Inferring Drug Set and Identifying the Mechanism of Drugs for PC3
Abstract
1. Introduction
2. Results
2.1. Inferring Pathways by Drug Clusters Using All Features
2.2. Drug Interaction with Clustering Using Feature Selection
3. Discussion
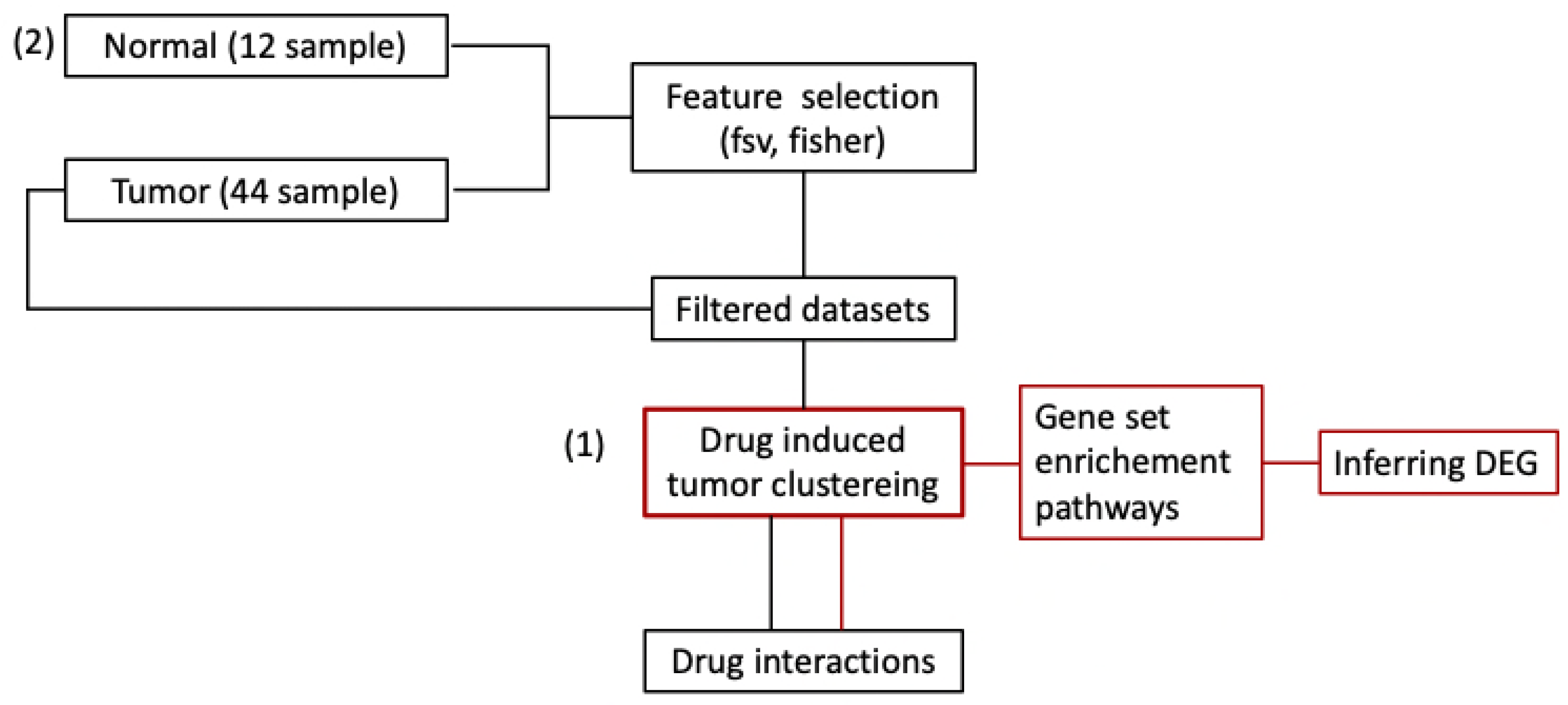
4. Materials and Methods
5. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, L.; Lu, B.; He, M.; Wang, Y.; Wang, Z.; Du, L. Prostate Cancer Incidence and Mortality: Global Status and Temporal Trends in 89 Countries From 2000 to 2019. Front Public Health 2022, 10, 811044. [Google Scholar] [CrossRef] [PubMed]
- Kim, S. Systemic Efficacy of Sirolimus via the ERBB Signaling Pathway in Breast Cancer. Processes 2022, 10, 552. [Google Scholar] [CrossRef]
- Ma, Y.; Hu, J.; Zhang, N.; Dong, X.; Li, Y.; Yang, B.; Tian, W.; Wang, X. Prediction of Candidate Drugs for Treating Pancreatic Cancer by Using a Combined Approach. PLoS ONE 2016, 11, e0149896. [Google Scholar] [CrossRef] [PubMed]
- Badkas, A.; De Landtsheer, S.; Sauter, T. Topological network measures for drug repositioning. Brief. Bioinform. 2021, 22, bbaa357. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Raj, M.R.; Sreeja, A. Analysis of computational gene prioritization approaches. Procedia Comput. Sci. 2018, 143, 395–410. [Google Scholar] [CrossRef]
- Horoszewicz, J.S.; Leong, S.S.; Chu, T.M.; Wajsman, Z.L.; Friedman, M.; Papsidero, L.; Kim, U.; Chai, L.S.; Kakati, S.; Arya, S.K.; et al. The LNCaP cell line–A new model for studies on human prostatic carcinoma. Prog. Clin. Biol. Res. 1980, 37, 115–132. [Google Scholar]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A k-means clustering algorithm. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Kim, S.; Kang, M.; Lee, S.; Bae, S.; Han, S.; Jang, J.Y.; Park, T. Identifying molecular subtypes related to clinicopathologic factors in pancreatic cancer. Biomed. Eng. Online 2014, 13 (Suppl. 2), S5. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural Networks for Pattern Recognition; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Sun, L.; Zhang, X.; Qian, Y.; Xu, J.; Zhang, S. Feature selection using neighborhood entropy-based uncertainty measures for gene expression data classification. Inf. Sci. 2019, 502, 18–41. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Matheson, A.J.; Figgitt, D.P. Rofecoxib: A review of its use in the management of osteoarthritis, acute pain and rheumatoid arthritis. Drugs 2001, 61, 833–865. [Google Scholar] [CrossRef] [PubMed]
- van Adelsberg, J.; Gann, P.; Ko, A.T.; Damber, J.E.; Logothetis, C.; Marberger, M.; Schmitz-Drager, B.J.; Tubaro, A.; Harms, C.J.; Roehrborn, C. The VIOXX in Prostate Cancer Prevention study: Cardiovascular events observed in the rofecoxib 25 mg and placebo treatment groups. Curr. Med. Res. Opin. 2007, 23, 2063–2070. [Google Scholar] [CrossRef] [PubMed]
- Di Silverio, F.; Bosman, C.; Salvatori, M.; Albanesi, L.; Pannunzi, L.P.; Ciccariello, M.; Cardi, A.; Salvatori, G.; Sciarra, A. Combination therapy with rofecoxib and finasteride in the treatment of men with lower urinary tract symptoms (LUTS) and benign prostatic hyperplasia (BPH). Eur. Urol. 2005, 47, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.I.; Subbaramaiah, K.; Du, B.; Chang, M.; Yang, P.; Newman, R.A.; Cordon-Cardo, C.; Thaler, H.T.; Dannenberg, A.J. Celecoxib inhibits prostate cancer growth: Evidence of a cyclooxygenase-2-independent mechanism. Clin. Cancer Res. 2005, 11, 1999–2007. [Google Scholar] [CrossRef] [PubMed]
- Kearney, P.M.; Baigent, C.; Godwin, J.; Halls, H.; Emberson, J.R.; Patrono, C. Do selective cyclo-oxygenase-2 inhibitors and traditional non-steroidal anti-inflammatory drugs increase the risk of atherothrombosis? Meta-analysis of randomised trials. BMJ 2006, 332, 1302–1308. [Google Scholar] [CrossRef]
- Solomon, D.H.; Avorn, J.; Sturmer, T.; Glynn, R.J.; Mogun, H.; Schneeweiss, S. Cardiovascular outcomes in new users of coxibs and nonsteroidal antiinflammatory drugs: High-risk subgroups and time course of risk. Arthritis Rheum. 2006, 54, 1378–1389. [Google Scholar] [CrossRef]
- Haroon, N.; Ather, M.H.; Khan, S.; Kumar, P.; Salam, B. Diclofenac suppository as a preemptive analgesia in ultrasound-guided biopsy of prostate: Randomized controlled trial. Urology 2015, 86, 682–685. [Google Scholar] [CrossRef]
- Montgomery, B.; Nelson, P.S.; Vessella, R.; Kalhorn, T.; Hess, D.; Corey, E. Estradiol suppresses tissue androgens and prostate cancer growth in castration resistant prostate cancer. BMC Cancer 2010, 10, 244. [Google Scholar] [CrossRef]
- Hall, J.M.; Couse, J.F.; Korach, K.S. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J. Biol. Chem. 2001, 276, 36869–36872. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.M. Estradiol and the developing brain. Physiol. Rev. 2008, 88, 91–124. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Segura, L.M.; Azcoitia, I.; DonCarlos, L.L. Neuroprotection by estradiol. Prog. Neurobiol. 2001, 63, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map. Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
- Sokal, R.R.; Rohlf, F.J. The comparison of dendrograms by objective methods. Taxonomy 1962, 11, 33–40. [Google Scholar] [CrossRef]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef]
- Giorgio. Feature Selection Library. MATLAB Central File Exchange. 2024. Available online: https://www.mathworks.com/matlabcentral/fileexchange/56937-feature-selection-library (accessed on 4 January 2024).
- Bradley, P.S.; Mangasarian, O.L. Feature Selection via Concave Minimization and Support Vector Machines. In Proceedings of the Fifteenth International Conference on Machine Learning, Madison, WI, USA, 24–27 July 1998; pp. 82–90. [Google Scholar]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef] [PubMed]
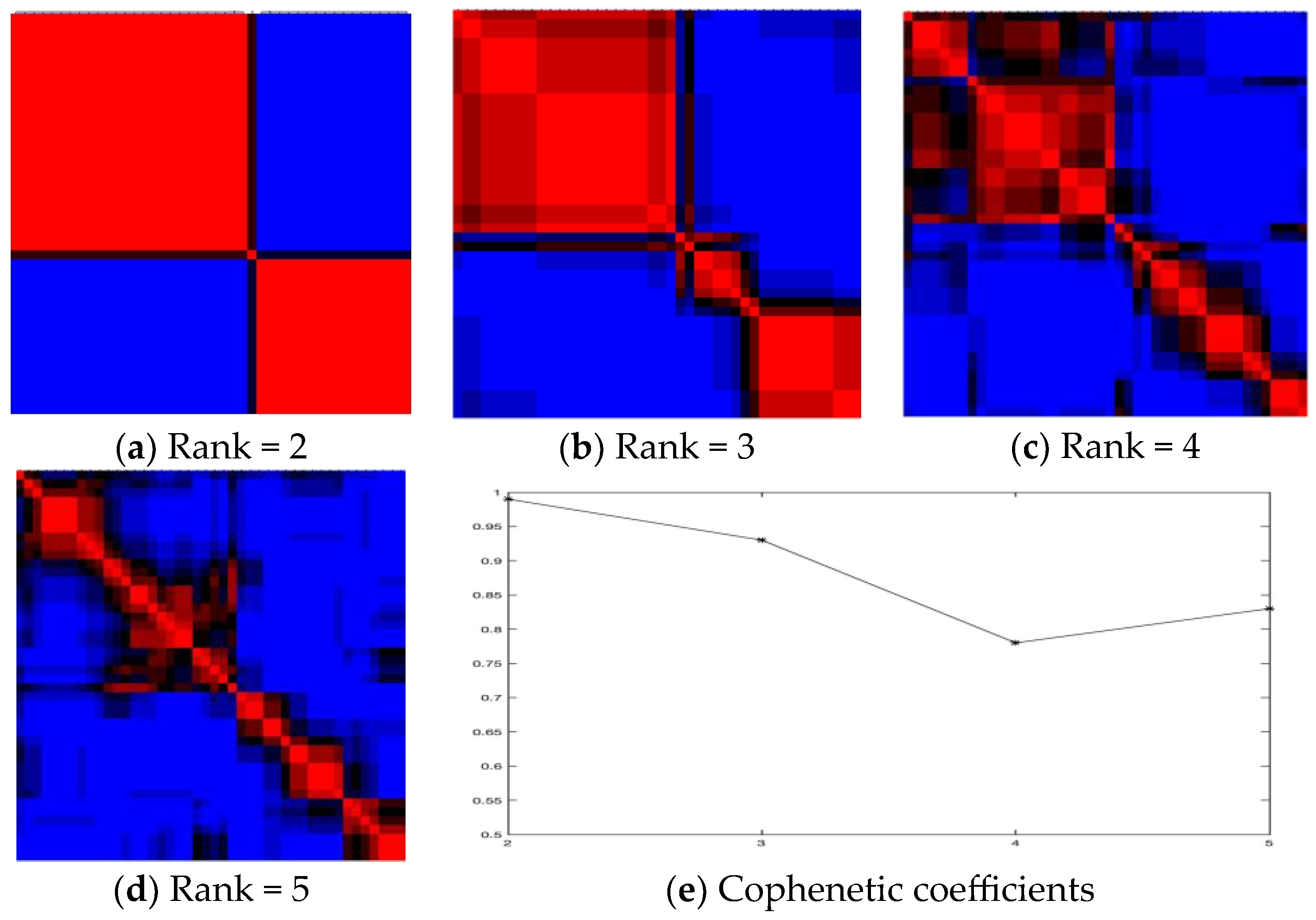
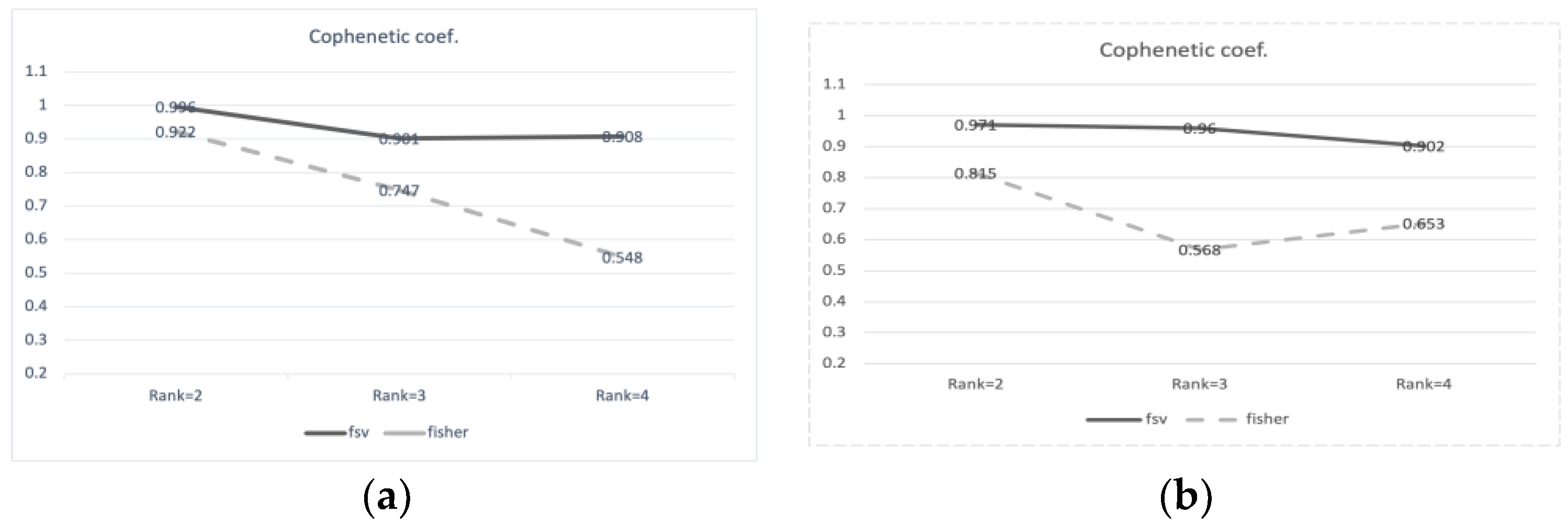
| Drugs in Cluster 1 | Drugs in Cluster 2 |
|---|---|
| LY-294002 | fulvestrant |
| Rosiglitazone | genistein |
| Troglitazone | alpha-estradiol |
| 17-allylamino-geldanamycin | monastrol |
| Valproic acid | mercaptopurine |
| Sodium phenylbutyrate | butirosin |
| Novobiocin | estradiol |
| Fasudil | docosahexaenoicacidethylester |
| Diclofenac | U0125 |
| 15-deltaprostaglandin J2 | resveratrol |
| Tretinoin | splitomicin |
| Trichostatin A | dimethyloxalylglycine |
| Monorden | HNMPA-(AM)3 |
| TTNPB | butein |
| indomethacin | fisetin |
| Tetraethylenepentamine | 4,5-dianilinophthalimide |
| Rofecoxib | deferoxamine |
| Copper sulfate | |
| Celecoxib | |
| Imatinib | |
| Pirinixic acid |
| (A) | ||
| Cluster 1 | Cluster 2 | |
| HSA00531_GLYCOSAMINOGLYCAN_DEGRADATION | HSA00040_PENTOSE_AND_GLUCURONATE_INTERCONVERSIONS | |
| HSA00910_NITROGEN_METABOLISM | HSA03050_PROTEASOME | |
| HSA04950_MATURITY_ONSET_DIABETES_OF_THE_YOUNG | HSA03010_RIBOSOME | |
| HSA00565_ETHER_LIPID_METABOLISM | HSA03060_PROTEIN_EXPORT | |
| HSA04080_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | HSA00680_METHANE_METABOLISM | |
| HSA04742_TASTE_TRANSDUCTION | HSA00062_FATTY_ACID_ELONGATION_IN_MITOCHONDRIA | |
| (B) | ||
| Cluster 1 | Cluster 2 | Cluster 3 |
| HSA00563_GLYCOSYLPHOSPHATIDYLINOSITOL_ANCHOR_BIOSYNTHESIS | HSA00602_GLYCOSPHINGOLIPID_BIOSYNTHESIS_NEO_LACTOSERIES | HSA03010_RIBOSOME |
| HSA00531_GLYCOSAMINOGLYCAN_DEGRADATION | HSA04060_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | HSA00040_PENTOSE_AND_GLUCURONATE_INTERCONVERSIONS |
| HSA00910_NITROGEN_METABOLISM | HSA00430_TAURINE_AND_HYPOTAURINE_METABOLISM | HSA03060_PROTEIN_EXPORT |
| HSA04614_RENIN_ANGIOTENSIN_SYSTEM | HSA04740_OLFACTORY_TRANSDUCTION | HSA03050_PROTEASOME |
| HSA00565_ETHER_LIPID_METABOLISM | HSA04940_TYPE_I_DIABETES_MELLITUS | HSA00720_REDUCTIVE_CARBOXYLATE_CYCLE |
| (A) | |||||
| TPT1 | COX6A1 | ACTB | ACTB | CCDC72 | RPL7 |
| ACTG1(ACTB) | RPS2 (SNORA64) | RPL38 | GAPDH | S100A6 | EEF1A1 |
| RPS10 | HUWE1 | GAPDH | RPS18 | ACTB | FTHP1 |
| RPLP0 | HSPA1A | ACTG1 | ALDOA | RPL3 | ALDOA |
| RPL24 | RPL11 | ODC1 | UBC | ACTG1 | RPS24 |
| (B) | |||||
| RPL11 | CCNT1 | EIF4H | UBE2L6 | SLC36A1 | RPL23 |
| MUC6 | KLHL24 | RPL3 | TNXA | RPL30 | CAMLG |
| 216138_at * | DES | HADHA | ANKRD1 | FGF16 | POLG2 |
| 207756_at * | TBCD | RPL9 | NEBL | HTR5A | UBB |
| RPS18 | RPL32 | 217709_at * | LRP2BP | RPLP0 | EEF1DP5 |
| Overlapping Drugs in Cluster 1 | Overlapping Drugs in Cluster 2 | ||
|---|---|---|---|
| 17-allylamino-geldanamycin (2) | LY-294002 (2) | copper sulfate | mercaptopurine |
| 15-delta prostaglandin J2 | rosiglitazone | deferoxamine | dimethyloxalylglycine |
| troglitazone (2) | rofecoxib | splitomicin | |
| monorden (PGSC0003) (2) | valproic acid (2) | pirinixic acid (2) | alpha-estradiol |
| celecoxib | sodium phenylbutyrate | monastrol | genistein |
| novobiocin | 4,5-dianilinophthalimide | fulvestrant | |
| indomethacin | fasudil | resveratrol | |
| Docosahexaenoic acid ethyl ester | diclofenac | fisetin | U0125 |
| tretinoin | butein | estradiol | |
| tetraethylenepentamine | HNMPA-(AM)3 TTNPB | butirosin imatinib | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S. Inferring Drug Set and Identifying the Mechanism of Drugs for PC3. Int. J. Mol. Sci. 2024, 25, 765. https://doi.org/10.3390/ijms25020765
Kim S. Inferring Drug Set and Identifying the Mechanism of Drugs for PC3. International Journal of Molecular Sciences. 2024; 25(2):765. https://doi.org/10.3390/ijms25020765
Chicago/Turabian StyleKim, Shinuk. 2024. "Inferring Drug Set and Identifying the Mechanism of Drugs for PC3" International Journal of Molecular Sciences 25, no. 2: 765. https://doi.org/10.3390/ijms25020765
APA StyleKim, S. (2024). Inferring Drug Set and Identifying the Mechanism of Drugs for PC3. International Journal of Molecular Sciences, 25(2), 765. https://doi.org/10.3390/ijms25020765






