Disruption of the Arabidopsis Acyl-Activating Enzyme 3 Impairs Seed Coat Mucilage Accumulation and Seed Germination
Abstract
1. Introduction
2. Results and Discussion
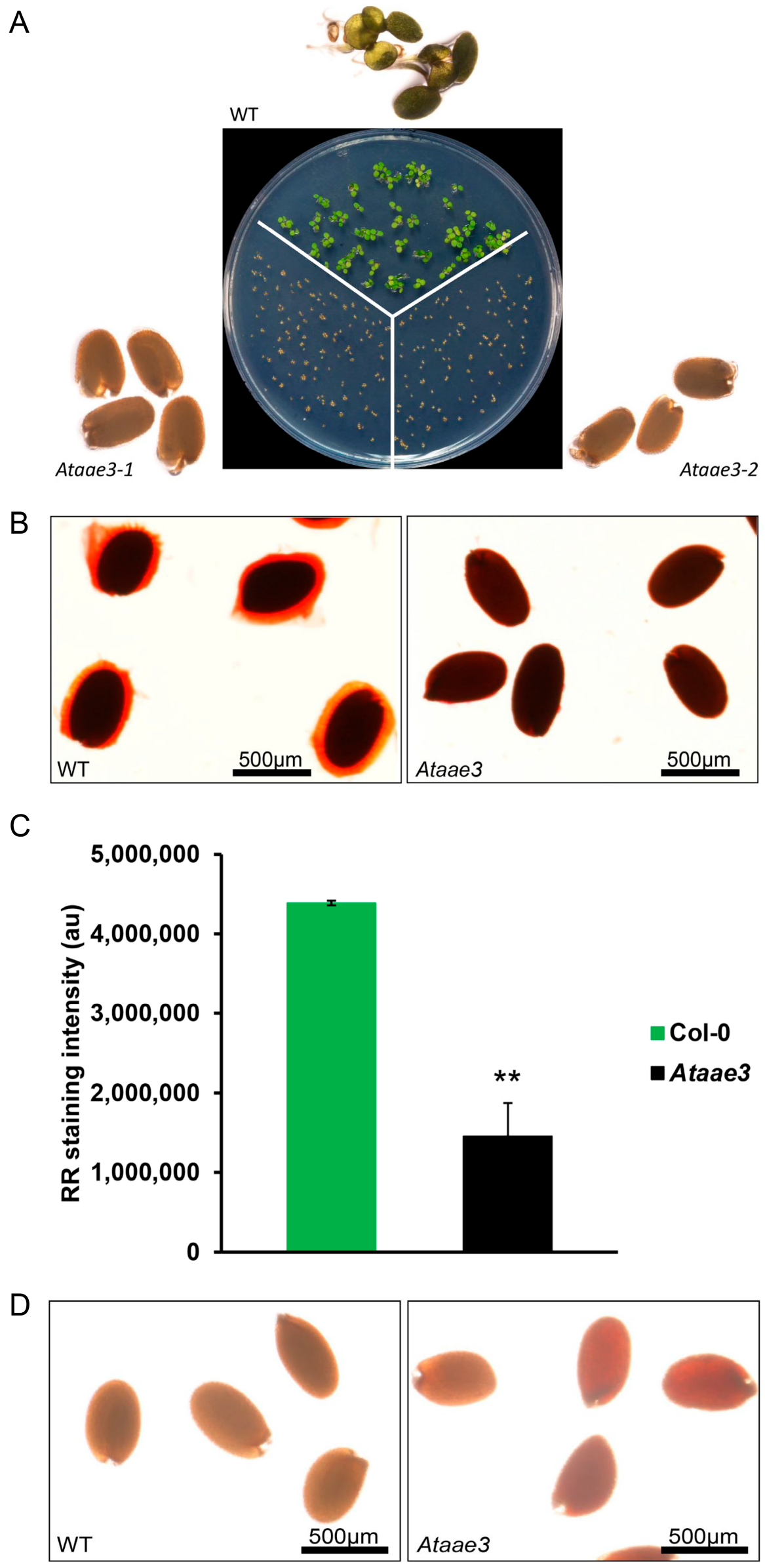
2.1. Disruption of AAE3 Reduces Seed Germination and Mucilage Accumulation
2.2. Disruption of AAE3 Reduces Mucilage-Related Gene Expression
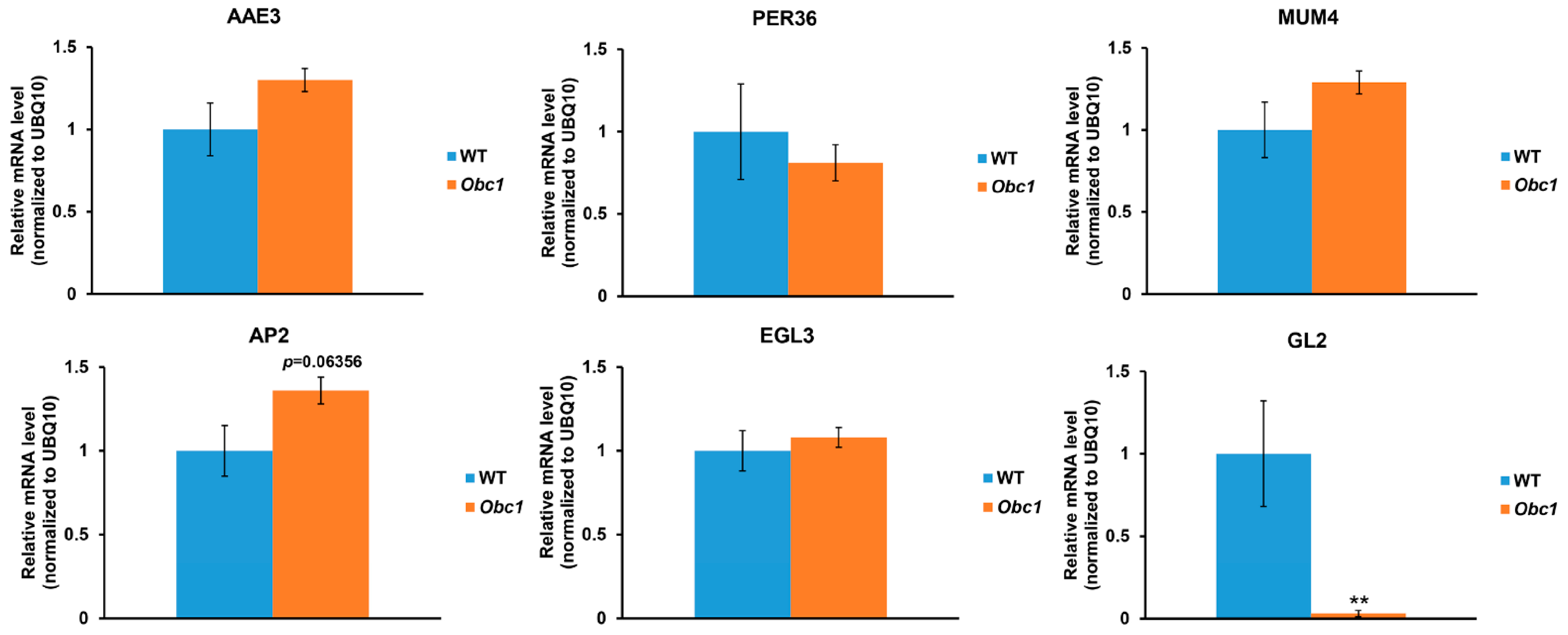
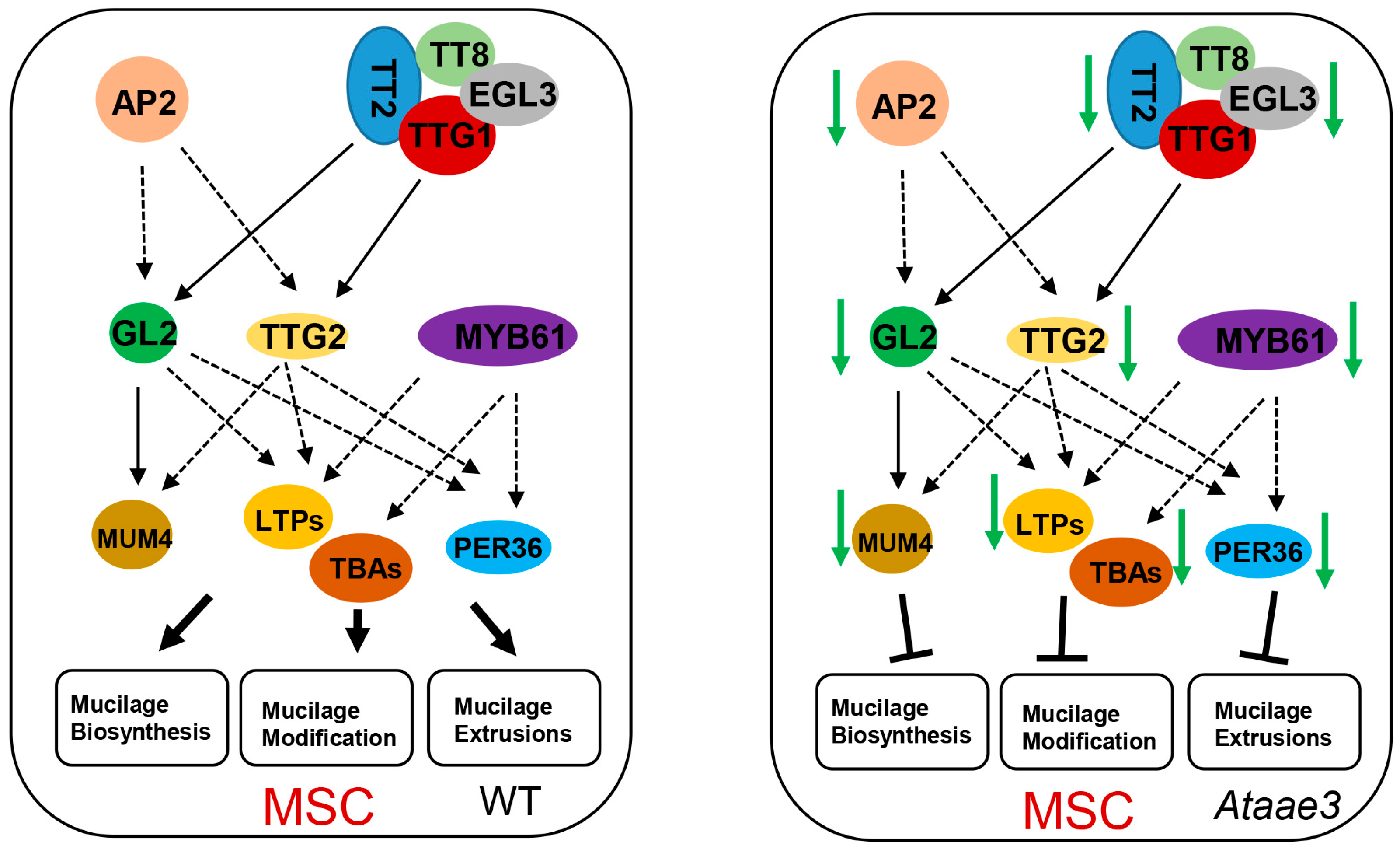
2.3. Disruption of AAE3 Reduces Mucilage Regulatory Gene Expression
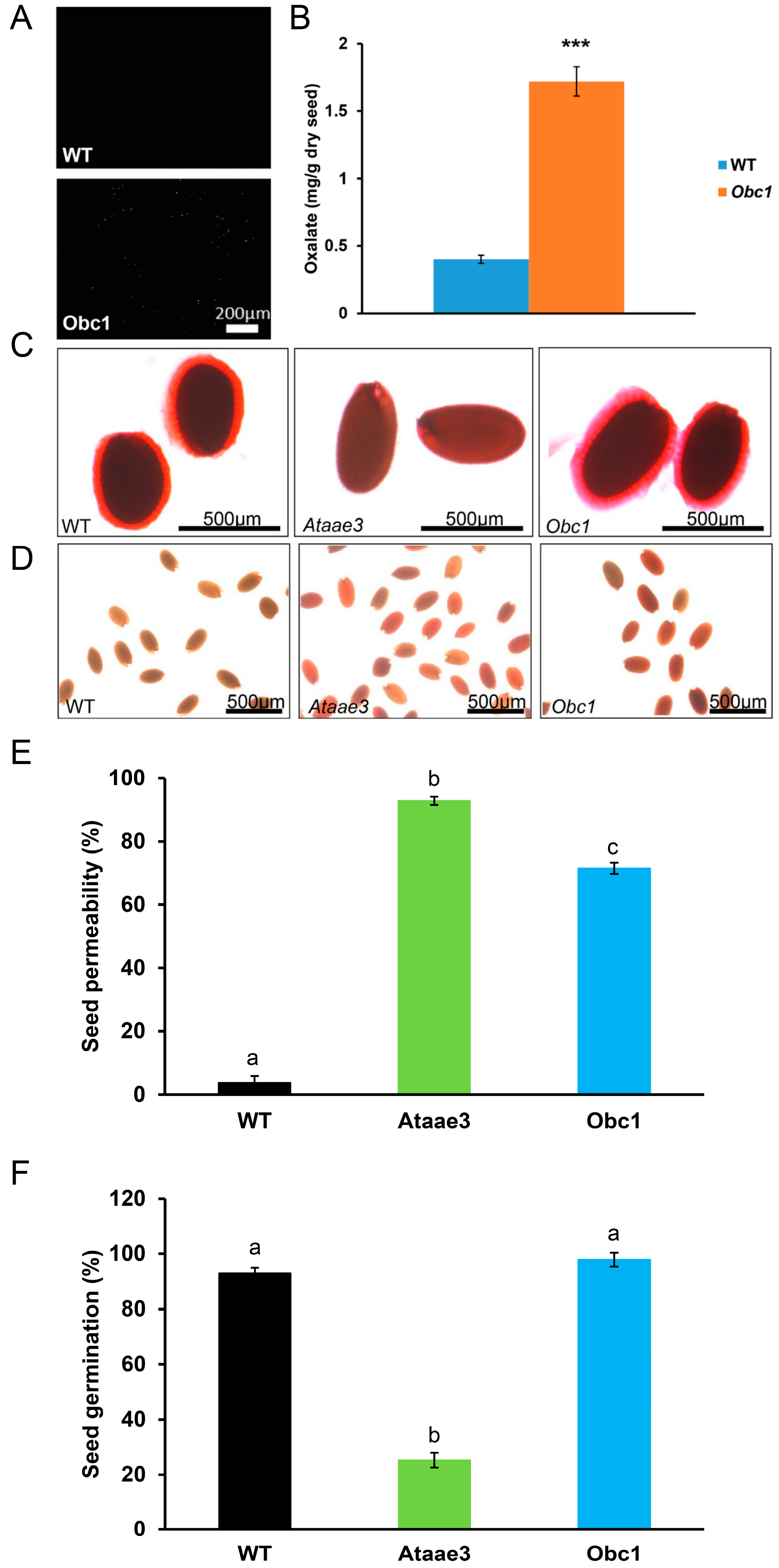
2.4. An Increase in Oxalate Accumulation Affects Seed Coat Development, but Not Mucilage Accumulation
2.5. AAE3 Is Critical for Seed Mucilage Accumulation
3. Materials and Methods
3.1. Reagents
3.2. Plant Materials and Growth Conditions
3.3. Dry Seed Mucilage Extraction and Measurement
3.4. Analysis of Seed Phenotypes
3.5. Microscopic Analysis of Calcium Oxalate Crystal in Arabidopsis Leaves
3.6. Oxalate Measurements
3.7. RNA Isolation, cDNA Synthesis, and qRT-PCR Analysis
3.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Western, T.L. The sticky tale of seed coat mucilages: Production, genetics, and role in seed germination and dispersal. Seed Sci. Res. 2012, 22, 1–25. [Google Scholar] [CrossRef]
- Western, T.L.; Skinner, D.J.; Haughn, G.W. Differentiation of Mucilage Secretory Cells of the Arabidopsis Seed Coat1. Plant Physiol. 2000, 122, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Beeckman, T.; De Rycke, R.; Viane, R.; Inzé, D. Histological Study of Seed Coat Development in Arabidopsis thaliana. J. Plant Res. 2000, 113, 139–148. [Google Scholar] [CrossRef]
- Haughn, G.; Western, T. Arabidopsis Seed Coat Mucilage is a Specialized Cell Wall that Can be Used as a Model for Genetic Analysis of Plant Cell Wall Structure and Function. Front. Plant Sci. 2012, 3, 64. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.Y.-L.; Kunieda, T.; Rogalski, J.; Foster, L.J.; Ellis, B.E.; Haughn, G.W. Identification and Characterization of Arabidopsis Seed Coat Mucilage Proteins. Plant Physiol. 2016, 173, 1059–1074. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, J.S.; North, H.M. Sticking to cellulose: Exploiting Arabidopsis seed coat mucilage to understand cellulose biosynthesis and cell wall polysaccharide interactions. New Phytol. 2017, 214, 959–966. [Google Scholar] [CrossRef]
- Phan, J.L.; Burton, R.A. New Insights into the Composition and Structure of Seed Mucilage. Annu. Plant Rev. Online 2018, 1, 63–104. [Google Scholar] [CrossRef]
- Šola, K.; Dean, G.H.; Haughn, G.W. Arabidopsis Seed Mucilage: A Specialised Extracellular Matrix that Demonstrates the Structure–Function Versatility of Cell Wall Polysaccharides. Annu. Plant Rev. Online 2019, 2, 1085–1116. [Google Scholar] [CrossRef]
- Tsai, A.Y.-L.; McGee, R.; Dean, G.H.; Haughn, G.W.; Sawa, S. Seed Mucilage: Biological Functions and Potential Applications in Biotechnology. Plant Cell Physiol. 2021, 62, 1847–1857. [Google Scholar] [CrossRef]
- Yang, X.; Baskin, J.M.; Baskin, C.C.; Huang, Z. More than just a coating: Ecological importance, taxonomic occurrence and phylogenetic relationships of seed coat mucilage. Perspect. Plant Ecol. Evol. Syst. 2012, 14, 434–442. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, R.; Li, S. Regulation of seed coat mucilage production and modification in Arabidopsis. Plant Sci. 2023, 328, 111591. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.Y.-L.; Higaki, T.; Nguyen, C.-N.; Perfus-Barbeoch, L.; Favery, B.; Sawa, S. Regulation of Root-Knot Nematode Behavior by Seed-Coat Mucilage-Derived Attractants. Mol. Plant 2019, 12, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.Y.-L.; Iwamoto, Y.; Tsumuraya, Y.; Oota, M.; Konishi, T.; Ito, S.; Kotake, T.; Ishikawa, H.; Sawa, S. Root-knot nematode chemotaxis is positively regulated by L-galactose sidechains of mucilage carbohydrate rhamnogalacturonan-I. Sci. Adv. 2021, 7, eabh4182. [Google Scholar] [CrossRef] [PubMed]
- Western, T.L.; Burn, J.; Tan, W.L.; Skinner, D.J.; Martin-McCaffrey, L.; Moffatt, B.A.; Haughn, G.W. Isolation and Characterization of Mutants Defective in Seed Coat Mucilage Secretory Cell Development in Arabidopsis. Plant Physiol. 2001, 127, 998–1011. [Google Scholar] [CrossRef]
- Haughn, G.; Chaudhury, A. Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci. 2005, 10, 472–477. [Google Scholar] [CrossRef]
- Francoz, E.; Ranocha, P.; Burlat, V.; Dunand, C. Arabidopsis seed mucilage secretory cells: Regulation and dynamics. Trends Plant Sci. 2015, 20, 515–524. [Google Scholar] [CrossRef]
- Golz, J.F.; Allen, P.J.; Li, S.F.; Parish, R.W.; Jayawardana, N.U.; Bacic, A.; Doblin, M.S. Layers of regulation—Insights into the role of transcription factors controlling mucilage production in the Arabidopsis seed coat. Plant Sci. 2018, 272, 179–192. [Google Scholar] [CrossRef]
- Radchuk, V.; Borisjuk, L. Physical, metabolic and developmental functions of the seed coat. Front. Plant Sci. 2014, 5, 510. [Google Scholar] [CrossRef]
- Li, P.; Liu, C.; Luo, Y.; Shi, H.; Li, Q.; PinChu, C.; Li, X.; Yang, J.; Fan, W. Oxalate in Plants: Metabolism, Function, Regulation, and Application. J. Agric. Food Chem. 2022, 70, 16037–16049. [Google Scholar] [CrossRef]
- Ilarslan, H.; Palmer, R.G.; Horner, H.T. Calcium Oxalate Crystals in Developing Seeds of Soybean. Ann. Bot. 2001, 88, 243–257. [Google Scholar] [CrossRef]
- Shockey, J.M.; Fulda, M.S.; Browse, J. Arabidopsis Contains a Large Superfamily of Acyl-Activating Enzymes. Phylogenetic and Biochemical Analysis Reveals a New Class of Acyl-Coenzyme A Synthetases. Plant Physiol. 2003, 132, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.; Kim, H.U.; Nakata, P.A.; Browse, J. A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell 2012, 24, 1217–1229. [Google Scholar] [CrossRef]
- Foster, J.; Luo, B.; Nakata, P.A. An Oxalyl-CoA Dependent Pathway of Oxalate Catabolism Plays a Role in Regulating Calcium Oxalate Crystal Accumulation and Defending against Oxalate-Secreting Phytopathogens in Medicago truncatula. PLoS ONE 2016, 11, e0149850. [Google Scholar] [CrossRef]
- Cheng, N.; Foster, J.; Mysore, K.S.; Wen, J.; Rao, X.; Nakata, P.A. Effect of Acyl Activating Enzyme (AAE) 3 on the growth and development of Medicago truncatula. Biochem. Biophys. Res. Commun. 2018, 505, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.Q.; Fan, W.; Xu, J.M.; Gong, Y.L.; Jin, J.F.; Chen, W.W.; Liu, L.Y.; Hai, M.R.; Yang, J.L.; Zheng, S.J. An Oxalyl-CoA Synthetase Is Involved in Oxalate Degradation and Aluminum Tolerance. Plant Physiol. 2016, 172, 1679–1690. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Liang, X.; Liu, E.E.; Zhang, J.J.; Peng, X.X. The oxalyl-CoA synthetase-regulated oxalate and its distinct effects on resistance to bacterial blight and aluminium toxicity in rice. Plant Biol. 2017, 19, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Fan, W.; Lou, H.Q.; Yang, J.L.; Zheng, S.J. Regulating cytoplasmic oxalate homeostasis by Acyl activating enzyme3 is critical for plant Al tolerance. Plant Signal. Behav. 2017, 12, e1276688. [Google Scholar] [CrossRef]
- Xian, P.; Cai, Z.; Cheng, Y.; Lin, R.; Lian, T.; Ma, Q.; Nian, H. Wild Soybean Oxalyl-CoA Synthetase Degrades Oxalate and Affects the Tolerance to Cadmium and Aluminum Stresses. Int. J. Mol. Sci. 2020, 21, 8869. [Google Scholar] [CrossRef]
- Li, P.; He, Q.; Jin, J.; Liu, Y.; Wen, Y.; Zhao, K.; Mao, G.; Fan, W.; Yang, J. Tomato Oxalyl-CoA Synthetase Degrades Oxalate and Affects Fruit Quality. Front. Plant Sci. 2022, 13, 951386. [Google Scholar] [CrossRef]
- Bewley, J.D. Seed Germination and Dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef]
- Longo, C.; Holness, S.; De Angelis, V.; Lepri, A.; Occhigrossi, S.; Ruta, V.; Vittorioso, P. From the Outside to the Inside: New Insights on the Main Factors That Guide Seed Dormancy and Germination. Genes 2020, 12, 52. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, L.; Lu, X.; Lan, X.; Shen, M.; Lu, C. Role of mucilage during achene germination and sprout growth of the endangered Tibetan medicinal herb Mirabilis himalaica (Nyctaginaceae) exposed to abiotic stresses. J. Plant Ecol. 2017, 11, 328–337. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, S.; Baskin, J.M.; Baskin, C.C.; Wang, Z.; Liu, R.; Du, J.; Yang, X.; Huang, Z. Seed mucilage interacts with soil microbial community and physiochemical processes to affect seedling emergence on desert sand dunes. Plant Cell Environ. 2019, 42, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Junior, A.G.; Santos, M.T.A.; Hass, J.; Paschoal, B.S.M.; De-Paula, O.C. What kind of seed dormancy occurs in the legume genus Cassia? Sci. Rep. 2020, 10, 12194. [Google Scholar] [CrossRef]
- Western, T.L.; Young, D.S.; Dean, G.H.; Tan, W.L.; Samuels, A.L.; Haughn, G.W. MUCILAGE-MODIFIED4 Encodes a Putative Pectin Biosynthetic Enzyme Developmentally Regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis Seed Coat. Plant Physiol. 2004, 134, 296–306. [Google Scholar] [CrossRef]
- Usadel, B.R.; Kuschinsky, A.M.; Rosso, M.G.; Eckermann, N.; Pauly, M. RHM2 Is Involved in Mucilage Pectin Synthesis and Is Required for the Development of the Seed Coat in Arabidopsis. Plant Physiol. 2004, 134, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Macquet, A.; Ralet, M.-C.; Loudet, O.; Kronenberger, J.; Mouille, G.; Marion-Poll, A.; North, H.M. A Naturally Occurring Mutation in an Arabidopsis Accession Affects a β-d-Galactosidase That Increases the Hydrophilic Potential of Rhamnogalacturonan I in Seed Mucilage. Plant Cell 2007, 19, 3990–4006. [Google Scholar] [CrossRef]
- Dean, G.H.; Zheng, H.; Tewari, J.; Huang, J.; Young, D.S.; Hwang, Y.T.; Western, T.L.; Carpita, N.C.; McCann, M.C.; Mansfield, S.D.; et al. The Arabidopsis MUM2 Gene Encodes a β-Galactosidase Required for the Production of Seed Coat Mucilage with Correct Hydration Properties. Plant Cell 2007, 19, 4007–4021. [Google Scholar] [CrossRef]
- Arsovski, A.A.; Popma, T.M.; Haughn, G.W.; Carpita, N.C.; McCann, M.C.; Western, T.L. AtBXL1 Encodes a Bifunctional β-d-Xylosidase/α-l-Arabinofuranosidase Required for Pectic Arabinan Modification in Arabidopsis Mucilage Secretory Cells. Plant Physiol. 2009, 150, 1219–1234. [Google Scholar] [CrossRef]
- Kunieda, T.; Shimada, T.; Kondo, M.; Nishimura, M.; Nishitani, K.; Hara-Nishimura, I. Spatiotemporal Secretion of PEROXIDASE36 Is Required for Seed Coat Mucilage Extrusion in Arabidopsis. Plant Cell 2013, 25, 1355–1367. [Google Scholar] [CrossRef]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef]
- Gonzalez, A.; Mendenhall, J.; Huo, Y.; Lloyd, A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009, 325, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Allen, P.J.; Napoli, R.S.; Browne, R.G.; Pham, H.; Parish, R.W. MYB–bHLH–TTG1 Regulates Arabidopsis Seed Coat Biosynthesis Pathways Directly and Indirectly via Multiple Tiers of Transcription Factors. Plant Cell Physiol. 2020, 61, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Meissner, R.C.; Shoue, D.A.; Carpita, N.C.; Bevan, M.W. MYB61 Is Required for Mucilage Deposition and Extrusion in the Arabidopsis Seed Coat. Plant Cell 2001, 13, 2777–2791. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; DeBowles, D.; Esfandiari, E.; Dean, G.; Carpita, N.C.; Haughn, G.W. The Arabidopsis Transcription Factor LUH/MUM1 Is Required for Extrusion of Seed Coat Mucilage. Plant Physiol. 2011, 156, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.; Tehseen, M.; Doblin, M.S.; Pettolino, F.A.; Wilson, S.M.; Bacic, A.; Golz, J.F. The Transcriptional Regulator LEUNIG_HOMOLOG Regulates Mucilage Release from the Arabidopsis Testa. Plant Physiol. 2011, 156, 46–60. [Google Scholar] [CrossRef]
- Nakata, P.A. An Assessment of Engineered Calcium Oxalate Crystal Formation on Plant Growth and Development as a Step toward Evaluating Its Use to Enhance Plant Defense. PLoS ONE 2015, 10, e0141982. [Google Scholar] [CrossRef]
- Nakata, P.A. The oxalic acid biosynthetic activity of Burkholderia mallei is encoded by a single locus. Microbiol. Res. 2011, 166, 531–538. [Google Scholar] [CrossRef]
- Yang, J.; Fu, M.; Ji, C.; Huang, Y.; Wu, Y. Maize oxalyl-CoA decarboxylase1 degrades oxalate and affects the seed metabolome and nutritional quality. Plant Cell 2018, 30, 2447–2462. [Google Scholar] [CrossRef]
- Foster, J.; Cheng, N.; Paris, V.; Wang, L.; Wang, J.; Wang, X.; Nakata, P.A. An Arabidopsis Oxalyl-CoA Decarboxylase, AtOXC, Is Important for Oxalate Catabolism in Plants. Int. J. Mol. Sci. 2021, 22, 3266. [Google Scholar] [CrossRef] [PubMed]
- Shockey, J.; Browse, J. Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J. 2011, 66, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Molina, I.; Shockey, J.; Browse, J. Organ fusion and defective cuticle function in a lacs1lacs2 double mutant of Arabidopsis. Planta 2010, 231, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Kim, H.U.; Weng, H.; Browse, J. Malonyl-CoA Synthetase, Encoded by ACYL ACTIVATING ENZYME13, Is Essential for Growth and Development of Arabidopsis. Plant Cell 2011, 23, 2247–2262. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Liu, H.; Wei, Q.; Zhao, H.; Liu, J.; Yu, Y. The acyl-activating enzyme PhAAE13 is an alternative enzymatic source of precursors for anthocyanin biosynthesis in petunia flowers. J. Exp. Bot. 2017, 68, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, X.; Huang, H.; Qiao, R.; Jenks, M.A.; Zhao, H.; Lu, S. Arabidopsis ACYL-ACTIVATING ENZYME 9 (AAE9) encoding an isobutyl-CoA synthetase is a key factor connecting branched-chain amino acid catabolism with iso-branched wax biosynthesis. New Phytol. 2022, 233, 2458–2470. [Google Scholar] [CrossRef]
- Srividya, N.; Lange, I.; Hartmann, M.; Li, Q.; Mirzaei, M.; Lange, B.M. Biochemical characterization of acyl activating enzymes for side chain moieties of Taxol and its analogs. J. Biol. Chem. 2020, 295, 4963–4973. [Google Scholar] [CrossRef]
- Edwards, A.; Njaci, I.; Sarkar, A.; Jiang, Z.; Kaithakottil, G.G.; Moore, C.; Cheema, J.; Stevenson, C.E.M.; Rejzek, M.; Novák, P.; et al. Genomics and biochemical analyses reveal a metabolon key to β-L-ODAP biosynthesis in Lathyrus sativus. Nat. Commun. 2023, 14, 876. [Google Scholar] [CrossRef]
- Hoffmann, E.M.; Bauknecht, N. A Dye Binding Assay for the Quantification of Soluble and Cell-Bound Acidic Polysaccharides Produced by Red Algae. Anal. Biochem. 1999, 267, 245–251. [Google Scholar] [CrossRef]
- Cheng, N.; Paris, V.; Rao, X.; Wang, X.; Nakata, P.A. A conserved oxalyl-coenzyme A decarboxylase in oxalate catabolism. Plant Signal. Behav. 2022, 17, 2062555. [Google Scholar] [CrossRef]
- Mizzotti, C.; Rotasperti, L.; Moretto, M.; Tadini, L.; Resentini, F.; Galliani, B.M.; Galbiati, M.; Engelen, K.; Pesaresi, P.; Masiero, S. Time-Course Transcriptome Analysis of Arabidopsis Siliques Discloses Genes Essential for Fruit Development and Maturation. Plant Physiol. 2018, 178, 1249–1268. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, N.; Nakata, P.A. Disruption of the Arabidopsis Acyl-Activating Enzyme 3 Impairs Seed Coat Mucilage Accumulation and Seed Germination. Int. J. Mol. Sci. 2024, 25, 1149. https://doi.org/10.3390/ijms25021149
Cheng N, Nakata PA. Disruption of the Arabidopsis Acyl-Activating Enzyme 3 Impairs Seed Coat Mucilage Accumulation and Seed Germination. International Journal of Molecular Sciences. 2024; 25(2):1149. https://doi.org/10.3390/ijms25021149
Chicago/Turabian StyleCheng, Ninghui, and Paul A. Nakata. 2024. "Disruption of the Arabidopsis Acyl-Activating Enzyme 3 Impairs Seed Coat Mucilage Accumulation and Seed Germination" International Journal of Molecular Sciences 25, no. 2: 1149. https://doi.org/10.3390/ijms25021149
APA StyleCheng, N., & Nakata, P. A. (2024). Disruption of the Arabidopsis Acyl-Activating Enzyme 3 Impairs Seed Coat Mucilage Accumulation and Seed Germination. International Journal of Molecular Sciences, 25(2), 1149. https://doi.org/10.3390/ijms25021149





