Genome-Wide Analysis of R2R3-MYB Genes and Functional Characterization of SmMYB75 in Eggplant Fruit Implications for Crop Improvement and Nutritional Enhancement
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Property Analysis of the SmR2R3-MYB Genes
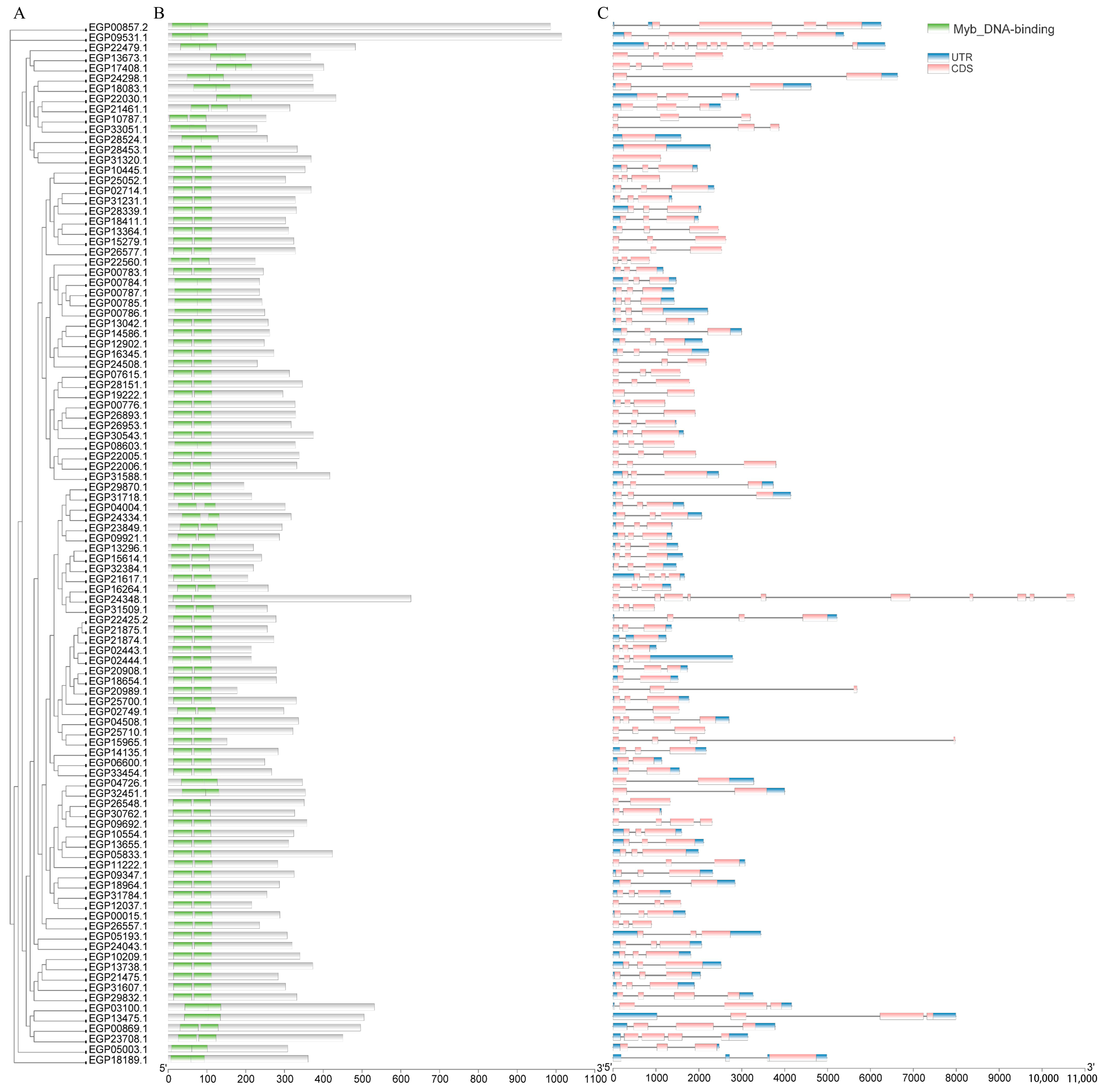
2.2. Phylogenetic Analysis and Classification of the SmR2R3-MYB Gene Family
2.3. Motif, Conserved Domain, and Structure Analyses of SmR2R3-MYBs
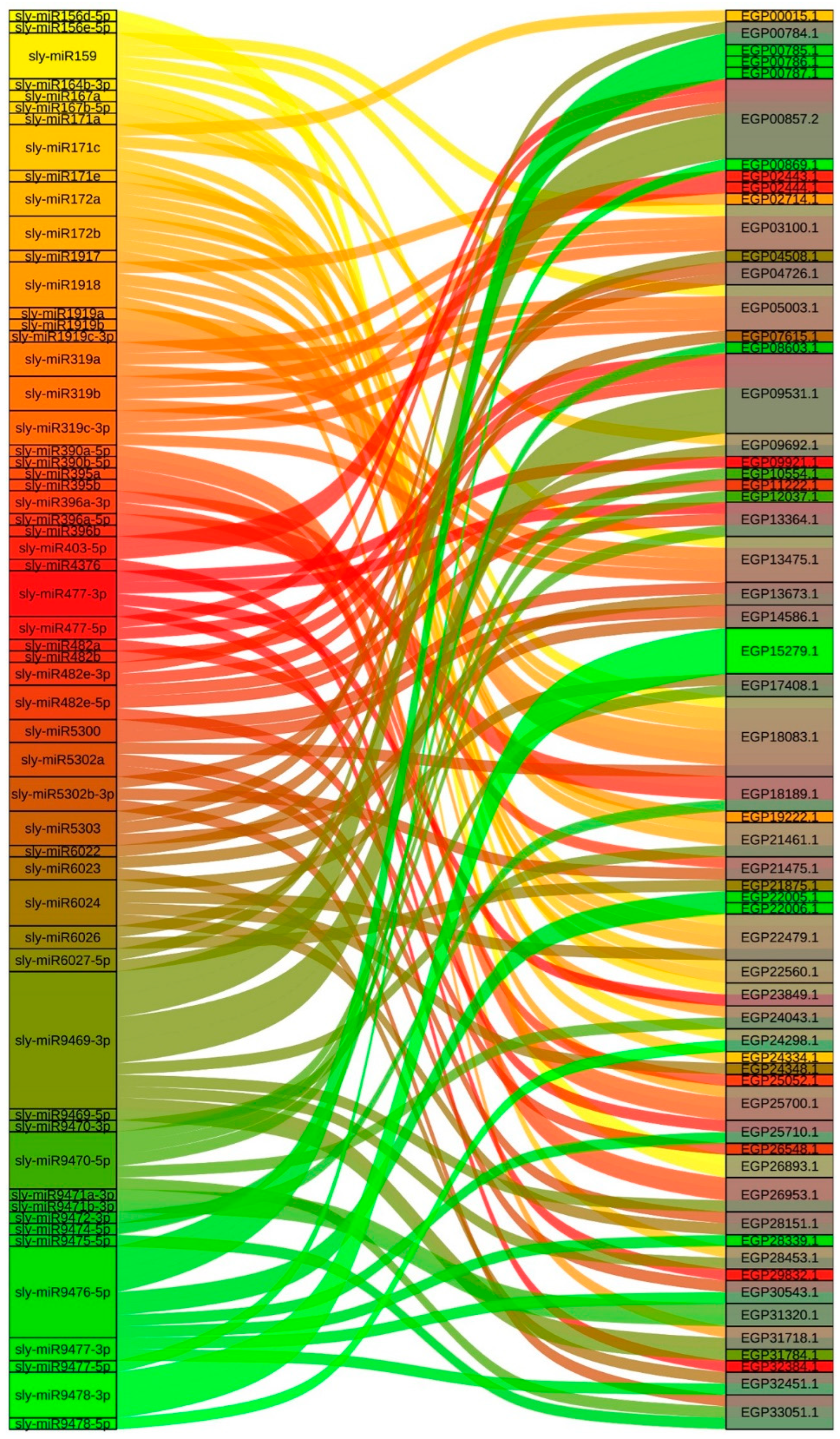
2.4. miRNAs That Target Regulate SmR2R3-MYB Genes
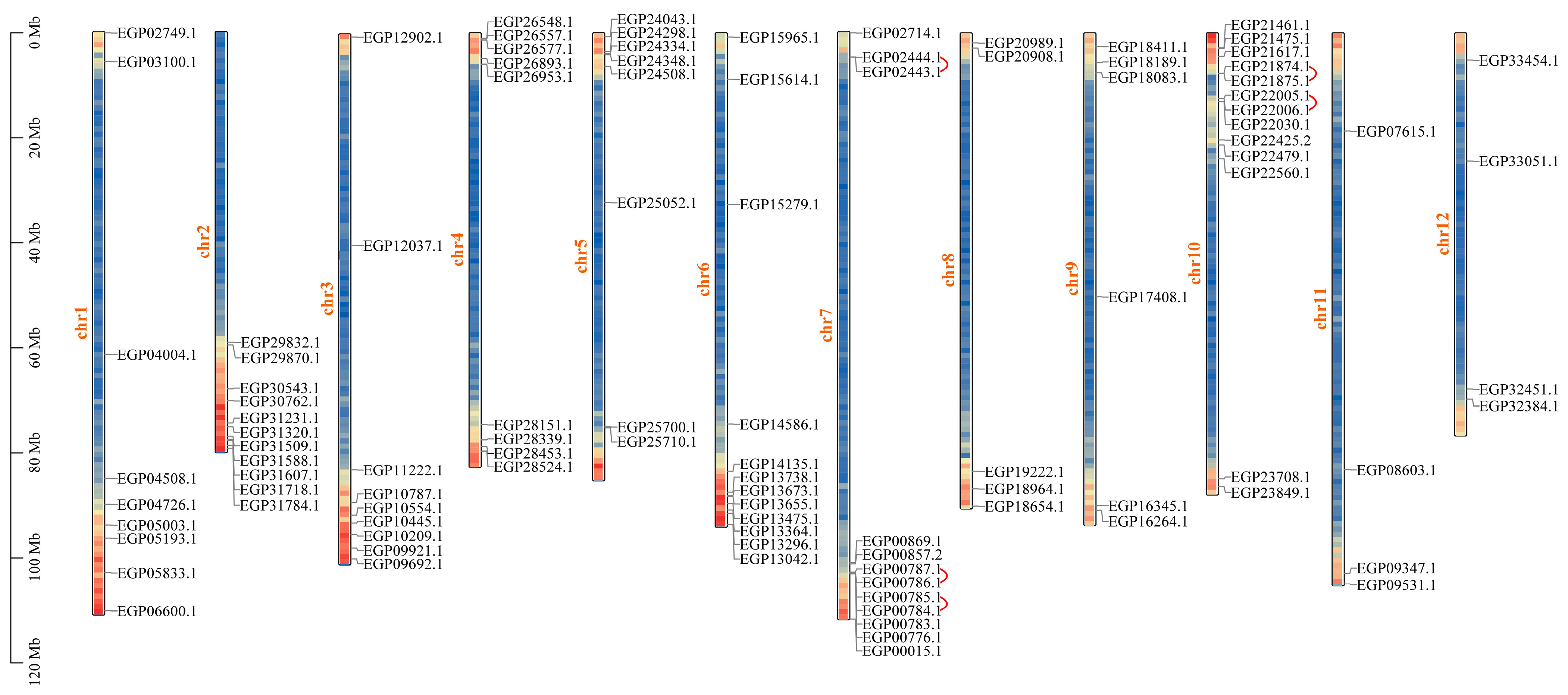
2.5. Chromosomal Distribution and Repeat Events of R2R3-MYB Genes in Eggplant Fruit
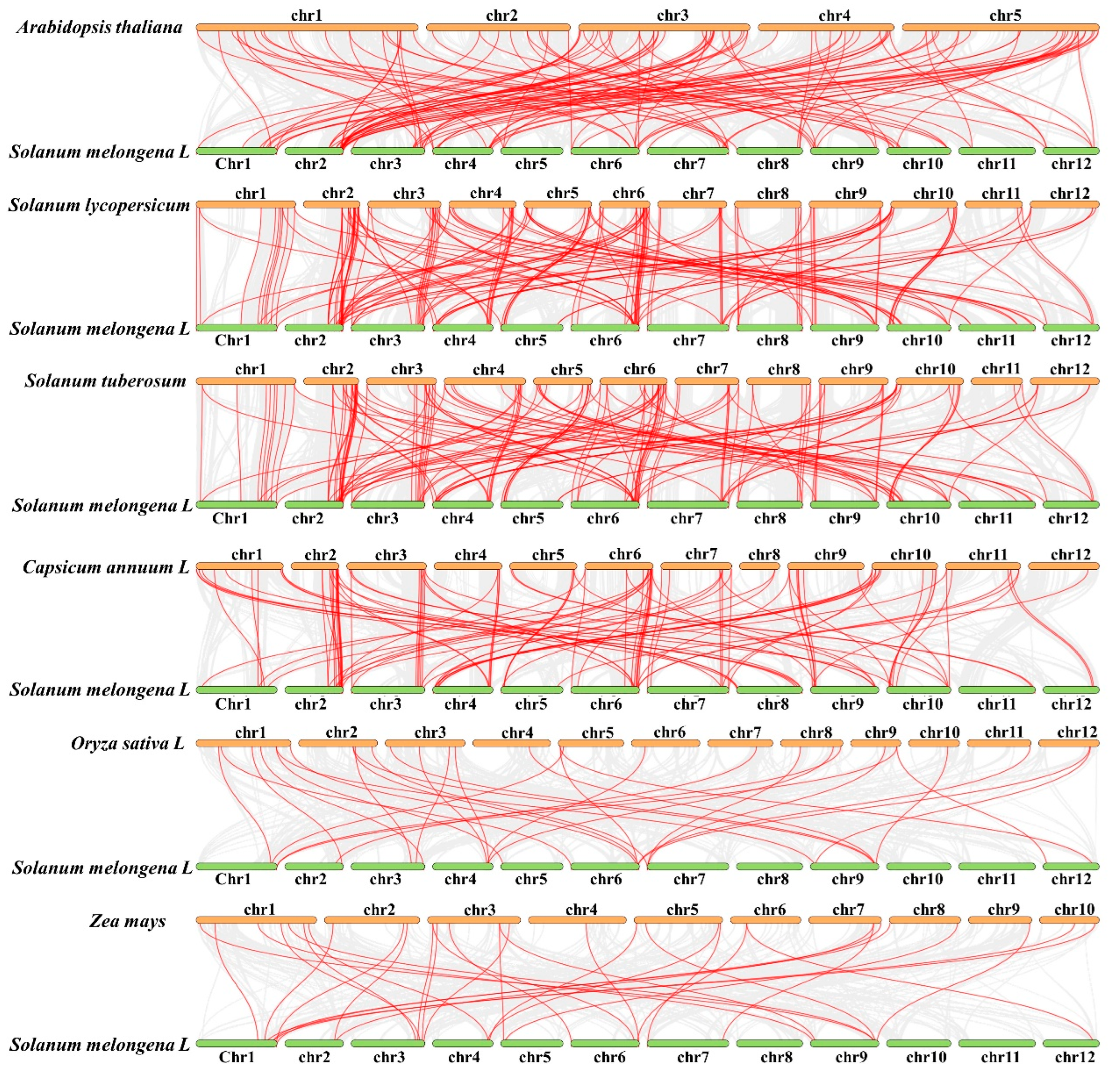
2.6. Comparative Synteny Analysis of R2R3-MYB Genes between Eggplants and Other Species
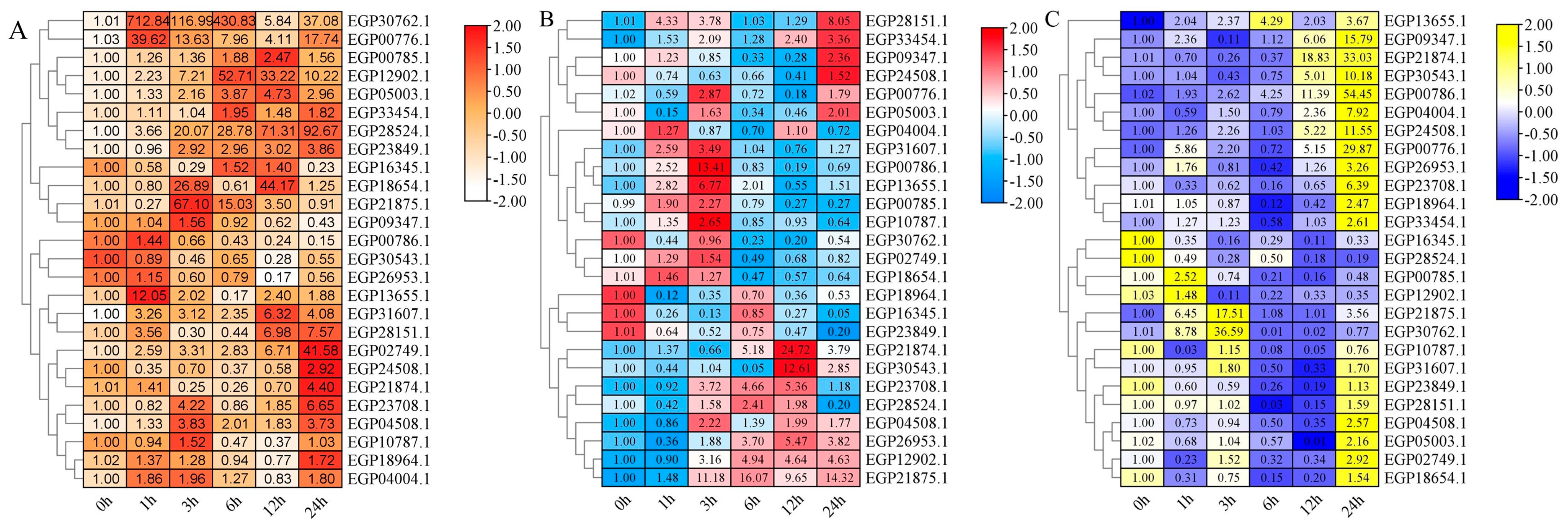
2.7. Expression Pattern of SmR2R3-MYBs under Diverse Treatments
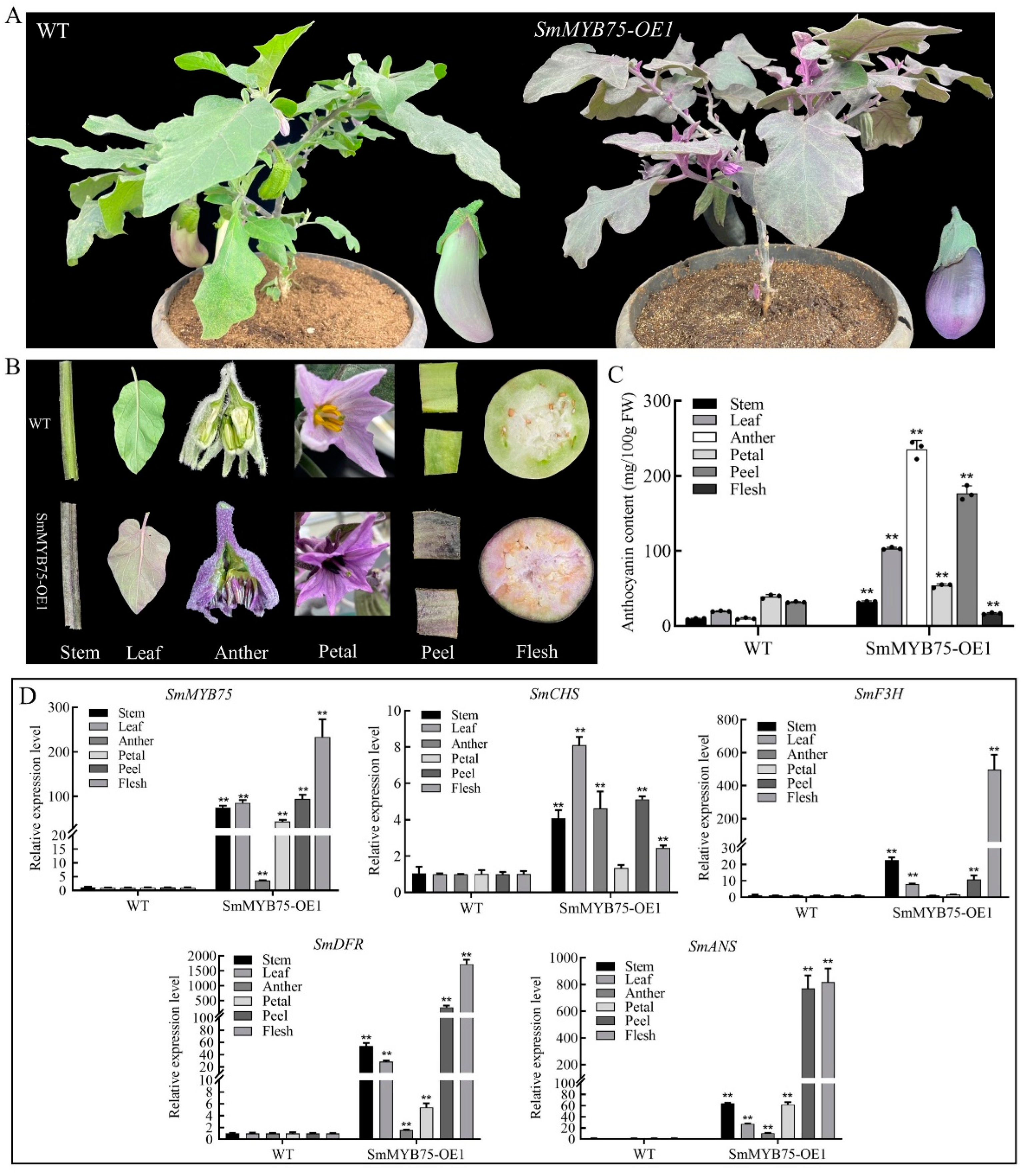
2.8. Generation and Characterization of SmMYB75-OE Transgenic Eggplant Fruit
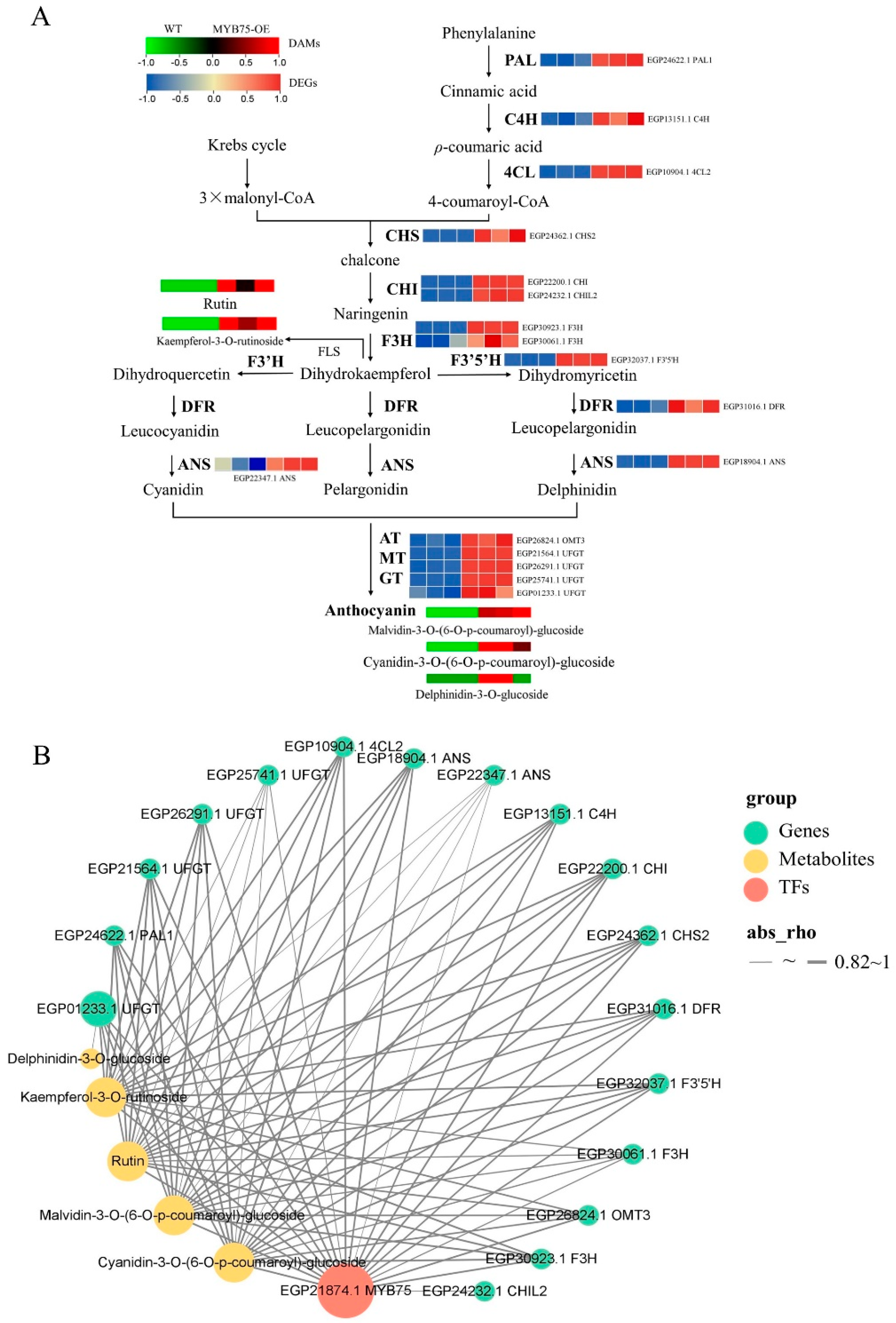
2.9. Anthocyanin-Targeted Metabolomic and Transcriptomic Analysis of Eggplant Flesh
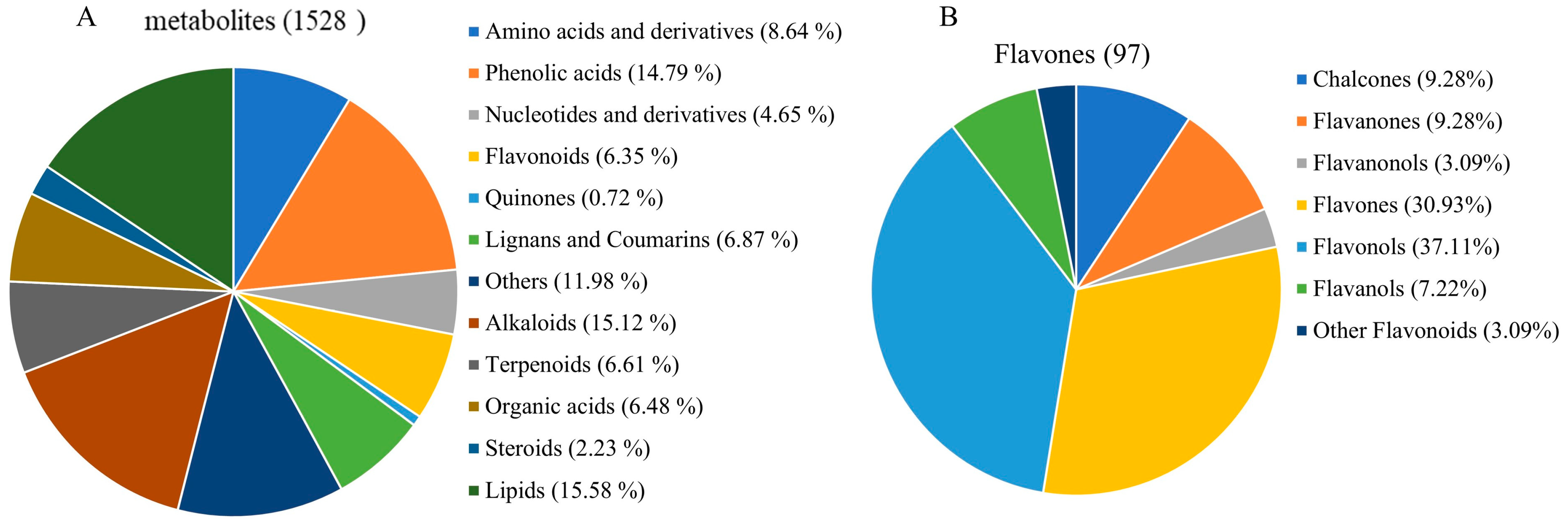
2.10. Comprehensive Targeted Metabolic Profiling
3. Discussion
4. Materials and Methods
4.1. Identification and Classification of R2R3-MYB Genes in Eggplants
4.2. Phylogenetic Tree Analysis and Classification of R2R3-MYB Genes in Eggplants
4.3. Gene Structure, Conserved Domain, and Motif Analysis
4.4. miRNA-R2R3-MYBs Prediction
4.5. Chromosomal Localization, Gene Duplication, and Syntenic Analysis
4.6. Expression Profiles of Abiotic Stress-Responsive Results and qRT-PCR
4.7. Vector Construction and Plant Transformation
4.8. RNA-Seq Analysis
4.9. Extraction and Measurement of Total Anthocyanins
4.10. Metabolite Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD Repeat (MBW) Pigment Regulatory Model. Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, B.; Yang, T.; Zhang, J.; Liu, B.; Zhan, X.; Liang, Y. The GAMYB-like gene SlMYB33 mediates flowering and pollen development in tomato. Hortic. Res. 2020, 7, 133. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.; Yu, W.; Yao, H.; Zhou, P.; Allan, A.C.; Zeng, L. NtMYB3, an R2R3-MYB from Narcissus.; Regulates Flavonoid Biosynthesis. Int. J. Mol. Sci. 2019, 20, 5456. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ma, H.; Duan, X.; Gao, S.; Zhou, X.; Cheng, Y. The R2R3-MYB gene PsMYB58 positively regulates anthocyanin biosynthesis in tree peony flowers. Plant Physiol. Biochem. 2021, 164, 279–288. [Google Scholar] [CrossRef]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef]
- Fang, Q.; Jiang, T.; Xu, L.; Liu, H.; Mao, H.; Wang, X.; Jiao, B.; Duan, Y.; Wang, Q. A salt-stress-regulator from the Poplar R2R3 MYB family integrates the regulation of lateral root emergence and ABA signaling to mediate salt stress tolerance in Arabidopsis. Plant Physiol. Biochem. 2017, 114, 100–110. [Google Scholar] [CrossRef]
- Gao, F.; Zhou, J.; Deng, R.Y.; Zhao, H.X.; Li, C.L.; Chen, H.; Suzuki, T.; Park, S.U.; Wu, Q. Overexpression of a tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis. J. Plant Physiol. 2017, 214, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, Z.; Ding, Y.; Liu, L.; Han, X.; Zhan, J.; Wei, X.; Diao, Y.; Qin, W. Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 2019, 286, 28–36. [Google Scholar] [CrossRef]
- Song, Y.; Yang, W.; Fan, H.; Zhang, X.; Sui, N. TaMYB86B encodes a R2R3-type MYB transcription factor and enhances salt tolerance in wheat. Plant Sci. 2020, 300, 110624. [Google Scholar] [CrossRef]
- Fan, H.; Cui, M.; Li, N.; Li, X.; Liang, Y.; Liu, L.; Cai, Y.; Lin, Y. Genome-wide identification and expression analyses of R2R3-MYB transcription factor genes from two Orchid species. PeerJ 2020, 8, e9781. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; He, Y.; Li, S.; Shi, S.; Li, L.; Liu, Y.; Chen, H. Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena L.). Plant Physiol. Biochem. 2021, 167, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Liu, J.Y.; Zhou, M.Q.; Li, H.Z. Chromosomal Localization of Anthocyanin Biosynthetic Genes bz1.; bz2 in Lotus. J. Wuhan Bot. Res. 2004, 22, 380–384. [Google Scholar]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Zong, Y.; Li, S.; Xi, X.; Cao, D.; Liu, B. Comprehensive Influences of Overexpression of a MYB Transcriptor Regulating Anthocyanin Biosynthesis on Transcriptome and Metabolome of Tobacco Leaves. Int. J. Mol. Sci. 2019, 20, 5123. [Google Scholar] [CrossRef]
- Xu, H.; Zou, Q.; Yang, G.; Jiang, S.; Chen, X. MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic. Res. 2020, 7, 17. [Google Scholar] [CrossRef]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant. Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Martin, C.; PazAres, J. MYB transcription factors in plants. Trends Genet. 1997, 13, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef]
- Zhao, M. Regulation of Arabidopsis trichome Patterning and Anthocyanin Biosynthesis by the TTG1-bHLH-MYB Complex; The University of Texas at Austin: Austin, TX, USA, 2007. [Google Scholar]
- Panaud, O.; Jackson, S.A.; Wendel, J.F. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 32–144. [Google Scholar]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.; Koes, R.; Perata, P. Tomato R2R3-MYB Proteins SlANT1 and SlAN2. Same Protein Activity.; Different Roles. PLoS ONE 2015, 10, e0136365. [Google Scholar] [CrossRef] [PubMed]
- An, X.H.; Tian, Y.; Chen, K.Q.; Liu, X.J.; Liu, D.D.; Xie, X.B.; Cheng, C.G.; Cong, P.H.; Hao, Y.J. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef]
- Raigón, M.D.; Prohens, J.; Muñoz-Falcón, J.E.; Nuez, F. Comparison of eggplant landraces and commercial varieties for fruit content of phenolics, minerals, dry matter and protein. J. Food Compos. Anal. 2008, 21, 370–376. [Google Scholar] [CrossRef]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 22. [Google Scholar] [CrossRef]
- Jiang, M.; Liu, Y.; Ren, L.; Lian, H.; Chen, H. Molecular cloning and characterization of anthocyanin biosynthesis genes in eggplant (Solanum melongena L.). Acta Physiol. Plant. 2016, 38, 1–13. [Google Scholar] [CrossRef]
- Jiang, M.; Ren, L.; Lian, H.; Liu, Y.; Chen, H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016, 249, 46–58. [Google Scholar] [CrossRef]
- Shao, W.T.; Liu, Y.; Han, H.Q.; Chen, H.Y. Cloning and Expression Analysis of an Anthocyanin-related Transcription Factor Gene SmMYB in Eggplant. Acta Hortic. Sin. 2013, 40, 467–478. [Google Scholar]
- Zhang, Y.; Hu, Z.; Chu, G.; Huang, C.; Tian, S.; Zhao, Z.; Chen, G. Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (Solanum melongena L.). J. Agric. Food Chem. 2014, 62, 2906–2912. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Liu, Y.; He, Y.; Li, L.; Li, D.; Chen, H. R2R3-MYB transcription factor SmMYB75 promotes anthocyanin biosynthesis in eggplant (Solanum melongena L.). Sci. Hortic. 2021, 282, 110020. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Ge, H.; Shi, S.; Li, D.; Liu, Y.; Chen, H. A light-responsive transcription factor SmMYB35 enhances anthocyanin biosynthesis in eggplant (Solanum melongena L.). Planta 2021, 255, 12. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, Y.; Ge, H.; Liu, Y.; Chen, H. Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Sci. 2021, 302, 110696. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Zhao, P.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol. Genet. Genom. 2014, 289, 1183–1207. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.; Tang, B.; Dai, X.; Zou, X. Genome-Wide Identification and Capsaicinoid Biosynthesis-Related Expression Analysis of the R2R3-MYB Gene Family in Capsicum annuum L. Front. Genet. 2020, 11, 598183. [Google Scholar] [CrossRef]
- Yl, A.; Lw, B.; Zhen, L.A.; Acab, C.; Sq, A.; Jz, A.; Yl, A. Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.)—ScienceDirect. Int. J. Biol. Macromol. 2020, 148, 817–832. [Google Scholar]
- Yang, J.; Zhang, B.; Gu, G.; Yuan, J.; Shen, S.; Jin, L.; Lin, Z.; Lin, J.; Xie, X. Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.). BMC Genom. 2022, 23, 432. [Google Scholar] [CrossRef]
- Li, D.; Qian, J.; Li, W.; Jiang, Y.; Gan, G.; Li, W.; Chen, R.; Yu, N.; Li, Y.; Wu, Y.; et al. Genome sequence and analysis of the eggplant (Solanum melongena L.). bioRxiv 2019. [Google Scholar] [CrossRef]
- Mehan, M.R.; Freimer, N.B.; Ophoff, R.A. A genome-wide survey of segmental duplications that mediate common human genetic variation of chromosomal architecture. Hum. Genom. 2004, 1, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Guo, C.; Shi, H.; Zhao, J.; Ma, F.; An, W.; He, X.; Luo, Q.; Cao, Y.; Zhan, X. Genome-Wide Comparative Analysis of the R2R3-MYB Gene Family in Five Solanaceae Species and Identification of Members Regulating Carotenoid Biosynthesis in Wolfberry. Int. J. Mol. Sci. 2022, 23, 2259. [Google Scholar] [CrossRef] [PubMed]
- Boter, M.; Golz, J.F.; Gimenez-Ibanez, S.; Fernandez-Barbero, G.; Franco-Zorrilla, J.M.; Solano, R. FILAMENTOUS FLOWER Is a Direct Target of JAZ3 and Modulates Responses to Jasmonate. Plant Cell 2015, 27, 3160–3174. [Google Scholar] [CrossRef]
- Pandey, A.; Misra, P.; Bhambhani, S.; Bhatia, C.; Trivedi, P.K. Expression of Arabidopsis MYB transcription factor, AtMYB111, in tobacco requires light to modulate flavonol content. Sci. Rep. 2014, 4, 5018. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.J.; Zheng, Q.D.; Yao, Y.H.; Ou, Y.; Chen, J.Y.; Wang, M.J.; Lai, H.P.; Yan, L.; Liu, Z.J.; Ai, Y. Genome-Wide Identification of the MYB Gene Family in Cymbidiumensifolium and Its Expression Analysis in Different Flower Colors. Int. J. Mol. Sci. 2021, 22, 13245. [Google Scholar] [CrossRef]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Li, D.; Tang, X.; Dong, Y.; Wang, Y.; Shi, S.; Li, S.; Liu, Y.; Ge, H.; Chen, H. Comparative genomic investigation of TCP gene family in eggplant (Solanum melongena L.) and expression analysis under divergent treatments. Plant Cell Rep. 2022, 41, 2213–2228. [Google Scholar] [CrossRef]
- Li, Z.; Liu, W.; Chen, Q.; Zhang, S.; Mei, Z.; Yu, L.; Wang, C.; Mao, Z.; Chen, Z. Mdm-miR858 targets MdMYB9 and MdMYBPA1 to participate anthocyanin biosynthesis in red-fleshed apple. Plant J. 2023, 113, 1295–1309. [Google Scholar] [CrossRef]
- Dias, A.P.; Braun, E.L.; McMullen, M.D.; Grotewold, E. Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol. 2003, 131, 610–620. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.-H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [PubMed]
- Lippold, F.; Sanchez, D.H.; Musialak, M.; Schlereth, A.; Scheible, W.-R.; Hincha, D.K.; Udvardi, M.K. AtMyb41 Regulates Transcriptional and Metabolic Responses to Osmotic Stress in Arabidopsis. Plant Physiol. 2009, 149, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Li, L.; Wei, M.; Li, J.; Yang, F. SmMYB113 Is a Key Transcription Factor Responsible for Compositional Variation of Anthocyanin and Color Diversity Among Eggplant Peels. Front. Plant Sci. 2022, 13, 843996. [Google Scholar] [PubMed]
- Ku, Y.S.; Ng, M.S.; Cheng, S.S.; Lo, A.W.; Xiao, Z.; Shin, T.S.; Chung, G.; Lam, H.M. Understanding the Composition, Biosynthesis, Accumulation and Transport of Flavonoids in Crops for the Promotion of Crops as Healthy Sources of Flavonoids for Human Consumption. Nutrients 2020, 12, 1717. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; He, Y.; Li, L.; Li, D.; Chen, H. New insights on the regulation of anthocyanin biosynthesis in purple Solanaceous fruit vegetables. Sci. Hortic. 2022, 297, 110917. [Google Scholar]
- He, Y.; Li, D.; Li, S.; Liu, Y.; Chen, H. SmBICs Inhibit Anthocyanin Biosynthesis in Eggplant (Solanum melongena L.). Plant Cell Physiol. 2021, 62, 1001–1011. [Google Scholar] [CrossRef]
- Shi, J.; Simal-Gandara, J.; Mei, J.; Ma, W.; Peng, Q.; Shi, Y.; Xu, Q.; Lin, Z.; Lv, H. Insight into the pigmented anthocyanins and the major potential co-pigmented flavonoids in purple-coloured leaf teas. Food Chem. 2021, 363, 130278. [Google Scholar]
- Li, D.; Qian, J.; Li, W.; Yu, N.; Gan, G.; Jiang, Y.; Li, W.; Liang, X.; Chen, R. A high-quality genome assembly of the eggplant provides insights into the molecular basis of disease resistance and chlorogenic acid synthesis. Mol. Ecol. Resour. 2021, 21, 1274–1286. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools. An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, 493–496. [Google Scholar]
- Lu, S.; Wang, J.; Farideh, C.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Marc, G.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget, a plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Xia, R. A painless way to customize Circos plot: From data preparation to visualization using TBtools. iMeta 2022, 1, e35. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods A Companion Methods Enzymol. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, H.; Ma, W.; Guo, Z.; Li, X.; Li, X.; Zhang, Q. Determination of patulin in apple juice by single-drop liquid-liquid-liquid microextraction coupled with liquid chromatography-mass spectrometry. Food Chem. 2018, 257, 1–6. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, J.; Zheng, S.; Wang, Z.; Xu, C.; Zhang, Q.; Wu, J.; Lou, H. Metabolite profiling and transcriptome analyses reveal novel regulatory mechanisms of melatonin biosynthesis in hickory. Hortic. Res. 2021, 8, 1–13. [Google Scholar] [CrossRef]
- Liu, C.; Chen, S.; Wang, S.; Zhao, X.; Li, K.; Chen, S.; Qu, G.Z. A genome wide transcriptional study of Populus alba × P. tremula var. glandulosa in response to nitrogen deficiency stress. Physiol. Mol. Biol. Plants 2021, 27, 1277–1293. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables. A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Zhang, S.; Ying, H.; Pingcuo, G.; Wang, S.; Zeng, X. Identification of Potential Metabolites Mediating Bird’s Selective Feeding on Prunus mira Flowers. BioMed Res. Int. 2019, 2019, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zeng, X.; Shi, J.; Xu, Q.; Wang, Y.; Jabu, D.; Sang, Z.; Nyima, T. Time-Course Comparative Metabolite Profiling under Osmotic Stress in Tolerant and Sensitive Tibetan Hulless Barley. Biomed Res. Int. 2018, 2018, 9415409. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, S.; Li, D.; Li, S.; Zhao, N.; Liao, J.; Ge, H.; Liu, Y.; Chen, H. Genome-Wide Analysis of R2R3-MYB Genes and Functional Characterization of SmMYB75 in Eggplant Fruit Implications for Crop Improvement and Nutritional Enhancement. Int. J. Mol. Sci. 2024, 25, 1163. https://doi.org/10.3390/ijms25021163
Shi S, Li D, Li S, Zhao N, Liao J, Ge H, Liu Y, Chen H. Genome-Wide Analysis of R2R3-MYB Genes and Functional Characterization of SmMYB75 in Eggplant Fruit Implications for Crop Improvement and Nutritional Enhancement. International Journal of Molecular Sciences. 2024; 25(2):1163. https://doi.org/10.3390/ijms25021163
Chicago/Turabian StyleShi, Suli, Dalu Li, Shaohang Li, Na Zhao, Jielei Liao, Haiyan Ge, Yang Liu, and Huoying Chen. 2024. "Genome-Wide Analysis of R2R3-MYB Genes and Functional Characterization of SmMYB75 in Eggplant Fruit Implications for Crop Improvement and Nutritional Enhancement" International Journal of Molecular Sciences 25, no. 2: 1163. https://doi.org/10.3390/ijms25021163
APA StyleShi, S., Li, D., Li, S., Zhao, N., Liao, J., Ge, H., Liu, Y., & Chen, H. (2024). Genome-Wide Analysis of R2R3-MYB Genes and Functional Characterization of SmMYB75 in Eggplant Fruit Implications for Crop Improvement and Nutritional Enhancement. International Journal of Molecular Sciences, 25(2), 1163. https://doi.org/10.3390/ijms25021163




