in primary and recurrent colorectal cancers. Kosin Med. J. 2022, 37, 283–290. [Google Scholar] [CrossRef]
- Bae, S.J.; Shin, M.W.; Son, T.; Lee, H.S.; Chae, J.S.; Jeon, S.; Oh, G.T.; Kim, K.W. Ninjurin1 positively regulates osteoclast development by enhancing the survival of prefusion osteoclasts. Exp. Mol. Med. 2019, 51, 1–16. [Google Scholar] [CrossRef]
- Shim, J.; Kim, J. Considerations for experimental animal ethics in the research planning and evaluation process. Kosin Med. J. 2022, 37, 271–277. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).