Molecular Signature of Monocytes Shaped by the Shigella sonnei 1790-Generalized Modules for Membrane Antigens Vaccine
Abstract
1. Introduction
2. Results
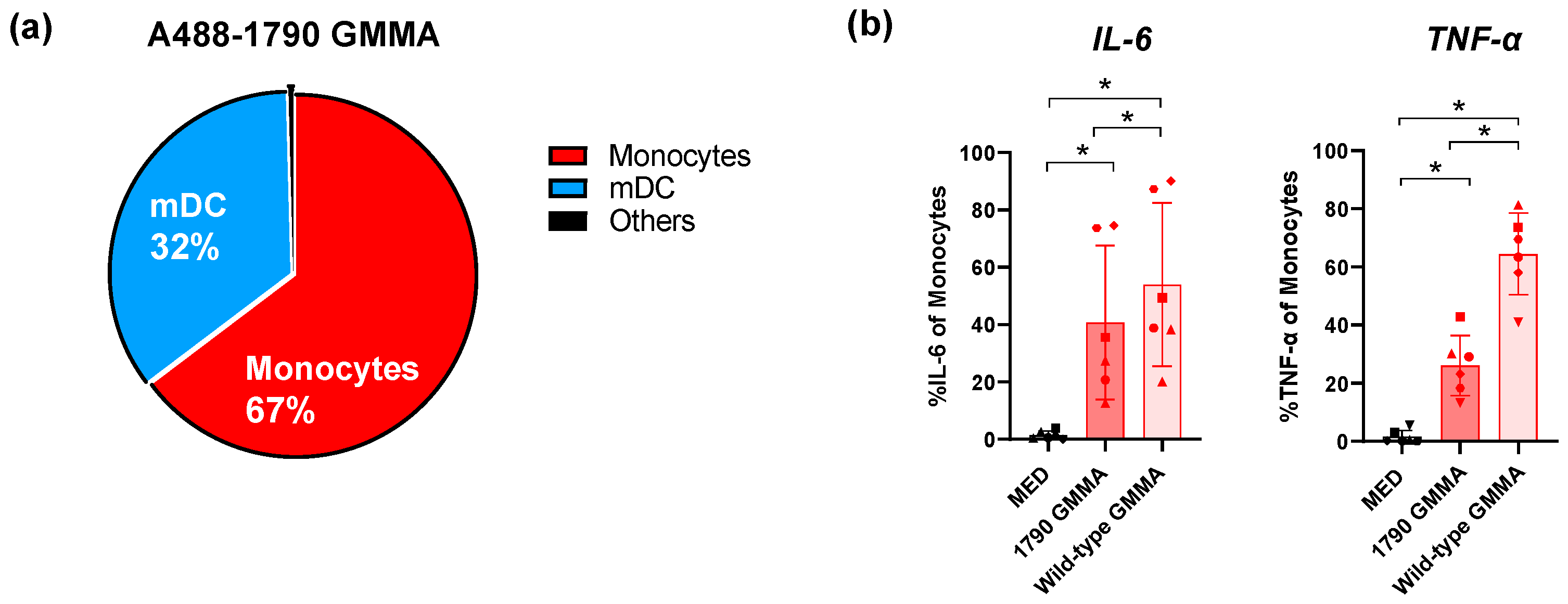
2.1. S. sonnei 1790-GMMA Directly Interact with and Activate Monocytes, Thus Promoting the Production of Pro-Inflammatory Cytokines
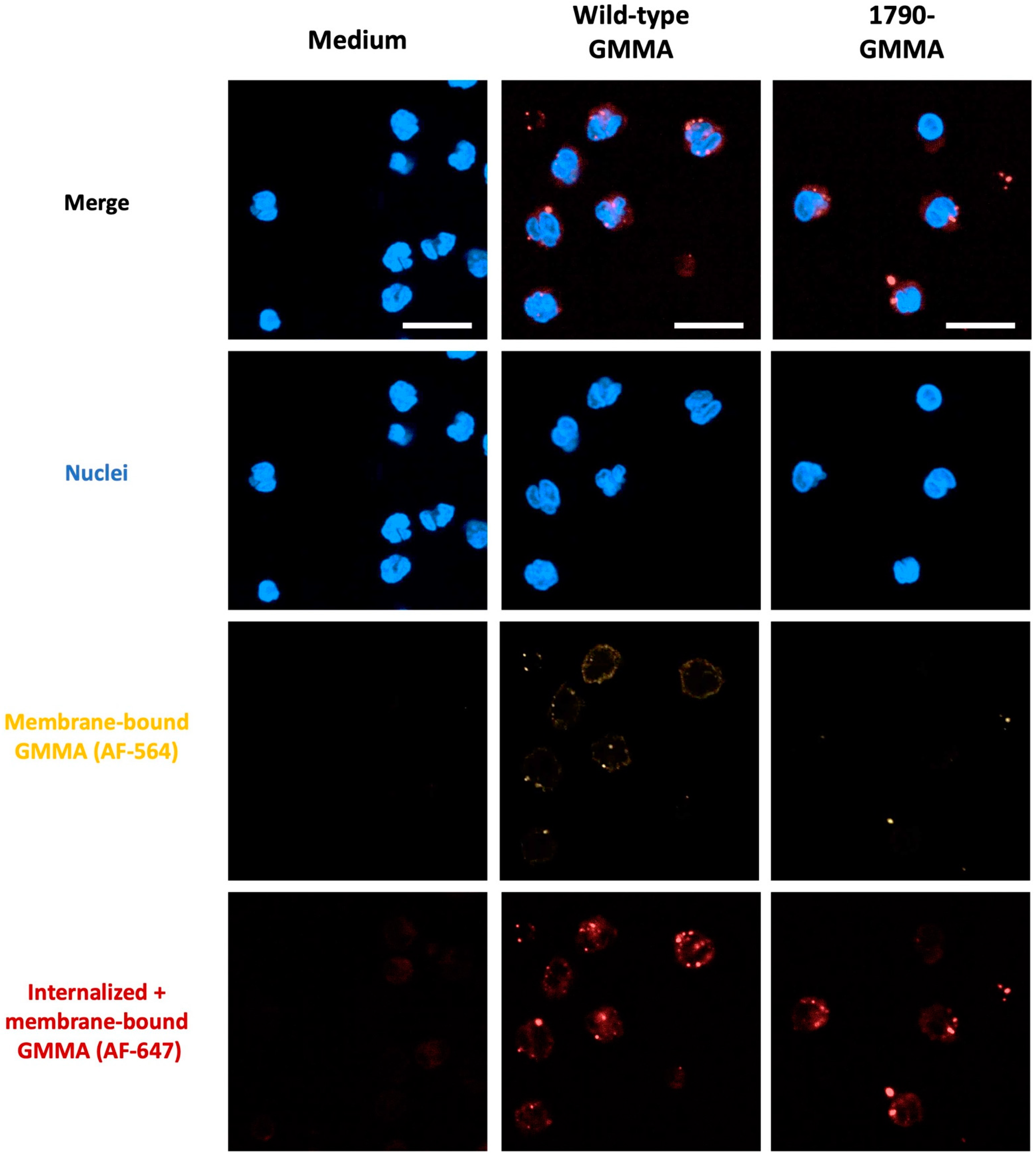
2.2. S. sonnei 1790-GMMA Are Internalized by Monocytes
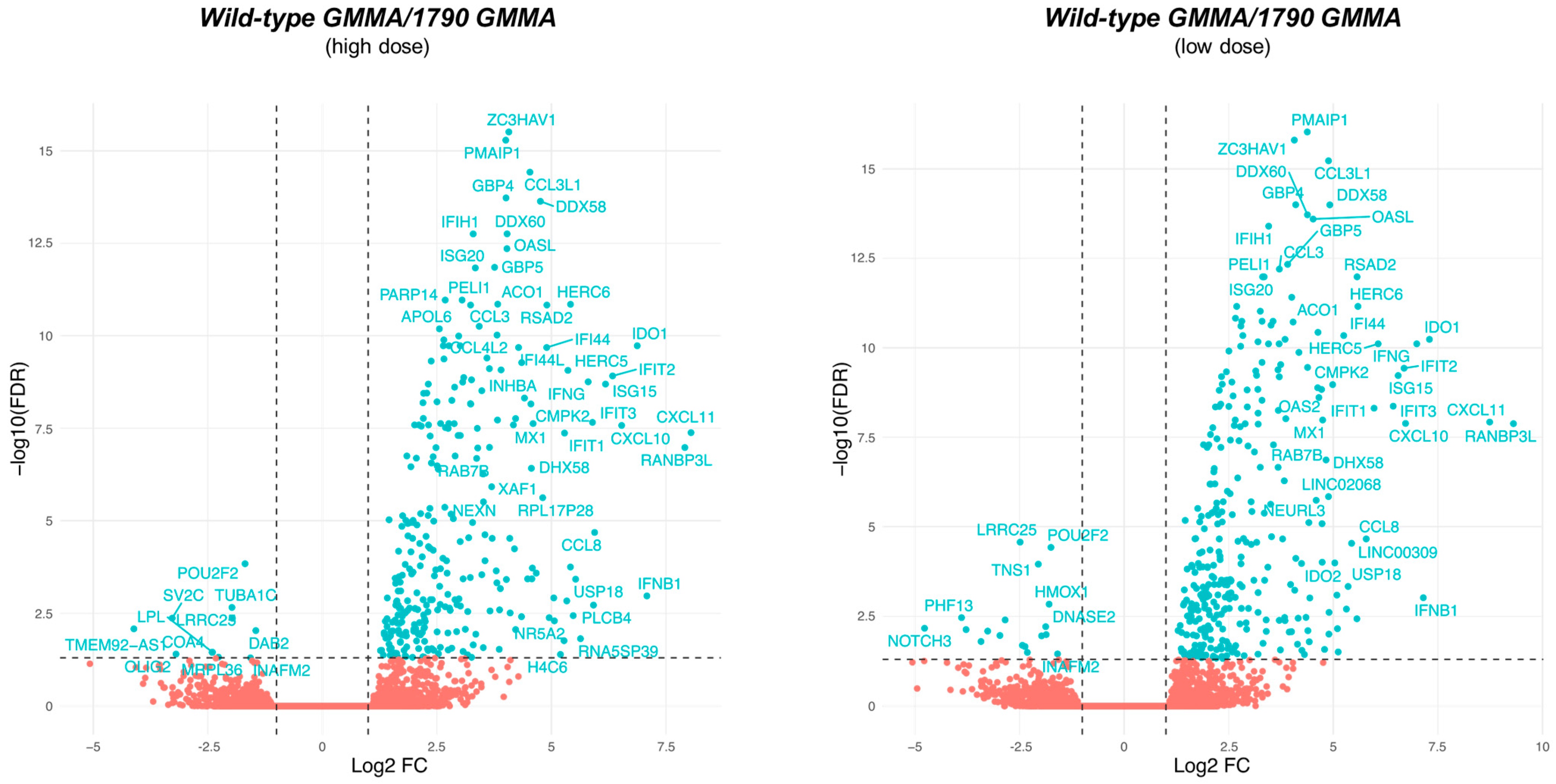
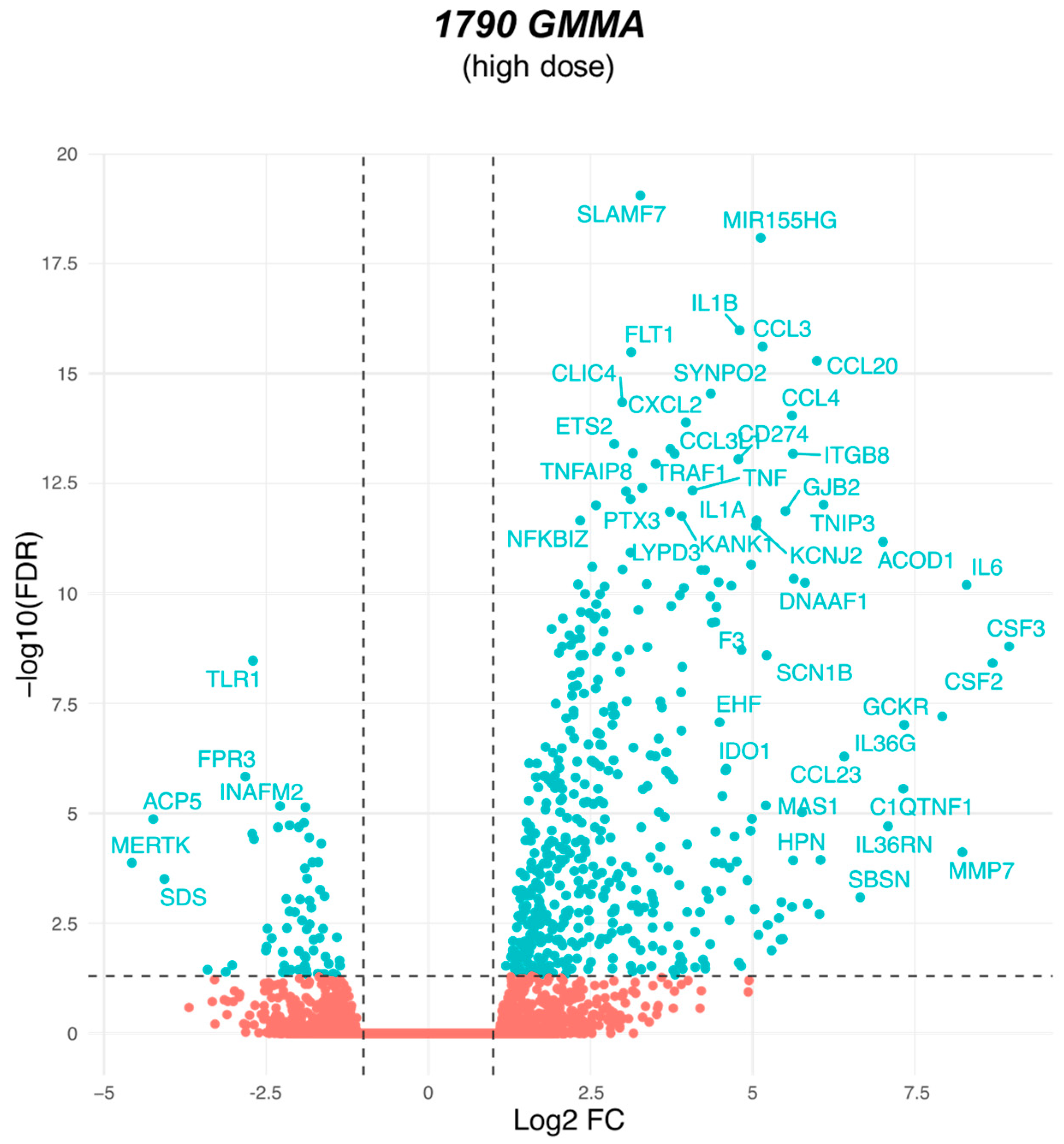
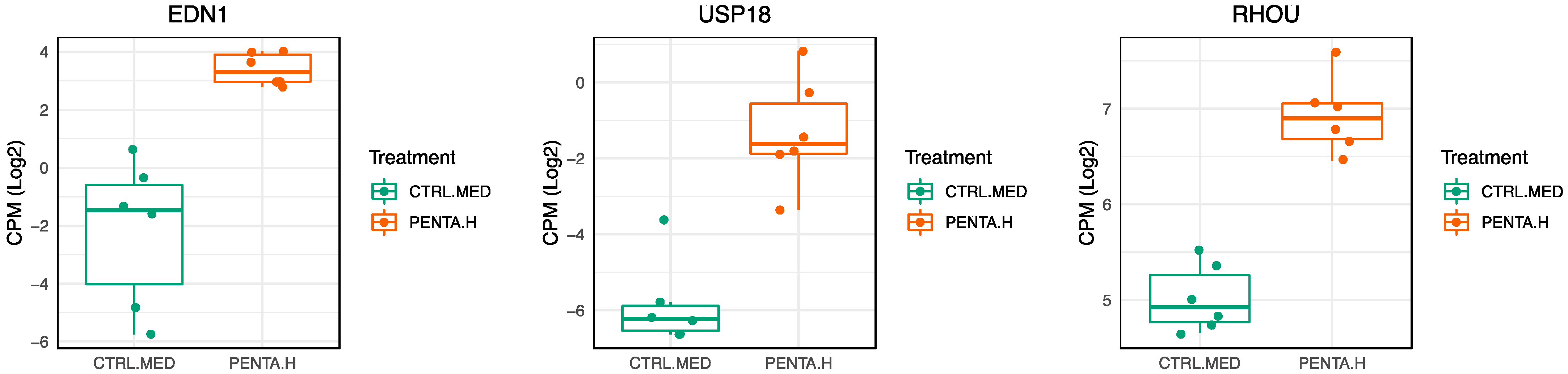
2.3. 1790-GMMA and Wild-Type GMMA Induced a Distinct Transcriptional Program in Monocytes
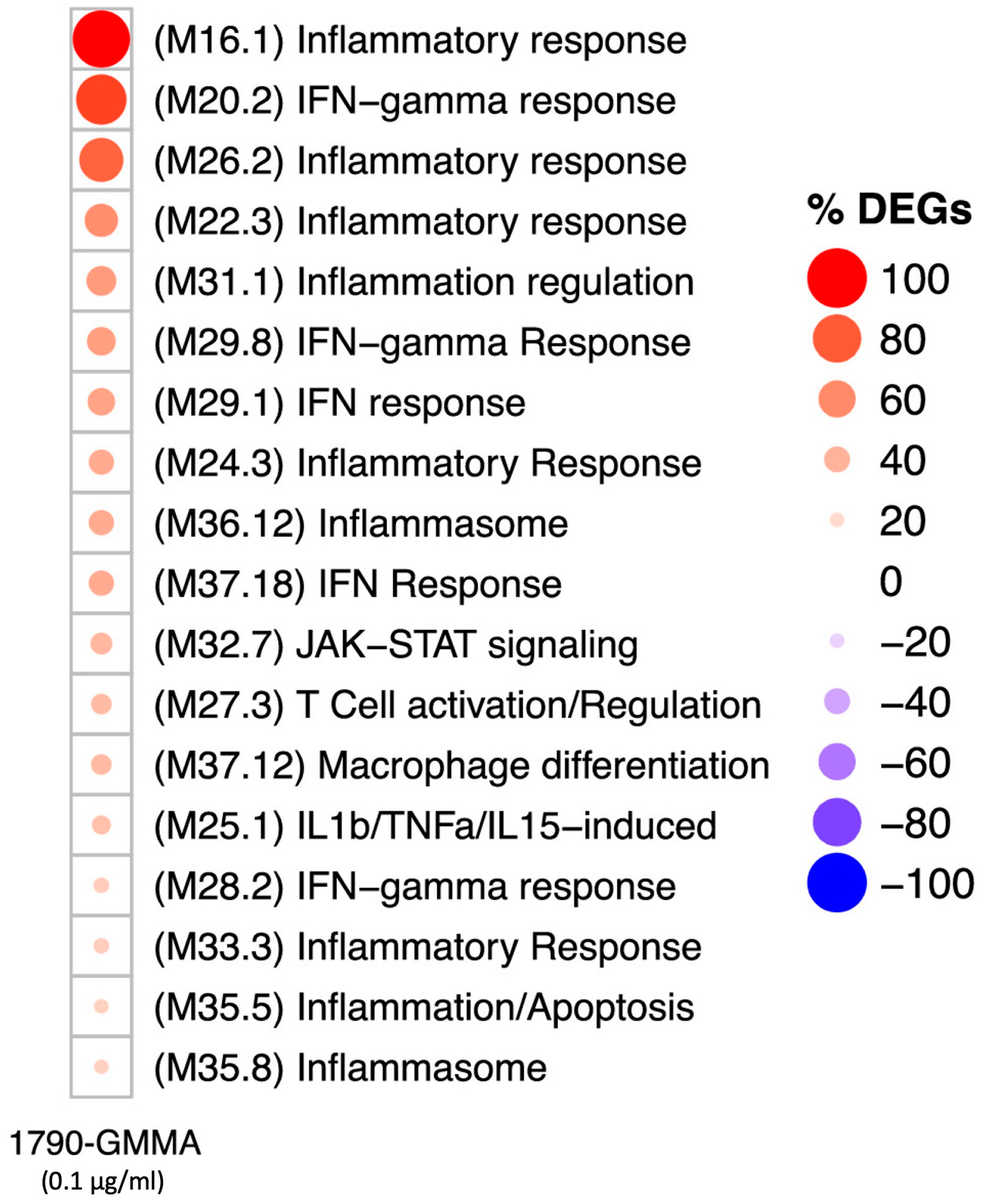
2.4. A Modular Framework Analysis Revealed the Transcriptional Fingerprinting of 1790-GMMA
3. Discussion
4. Materials and Methods
4.1. Isolation of hPBMCs and Monocytes from Healthy Donors
4.2. Thawing of hPBMCs
4.3. Labeled S. sonnei GMMA Preparation
4.4. hPBMC Treatments
4.5. hPBMC Flow Cytometry for Phenotypic Characterization and Intracellular Cytokine Determination
4.6. Confocal Microscopy Analysis of GMMA Localization in Monocytes
4.7. RNA Isolation and RNA Sequencing
4.8. Transcriptomic Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, J.; Pholwat, S.; Zhang, J.; Taniuchi, M.; Haque, R.; Alam, M.; Ochieng, J.B.; Jones, J.A.; Platts-Mills, J.A.; Tennant, S.M.; et al. Evaluation of Molecular Serotyping Assays for Shigella Flexneri Directly on Stool Samples. J. Clin. Microbiol. 2021, 59, e02455-20. [Google Scholar] [CrossRef] [PubMed]
- Picking, W.D.; Viswanathan, V.K.; Mariane, S.; Fleiszig, J.; Marteyn, B.S.; Anderson, M.; Sansonetti, P.J. Shigella Diversity and Changing Landscape: Insights for the Twenty-First Century. Front. Cell. Infect. Microbiol. 2016, 1, 45. [Google Scholar] [CrossRef]
- Wu, Y.; Lau, H.K.; Lee, T.; Lau, D.K.; Payne, J. In Silico Serotyping Based on Whole-Genome Sequencing Improves the Accuracy of Shigella Identification. Appl. Environ. Microbiol. 2019, 85, e00165-19. [Google Scholar] [CrossRef]
- Akhondi, H.; Simonsen, K.A. Bacterial Diarrhea. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Okamura, N.; Nakaya, R.; Suzuki, K.; Kondo, S.; Hisatsune, K.; Imagawa, Y.; Sagara, H.; Matsubara, Y. Differences among Shigella spp. in Susceptibility to the Bactericidal Activity of Human Serum. Microbiology 1988, 134, 2057–2065. [Google Scholar] [CrossRef] [PubMed]
- West, N.P.; Sansonetti, P.; Mounier, J.; Exley, R.M.; Parsot, C.; Guadagnini, S.; Prévost, M.-C.; Prochnicka-Chalufour, A.; Delepierre, M.; Tanguy, M. Optimization of Virulence Functions through Glucosylation of Shigella LPS. Science 2005, 307, 1313–1317. [Google Scholar] [CrossRef]
- Raqib, R.; Wretlind, B.; Andersson, J.; Lindberg, A.A. Cytokine Secretion in Acute Shigellosis Is Correlated to Disease Activity and Directed More to Stool than to Plasma. J. Infect. Dis. 1995, 171, 376–384. [Google Scholar] [CrossRef]
- Phalipon, A.; Sansonetti, P.J. Shigella’s Ways of Manipulating the Host Intestinal Innate and Adaptive Immune System: A Tool Box for Survival? Immunol. Cell Biol. 2007, 85, 119–129. [Google Scholar] [CrossRef]
- Bloom, D.E.; Black, S.; Salisbury, D.; Rappuoli, R. Antimicrobial Resistance and the Role of Vaccines. Proc. Natl. Acad. Sci. USA 2018, 115, 12868–12871. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Munk, P.; Njage, P.; Van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J. Global Monitoring of Antimicrobial Resistance Based on Metagenomics Analyses of Urban Sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Yang, C.; Xiang, Y.; Qiu, S. Resistance in Enteric Shigella and Nontyphoidal Salmonella: Emerging Concepts. Curr. Opin. Infect. Dis. 2023, 36, 360–365. [Google Scholar] [CrossRef]
- Ellis, T.N.; Kuehn, M.J. Virulence and Immunomodulatory Roles of Bacterial Outer Membrane Vesicles. Microbiol. Mol. Biol. Rev. 2010, 74, 81–94. [Google Scholar] [CrossRef]
- McBroom, A.J.; Johnson, A.P.; Vemulapalli, S.; Kuehn, M.J. Outer Membrane Vesicle Production by Escherichia coli Is Independent of Membrane Instability. J. Bacteriol. 2006, 188, 5385–5392. [Google Scholar] [CrossRef]
- Furuyama, N.; Sircili, M.P. Outer Membrane Vesicles (OMVs) Produced by Gram-Negative Bacteria: Structure, Functions, Biogenesis, and Vaccine Application. Biomed. Res. Int. 2021, 2021, 1490732. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Kulkarni, H.M.; Jagannadham, M. v Biogenesis and Multifaceted Roles of Outer Membrane Vesicles from Gram-Negative Bacteria. Microbiology 2014, 160, 2109–2121. [Google Scholar] [CrossRef]
- Antonelli, G.; Cappelli, L.; Cinelli, P.; Cuffaro, R.; Manca, B.; Nicchi, S.; Tondi, S.; Vezzani, G.; Viviani, V.; Delany, I. Strategies to Tackle Antimicrobial Resistance: The Example of Escherichia coli and Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 4943. [Google Scholar] [CrossRef]
- Mancini, F.; Rossi, O.; Necchi, F.; Micoli, F. OMV Vaccines and the Role of TLR Agonists in Immune Response. Int. J. Mol. Sci. 2020, 21, 4416. [Google Scholar] [CrossRef]
- Berlanda Scorza, F.; Colucci, A.M.; Maggiore, L.; Sanzone, S.; Rossi, O.; Ferlenghi, I.; Pesce, I.; Caboni, M.; Norais, N.; Di Cioccio, V. High Yield Production Process for Shigella Outer Membrane Particles. PLoS ONE 2012, 7, e35616. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, L.; Stork, M.; van der Ley, P. Outer Membrane Vesicles as Platform Vaccine Technology. Biotechnol. J. 2015, 10, 1689–1706. [Google Scholar] [CrossRef] [PubMed]
- Rossi, O.; Citiulo, F.; Mancini, F. Outer Membrane Vesicles: Moving within the Intricate Labyrinth of Assays That Can Predict Risks of Reactogenicity in Humans. Hum. Vaccines Immunother. 2021, 17, 601–613. [Google Scholar] [CrossRef] [PubMed]
- van der Ley, P.; van den Dobbelsteen, G. Next-Generation Outer Membrane Vesicle Vaccines against Neisseria meningitidis Based on Nontoxic LPS Mutants. Hum. Vaccines 2011, 7, 886–890. [Google Scholar] [CrossRef] [PubMed]
- Piccioli, D.; Bartolini, E.; Micoli, F. GMMA as a ‘Plug and Play’ Technology to Tackle Infectious Disease to Improve Global Health: Context and Perspectives for the Future. Expert Rev. Vaccines 2022, 21, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Mancini, F.; Micoli, F.; Necchi, F.; Pizza, M.; Berlanda Scorza, F.; Rossi, O. GMMA-Based Vaccines: The Known and the Unknown. Front. Immunol. 2021, 12, 715393. [Google Scholar] [CrossRef] [PubMed]
- Girardi, P.; Harutyunyan, S.; Neuhauser, I.; Glaninger, K.; Korda, O.; Nagy, G.; Nagy, E.; Szijártó, V.; Pall, D.; Szarka, K. Evaluation of the Safety, Tolerability and Immunogenicity of ShigETEC, an Oral Live Attenuated Shigella-ETEC Vaccine in Placebo-Controlled Randomized Phase 1 Trial. Vaccines 2022, 10, 340. [Google Scholar] [CrossRef] [PubMed]
- Rossi, O.; Pesce, I.; Giannelli, C.; Aprea, S.; Caboni, M.; Citiulo, F.; Valentini, S.; Ferlenghi, I.; MacLennan, C.A.; D’Oro, U. Modulation of Endotoxicity of Shigella Generalized Modules for Membrane Antigens (GMMA) by Genetic Lipid A Modifications: Relative Activation of TLR4 and TLR2 Pathways in Different Mutants. J. Biol. Chem. 2014, 289, 24922–24935. [Google Scholar] [CrossRef] [PubMed]
- Gerke, C.; Colucci, A.M.; Giannelli, C.; Sanzone, S.; Giorgina Vitali, C.; Sollai, L.; Rossi, O.; Martin, L.B.; Auerbach, J.; Di Cioccio, V.; et al. Production of a Shigella sonnei Vaccine Based on Generalized Modules for Membrane Antigens (GMMA), 1790GAHB. PLoS ONE 2015, 10, e0134478. [Google Scholar] [CrossRef]
- Clementz, T.; Bednarski, J.J.; Raetz, C.R.H. Function of the HtrB High Temperature Requirement Gene of Escherichia coli in the Acylation of Lipid A: HtrB Catalyzed Incorporation of Laurate (∗). J. Biol. Chem. 1996, 271, 12095–12102. [Google Scholar] [CrossRef]
- Rossi, O.; Citiulo, F.; Giannelli, C.; Cappelletti, E.; Gasperini, G.; Mancini, F.; Acquaviva, A.; Raso, M.M.; Sollai, L.; Alfini, R.; et al. A Next-Generation GMMA-Based Vaccine Candidate to Fight Shigellosis. NPJ Vaccines 2023, 8, 130. [Google Scholar] [CrossRef]
- Mancini, F.; Alfini, R.; Caradonna, V.; Monaci, V.; Carducci, M.; Gasperini, G.; Piccioli, D.; Biagini, M.; Giannelli, C.; Rossi, O. Exploring the Role of GMMA Components in the Immunogenicity of a 4-Valent Vaccine against Shigella. Int. J. Mol. Sci. 2023, 24, 2742. [Google Scholar] [CrossRef]
- Tondi, S.; Clemente, B.; Esposito, C.; Sammicheli, C.; Tavarini, S.; Martin, L.B.; Rossi, O.; Micoli, F.; Bartolini, E.; Brazzoli, M.; et al. Dissecting in Vitro the Activation of Human Immune Response Induced by Shigella Sonnei GMMA. Front. Cell Infect. Microbiol. 2022, 12, 767153. [Google Scholar] [CrossRef]
- O’donoghue, E.J.; Krachler, A.M. Mechanisms of Outer Membrane Vesicle Entry into Host Cells. Cell Microbiol. 2016, 18, 1508–1517. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, R.; Baldwin, N.; Cepika, A.-M.; Athale, S.; Xue, Y.; Yu, C.I.; Metang, P.; Cheruku, A.; Berthier, I.; Gayet, I.; et al. Transcriptional Specialization of Human Dendritic Cell Subsets in Response to Microbial Vaccines. Nat. Commun. 2014, 5, 5283. [Google Scholar] [CrossRef]
- Guerriero, J.L. Chapter Three–Macrophages: Their Untold Story in T Cell Activation and Function. In International Review of Cell and Molecular Biology; Galluzzi, L., Rudqvist, N.-P., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 342, ISBN 1937-6448. [Google Scholar]
- Zhang, J.; Zhao, W.; Xu, L.; Wang, X.; Li, X.; Yang, X. Endothelium-Specific Endothelin-1 Expression Promotes pro-Inflammatory Macrophage Activation by Regulating MiR-33/NR4A Axis. Exp. Cell Res. 2021, 399, 112443. [Google Scholar] [CrossRef] [PubMed]
- Helset, E.; Sildnes, T.; Seljelid, R.; Konopski, Z.S. Endothelin-1 Stimulates Human Monocytes in Vitro to Release TNF-α, IL-1β and IL-6. Mediat. Inflamm. 1993, 2, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, S.-K.; Schmitz, R.; Bergerhausen, M.; Lang, J.; Cham, L.B.; Duhan, V.; Häussinger, D.; Hardt, C.; Addo, M.; Prinz, M. Usp18 Expression in CD169+ Macrophages Is Important for Strong Immune Response after Vaccination with VSV-EBOV. Vaccines 2020, 8, 142. [Google Scholar] [CrossRef] [PubMed]
- Honke, N.; Shaabani, N.; Zhang, D.-E.; Hardt, C.; Lang, K.S. Multiple Functions of USP18. Cell Death Dis. 2016, 7, e2444. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho Proteins, PI 3-Kinases, and Monocyte/Macrophage Motility. FEBS Lett. 2001, 498, 168–171. [Google Scholar] [CrossRef]
- Muntjewerff, E.M.; Meesters, L.D.; van den Bogaart, G. Antigen Cross-Presentation by Macrophages. Front. Immunol. 2020, 11, 1276. [Google Scholar] [CrossRef]
- Puzari, M.; Sharma, M.; Chetia, P. Emergence of Antibiotic Resistant Shigella Species: A Matter of Concern. J. Infect. Public Health 2018, 11, 451–454. [Google Scholar] [CrossRef]
- Klontz, K.C.; Singh, N. Treatment of Drug-Resistant Shigella Infections. Expert Rev. Anti Infect. Ther. 2015, 13, 69–80. [Google Scholar] [CrossRef]
- Sharif, N.; Ahmed, S.N.; Khandaker, S.; Monifa, N.H.; Abusharha, A.; Vargas, D.L.R.; De la Torre Díez, I.; Castilla, A.G.K.; Talukder, A.A.; Parvez, A.K.; et al. Multidrug Resistance Pattern and Molecular Epidemiology of Pathogens among Children with Diarrhea in Bangladesh, 2019–2021. Sci. Rep. 2023, 13, 13975. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 9 September 2023).
- Micoli, F.; Alfini, R.; Di Benedetto, R.; Necchi, F.; Schiavo, F.; Mancini, F.; Carducci, M.; Palmieri, E.; Balocchi, C.; Gasperini, G. GMMA Is a Versatile Platform to Design Effective Multivalent Combination Vaccines. Vaccines 2020, 8, 540. [Google Scholar] [CrossRef] [PubMed]
- Piccioli, D.; Alfini, R.; Monaci, V.; Arato, V.; Carducci, M.; Aruta, M.G.; Rossi, O.; Necchi, F.; Anemona, A.; Bartolini, E. Antigen Presentation by Follicular Dendritic Cells to Cognate B Cells Is Pivotal for Generalised Modules for Membrane Antigens (GMMA) Immunogenicity. Vaccine 2022, 40, 6305–6314. [Google Scholar] [CrossRef] [PubMed]
- Frenck, R.W.; Conti, V.; Ferruzzi, P.; Ndiaye, A.G.W.; Parker, S.; McNeal, M.M.; Dickey, M.; Granada, J.P.; Cilio, G.L.; De Ryck, I. Efficacy, Safety, and Immunogenicity of the Shigella sonnei 1790GAHB GMMA Candidate Vaccine: Results from a Phase 2b Randomized, Placebo-Controlled Challenge Study in Adults. EClinicalMedicine 2021, 39, 101076. [Google Scholar] [CrossRef]
- Mancini, F.; Gasperini, G.; Rossi, O.; Aruta, M.G.; Raso, M.M.; Alfini, R.; Biagini, M.; Necchi, F.; Micoli, F. Dissecting the Contribution of O-Antigen and Proteins to the Immunogenicity of Shigella sonnei Generalized Modules for Membrane Antigens (GMMA). Sci. Rep. 2021, 11, 906. [Google Scholar] [CrossRef]
- Wen, Y.; Wang, X.; Cahya, S.; Anderson, P.; Velasquez, C.; Torres, C.; Ferrante, A.; Kaliyaperumal, A. Comparability Study of Monocyte Derived Dendritic Cells, Primary Monocytes, and THP1 Cells for Innate Immune Responses. J. Immunol. Methods 2021, 498, 113147. [Google Scholar] [CrossRef]
- Navegantes, K.C.; de Souza Gomes, R.; Pereira, P.A.T.; Czaikoski, P.G.; Azevedo, C.H.M.; Monteiro, M.C. Immune Modulation of Some Autoimmune Diseases: The Critical Role of Macrophages and Neutrophils in the Innate and Adaptive Immunity. J. Transl. Med. 2017, 15, 1–21. [Google Scholar] [CrossRef]
- Lee, H.K.; Iwasaki, A. Innate Control of Adaptive Immunity: Dendritic Cells and Beyond. Semin. Immunol. 2007, 19, 48–55. [Google Scholar] [CrossRef]
- Sun, Y.; Kuang, Y.; Zuo, Z. The Emerging Role of Macrophages in Immune System Dysfunction under Real and Simulated Microgravity Conditions. Int. J. Mol. Sci. 2021, 22, 2333. [Google Scholar] [CrossRef]
- Perfetto, S.P.; Ambrozak, D.; Nguyen, R.; Chattopadhyay, P.; Roederer, M. Quality Assurance for Polychromatic Flow Cytometry. Nat. Protoc. 2006, 1, 1522–1530. [Google Scholar] [CrossRef]
- Modi, A.; Vai, S.; Caramelli, D.; Lari, M. The Illumina Sequencing Protocol and the NovaSeq 6000 System. In Bacterial Pangenomics: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2021; pp. 15–42. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Mann, H.B.; Whitney, D.R. On a Test of Whether One of Two Random Variables Is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Daniel, W.W. Applied Nonparametric Statistics; PWS-Kent: Devon, UK, 1990. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tondi, S.; Siena, E.; Essaghir, A.; Bozzetti, B.; Bechtold, V.; Scaillet, A.; Clemente, B.; Marrocco, M.; Sammicheli, C.; Tavarini, S.; et al. Molecular Signature of Monocytes Shaped by the Shigella sonnei 1790-Generalized Modules for Membrane Antigens Vaccine. Int. J. Mol. Sci. 2024, 25, 1116. https://doi.org/10.3390/ijms25021116
Tondi S, Siena E, Essaghir A, Bozzetti B, Bechtold V, Scaillet A, Clemente B, Marrocco M, Sammicheli C, Tavarini S, et al. Molecular Signature of Monocytes Shaped by the Shigella sonnei 1790-Generalized Modules for Membrane Antigens Vaccine. International Journal of Molecular Sciences. 2024; 25(2):1116. https://doi.org/10.3390/ijms25021116
Chicago/Turabian StyleTondi, Serena, Emilio Siena, Ahmed Essaghir, Benoît Bozzetti, Viviane Bechtold, Aline Scaillet, Bruna Clemente, Mariateresa Marrocco, Chiara Sammicheli, Simona Tavarini, and et al. 2024. "Molecular Signature of Monocytes Shaped by the Shigella sonnei 1790-Generalized Modules for Membrane Antigens Vaccine" International Journal of Molecular Sciences 25, no. 2: 1116. https://doi.org/10.3390/ijms25021116
APA StyleTondi, S., Siena, E., Essaghir, A., Bozzetti, B., Bechtold, V., Scaillet, A., Clemente, B., Marrocco, M., Sammicheli, C., Tavarini, S., Micoli, F., Oldrini, D., Pezzicoli, A., Di Fede, M., Brazzoli, M., Ulivieri, C., & Schiavetti, F. (2024). Molecular Signature of Monocytes Shaped by the Shigella sonnei 1790-Generalized Modules for Membrane Antigens Vaccine. International Journal of Molecular Sciences, 25(2), 1116. https://doi.org/10.3390/ijms25021116







