Deciphering the Molecular Mechanism of the Intermediate Secondary Growth and Internode Elongation of the Castor Bean (Ricinus communis L.) by the Combined Analysis of the Transcriptome and Metabolome
Abstract
1. Introduction
2. Results
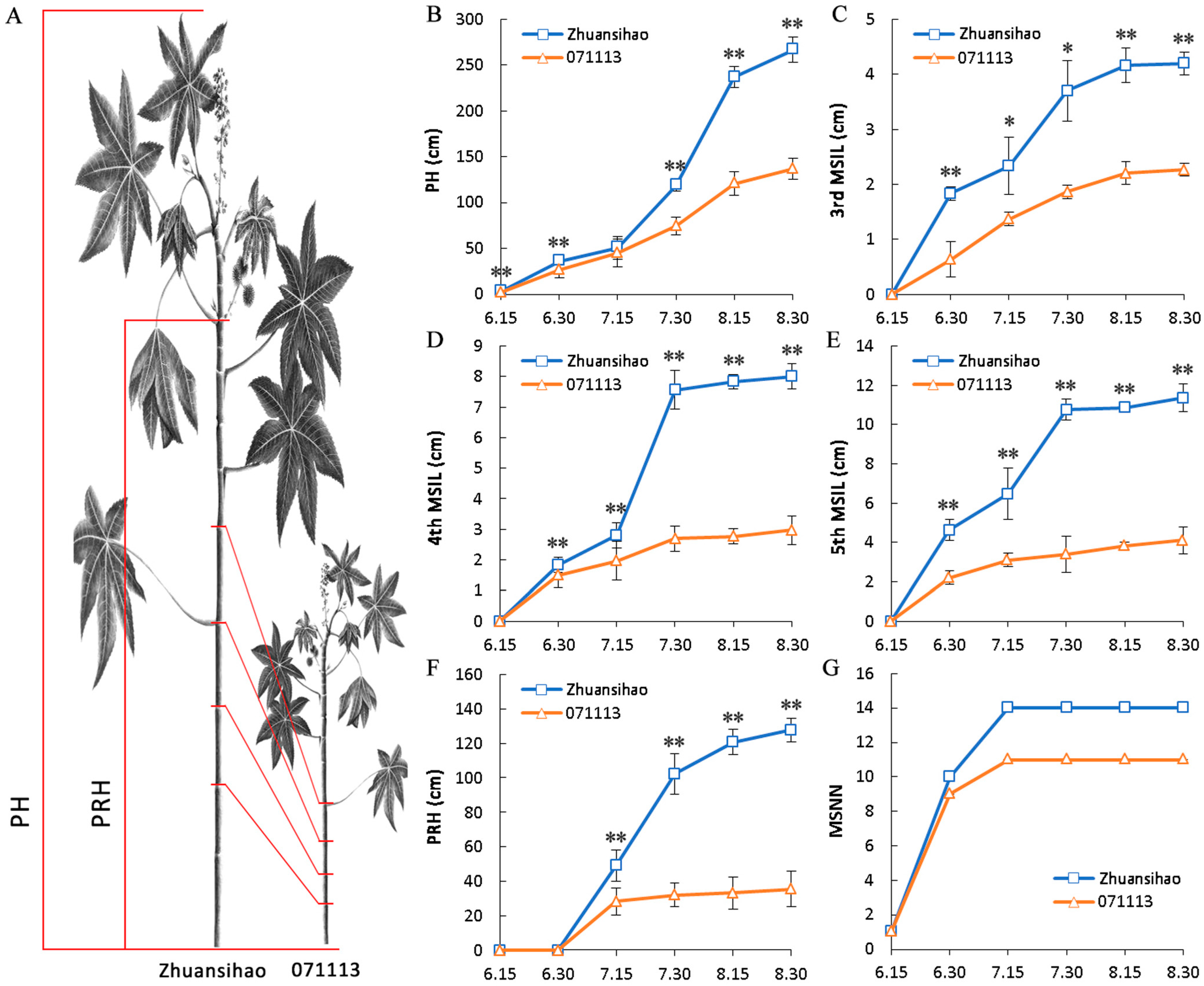
2.1. Data Associated with Plant Height
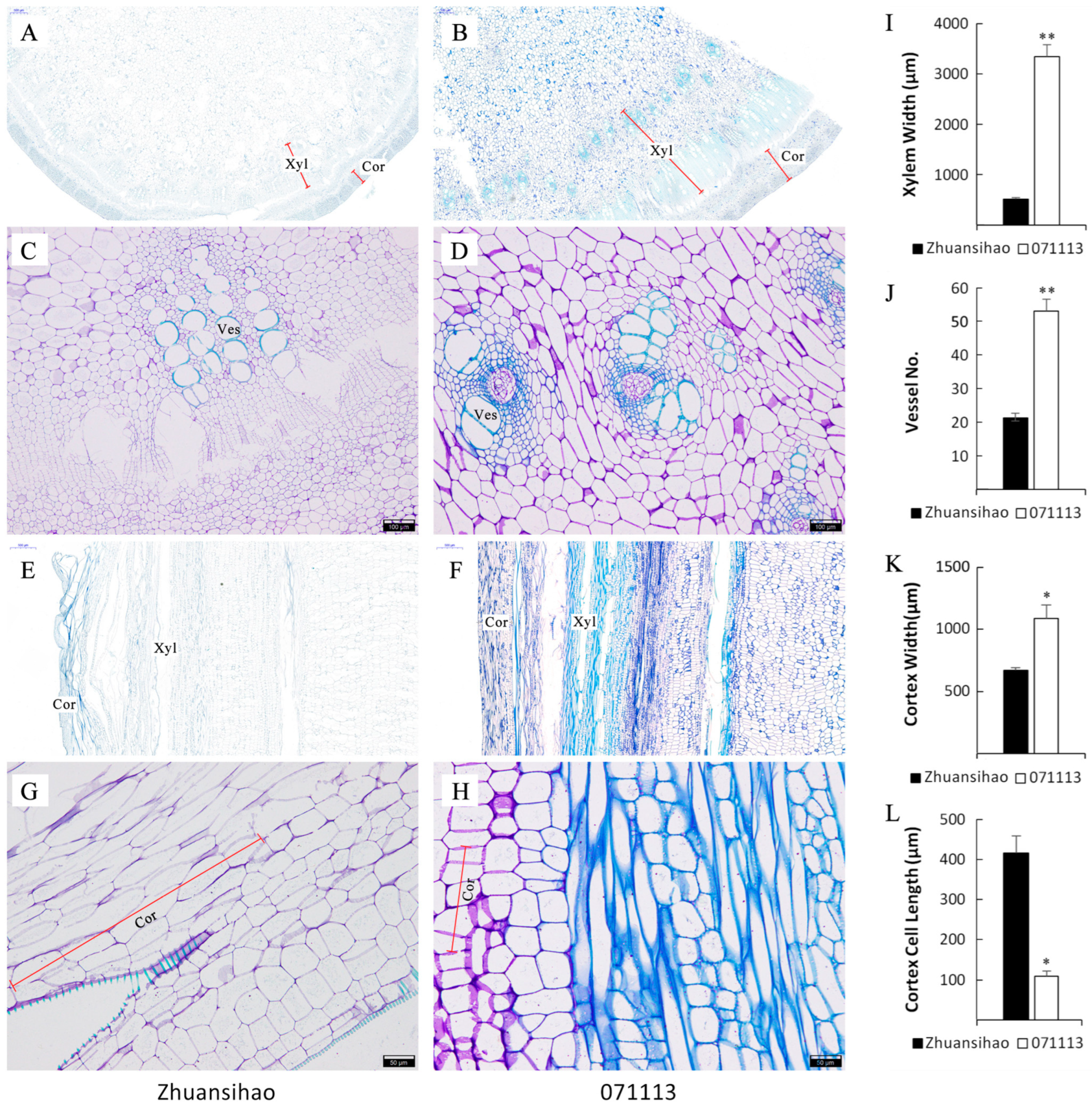
2.2. Examination of Cellular Characteristics
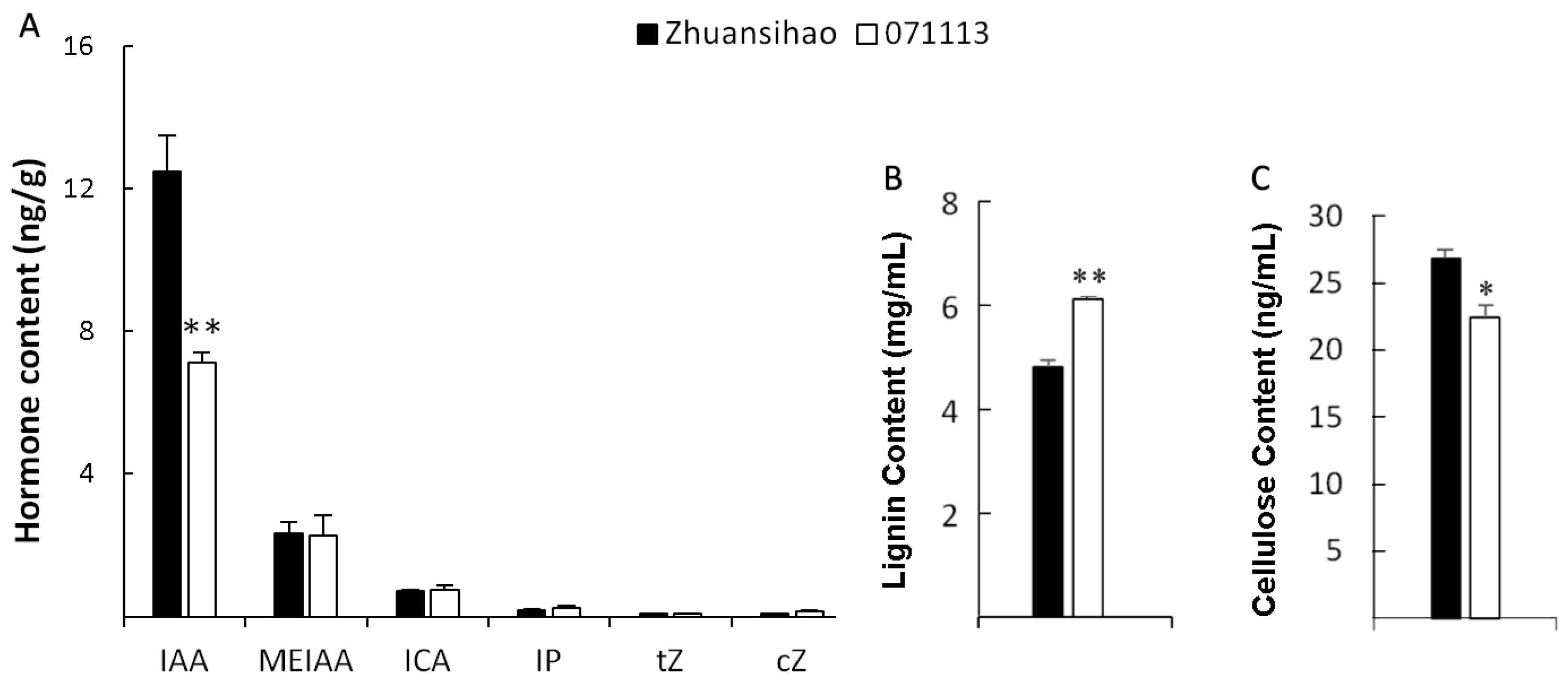
2.3. Content Determinations of Phytohormone, Lignin, and Cellulose
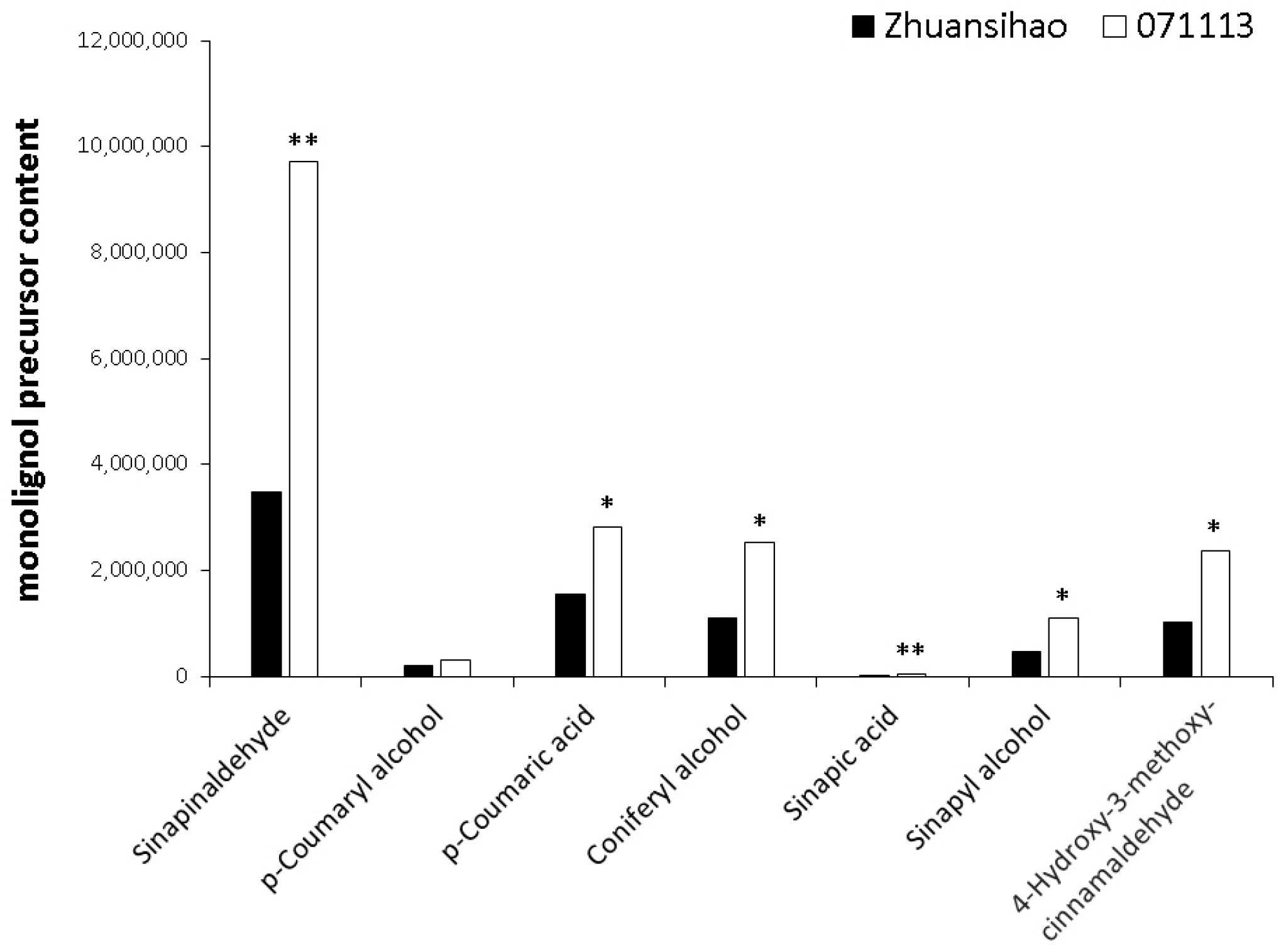
2.4. Metabolome Analysis
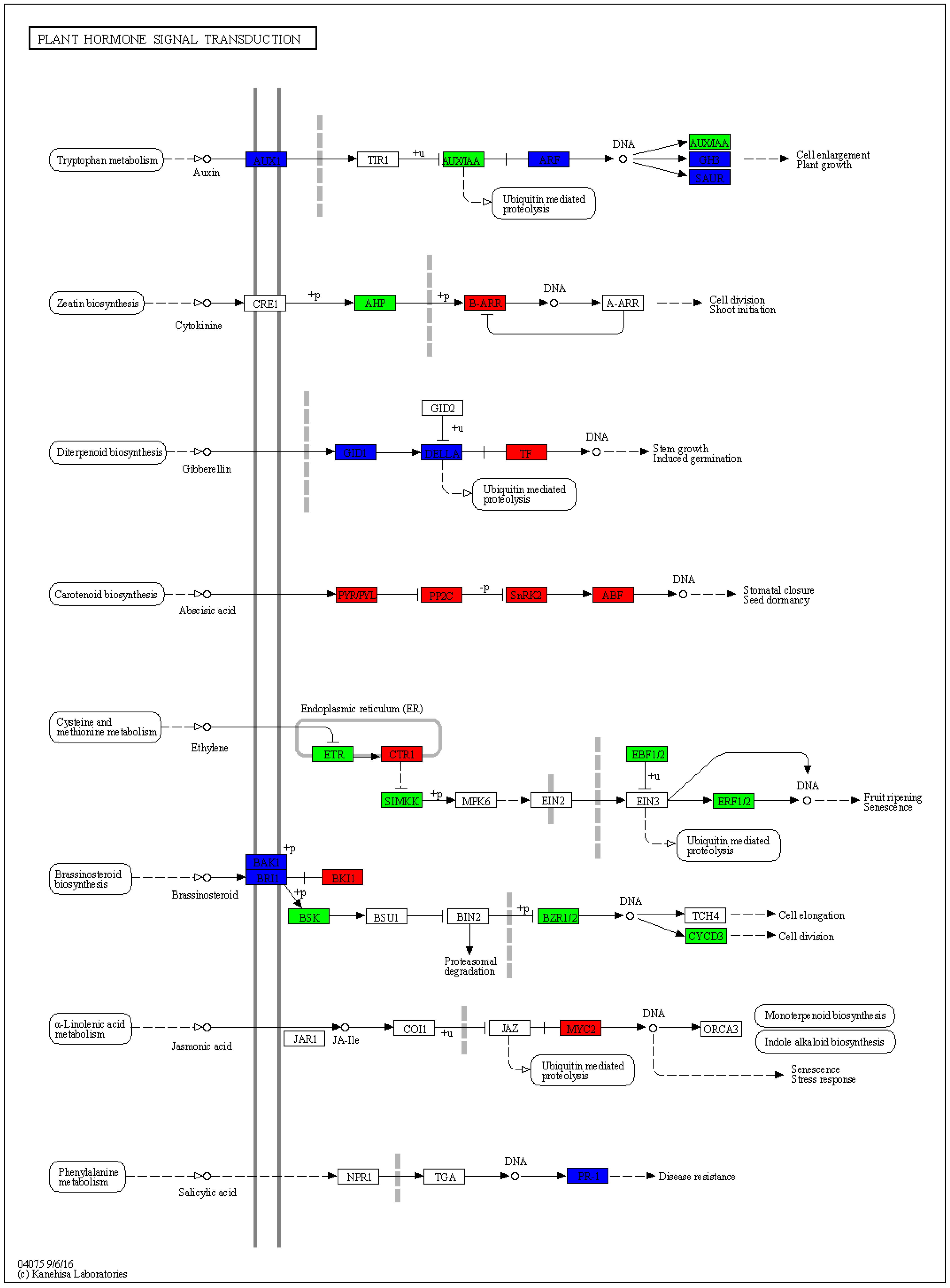
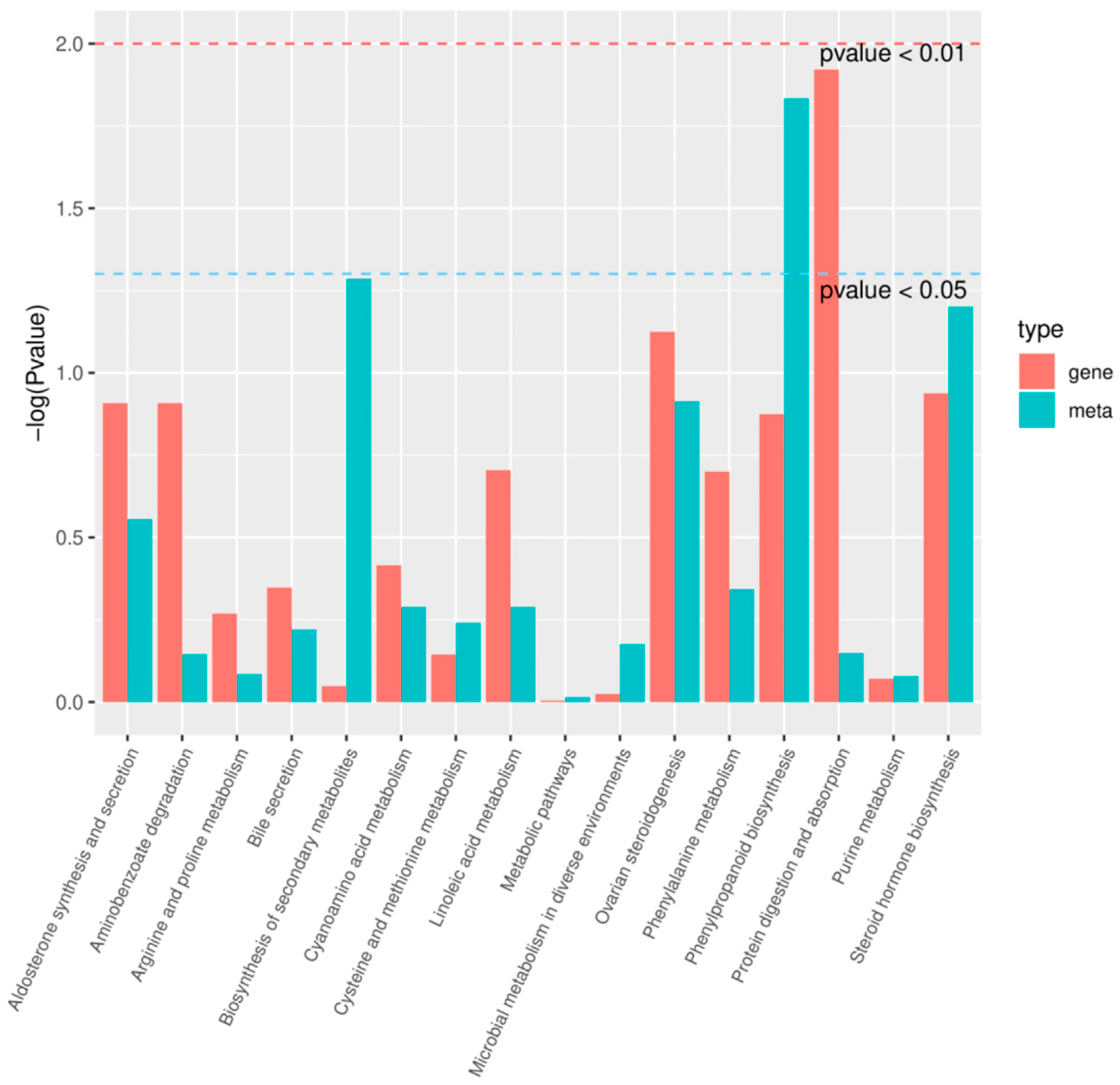
2.5. Transcriptome Sequencing
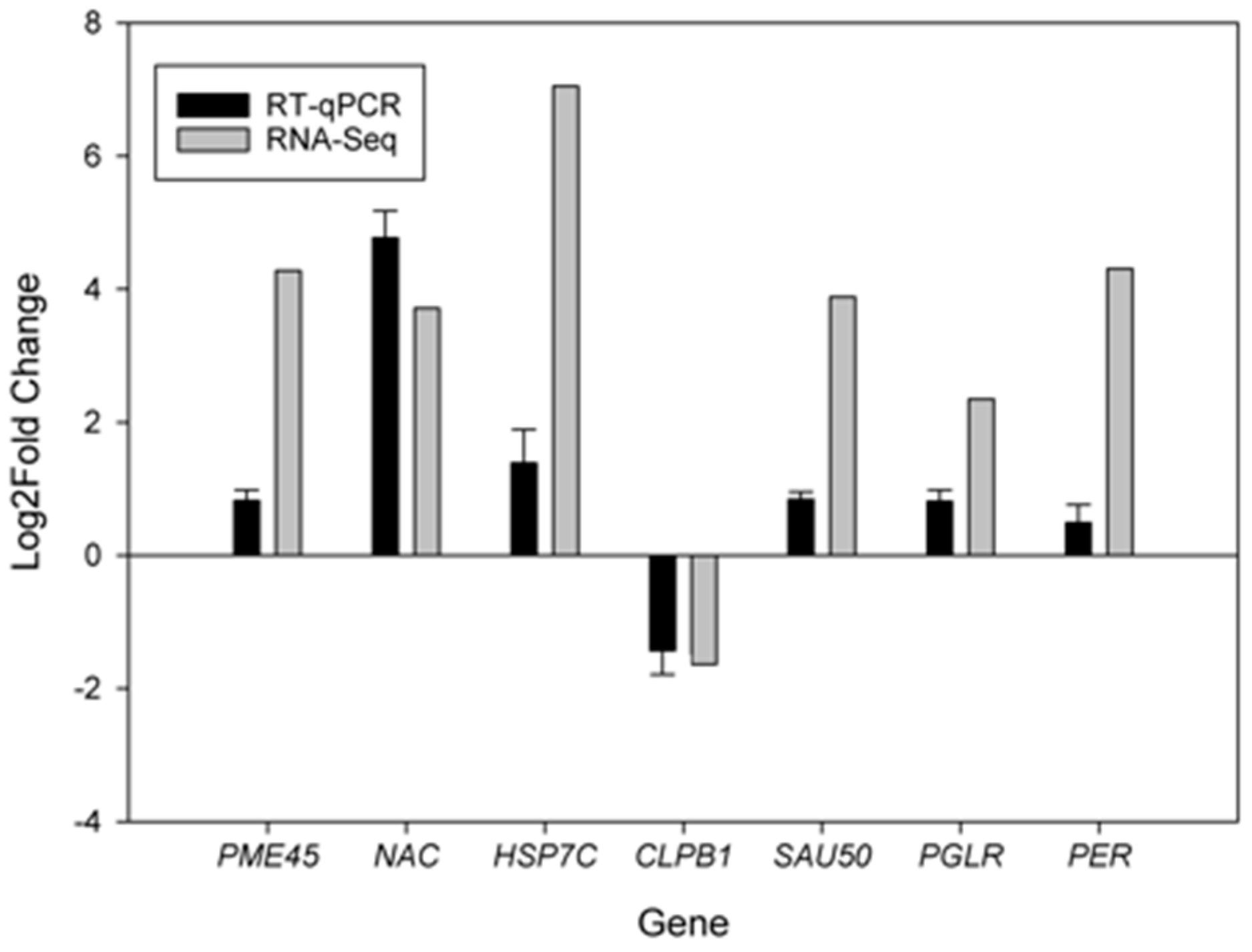
2.6. Verification of Differentially Expressed Genes
2.7. Metabolome and Transcriptome Correlation Analysis
3. Discussion
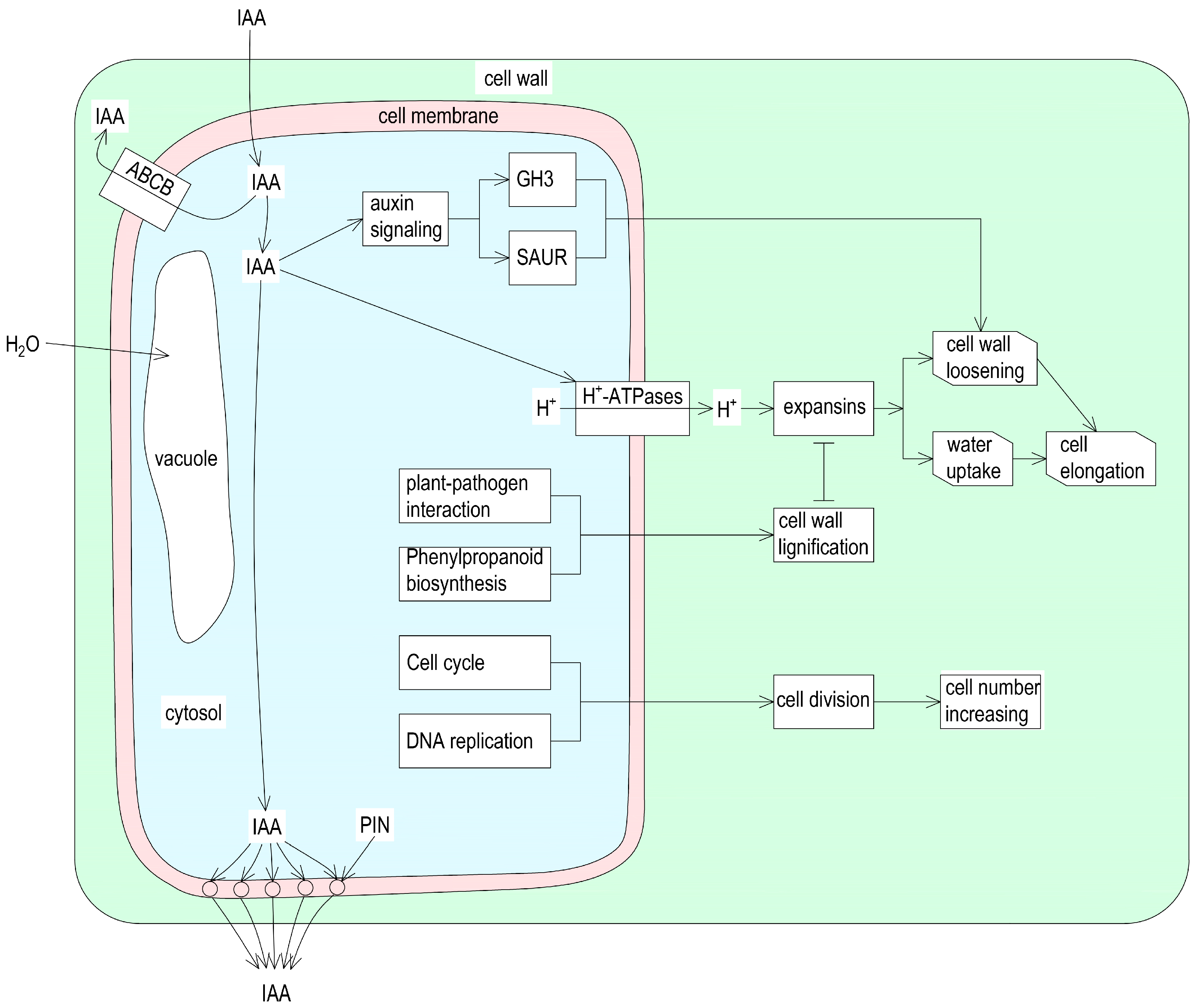
3.1. Auxin Is Required for Castor Internode Elongation
3.2. Secondary Cell Wall Lignification Hinders Internode Elongation
4. Materials and Methods
4.1. Plant Materials
4.2. Tissue Fixation, Staining, and Microscopy
4.3. Plant Hormone Measurement
4.4. Lignin and Cellulose Measurements
4.5. Metabolome Comparative Analysis
4.6. Transcriptome Comparative Analysis
4.7. Real-Time qPCR
4.8. Correlation Analysis of Transcriptom and Metabolomes
4.9. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef]
- Mao, L.; Zhang, L.; Evers, J.B.; van der Werf, W.; Liu, S.; Zhang, S.; Wang, B.; Li, Z. Yield components and quality of intercropped cotton in response to mepiquat chloride and plant density. Field Crops Res. 2015, 179, 63–71. [Google Scholar] [CrossRef]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Shan, F.; Wang, C.; Yan, C.; Dong, S.; Xu, Y.; Gong, Z.; Ma, C. Internode elongation pattern, internode diameter and hormone changes in soybean (Glycine max) under different shading conditions. Crop Pasture Sci. 2020, 71, 679–688. [Google Scholar] [CrossRef]
- Chen, R.; Fan, Y.; Yan, H.; Zhou, H.; Zhou, Z.; Weng, M.; Huang, X.; Lakshmanan, P.; Li, Y.; Qiu, L.; et al. Enhanced activity of genes associated with photosynthesis, phytohormone metabolism andcell wall synthesis is involved in gibberellin-mediated sugarcaneinternode growth. Front. Genet. 2020, 11, 570094. [Google Scholar] [CrossRef]
- Yang, D.-L.; Yao, J.; Mei, C.-S.; Tong, X.-H.; Zeng, L.-J.; Li, Q.; Xiao, L.-T.; Sun, T.-P.; Li, J.; Deng, X.-W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Esmon, C.A.; Tinsley, A.G.; Ljung, K.; Sandberg, G.; Hearne, L.B.; Liscum, E. A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc. Natl. Acad. Sci. USA 2006, 103, 236–241. [Google Scholar] [CrossRef]
- Ko, J.-H.; Han, K.-H.; Park, S.; Yang, J. Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol. 2004, 135, 1069–1083. [Google Scholar] [CrossRef]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a systems approach to understanding plant cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef]
- Bacic, A.; Harris, P.J.; Stone, B.A. Structure and function of plant cell walls. Biochem. Plants 1988, 14, 297–371. [Google Scholar]
- Zhang, B.; Gao, Y.; Zhang, L.; Zhou, Y. The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 2021, 63, 251–272. [Google Scholar] [CrossRef]
- Xu, X.; Backes, A.; Legay, S.; Berni, R.; Faleri, C.; Gatti, E.; Hausman, J.F.; Cai, G.; Guerriero, G. Cell wall composition and transcriptomics in stem tissues of stinging nettle (Urtica dioica L.): Spotlight on a neglected fibre crop. Plant Direct 2019, 3, e00151. [Google Scholar] [CrossRef]
- Mellerowicz, E.J.; Baucher, M.; Sundberg, B.; Boerjan, W. Unravelling cell wall formation in the woody dicot stem. Plant Mol. Biol. 2001, 47, 239–274. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wu, D.; Yang, T.; Sun, C.; Wang, Z.; Han, B.; Wu, S.; Yu, A.; Chapman, M.A.; Muraguri, S.; et al. Genomic insights into the origin, domestication and genetic basis of agronomic traits of castor bean. Genome Biol. 2021, 22, 113. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Shi, Y.; Li, W.; Chen, S.; Wang, Y.; He, X.; Yin, X. RcPAL, a key gene in lignin biosynthesis in Ricinus communis L. BMC Plant Biol. 2019, 19, 181. [Google Scholar] [CrossRef]
- Liu, C.; Lu, J.; Yin, X.; Bi, C.; Wen, D.; Zheng, J.; Liu, S.; Shi, Z.; Cheng, Y.J. Genetic analysis of traits related to plant height in Ricinus communis L. based on QTL mapping. Acta Agron. Sin. 2014, 40, 751–759. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; He, Z.; Jia, J.; Qiao, W. Dwarf castor-oil plant hybrids yield and the main agronomic characters of correlation and multiple regression analysis. J. Inn. Mong. Univ. Natl. 2010, 25, 40–43. [Google Scholar]
- Zhao, J.; Zhu, G.; He, Z.; Bao, H. Analysis of yield components for the main castor varieties in Tongliao region. J. Inn. Mong. Univ. Natl. 2014, 29, 407–413. [Google Scholar]
- Feng, L.; Li, G.; He, Z.; Han, W.; Sun, J.; Huang, F.; Di, J.; Chen, Y. The ARF, GH3, and Aux/IAA gene families in castor bean (Ricinus communis L.): Genome-wide identification and expression profiles in high-stalk and dwarf strains. Ind. Crops Prod. 2019, 141, 111804. [Google Scholar] [CrossRef]
- Mazur, E.; Kurczyńska, E.U.; Friml, J. Cellular events during interfascicular cambium ontogenesis in inflorescence stems of Arabidopsis. Protoplasma 2014, 251, 1125–1139. [Google Scholar] [CrossRef]
- Reid, J.B.; Ross, J.J. Regulation of tissue repair in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 17241–17242. [Google Scholar] [CrossRef] [PubMed]
- Fendrych, M.; Leung, J.; Friml, J. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 2016, 5, e19048. [Google Scholar] [CrossRef] [PubMed]
- Wulf, K.E.; Reid, J.B.; Foo, E. Auxin transport and stem vascular reconnection—Has our thinking become canalized? Ann. Bot. 2019, 123, 429–439. [Google Scholar] [CrossRef]
- Ju, F.; Liu, S.; Zhang, S.; Ma, H.; Chen, J.; Ge, C.; Shen, Q.; Zhang, X.; Zhao, X.; Zhang, Y.; et al. Transcriptome analysis and identification of genes associated with fruiting branch internode elongation in upland cotton. BMC Plant Biol. 2019, 19, 415. [Google Scholar] [CrossRef]
- McKay, M.J.; Ross, J.J.; Lawrence, N.L.; Cramp, R.E.; Beveridge, C.A.; Reid, J.B. Control of Internode Length in Pisum sativum (Further Evidence for the Involvement of Indole-3-Acetic Acid). Plant Physiol. 1994, 106, 1521–1526. [Google Scholar] [CrossRef] [PubMed]
- Sorce, C.; Picciarelli, P.; Calistri, G.; Lercari, B.; Ceccarelli, N. The involvement of indole-3-acetic acid in the control of stem elongation in dark- and light-grown pea (Pisum sativum) seedlings. Plant Physiol. 2007, 165, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Wei, Q.; Hu, Y.; Chen, M.; Chen, Z.; Zheng, S.; Ma, Q.; Luo, Z. Physiological and full-length transcriptome analyses reveal the dwarfing regulation in trifoliate orange (Poncirus trifoliata L.). Plants 2023, 12, 271. [Google Scholar] [CrossRef]
- Bai, Y.; Cai, M.; Mu, C.; Cheng, W.; Zheng, H.; Cheng, Z.; Li, J.; Mu, S.; Gao, J. New insights into the local auxin biosynthesis and its effects on the rapid growth of moso bamboo (Phyllostachys edulis). Front. Plant Sci. 2022, 13, 858686. [Google Scholar] [CrossRef]
- Wei, X.; Zhou, H.; Xie, D.; Li, J.; Yang, M.; Chang, T.; Wang, D.; Hu, L.; Xie, G.; Wang, J.; et al. Genome-wide association study in rice revealed a novel gene in determining plant height and stem development, by encoding a WRKY transcription factor. Int. J. Mol. Sci. 2021, 22, 8192. [Google Scholar] [CrossRef]
- Kebrom, T.H. A growing stem inhibits bud outgrowth—The overlooked theory of apical dominance. Front. Plant Sci. 2017, 8, 1874. [Google Scholar] [CrossRef] [PubMed]
- Barbier, F.F.; Dun, E.A.; Beveridge, C.A. Apical dominance. Curr. Biol. 2017, 27, R864–R865. [Google Scholar] [CrossRef]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Gray, W.M.; Muskett, P.R.; Chuang, H.W.; Parker, J.E. Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 2003, 15, 1310–1319. [Google Scholar] [CrossRef]
- Giri, J.; Bhosale, R.; Huang, G.; Pandey, B.K.; Parker, H.; Zappala, S.; Yang, J.; Dievart, A.; Bureau, C.; Ljung, K.; et al. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat. Commun. 2018, 9, 1408. [Google Scholar] [CrossRef]
- Noh, B.; Murphy, A.S.; Spalding, E.P. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 2001, 13, 2441–2454. [Google Scholar]
- Rayle, D.L.; Cleland, R. Enhancement of wall loosening and elongation by Acid solutions. Plant Physiol. 1970, 46, 250–253. [Google Scholar] [CrossRef]
- Rayle, D.L.; Cleland, R. Control of plant cell enlargement by hydrogen ions. Curr. Top. Dev. Biol. 1977, 11, 187–214. [Google Scholar]
- Sieburth, L.E. Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol. 1999, 121, 1179–1190. [Google Scholar] [CrossRef] [PubMed]
- Mattsson, J.; Sung, Z.R.; Berleth, T. Responses of plant vascular systems to auxin transport inhibition. Development 1999, 126, 2979–2991. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Liu, Y.; Li, L.; Wang, X.; Zhou, Y.; Zhang, M.; Li, Z.; Yi, F.; Duan, L. Deciphering transcriptional mechanisms of maize internodal elongation by regulatory network analysis. J. Exp. Bot. 2023, 74, 4503–4519. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Cui, D.; Ye, Z.-H. Secondary cell wall biosynthesis. New Phytol. 2019, 221, 1703–1723. [Google Scholar] [CrossRef] [PubMed]
- Ranocha, P.; Chabannes, M.; Chamayou, S.; Danoun, S.; Jauneau, A.; Boudet, A.-M.; Goffner, D. Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 2002, 129, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]


| #ID | Name | log2FC | p Value | VIP | Regulated |
|---|---|---|---|---|---|
| pma0294 | Chrysoeriol 5-O-hexoside | 1.77 | 0.01 | 1.59 | up |
| pma1751 | Nicotinic acid-hexoside | 0.90 | 0.03 | 1.53 | up |
| pmb0542 | Cyanidin 3-O-malonylhexoside | 0.62 | 0.03 | 1.45 | up |
| pmb0576 | Apigenin O-malonylhexoside | 1.27 | 0.03 | 1.52 | up |
| pmb0607 | Chrysoeriol 7-O-hexoside | 1.58 | 0.03 | 1.62 | up |
| pmb0608 | Chrysoeriol O-malonylhexoside | 1.79 | 0.04 | 1.59 | up |
| pmb0852 | LysoPC 18:2 | 1.20 | 0.03 | 1.47 | up |
| pmb1240 | Phellodensin F | 0.61 | 0.05 | 1.45 | up |
| pmb2406 | LysoPC 17:0 | 0.76 | 0.04 | 1.43 | up |
| pmb2556 | Syringaldehyde O-glucoside | −0.71 | 0.01 | 1.53 | down |
| pmb2601 | 7-hydroxycoumarin-beta-rhamnoside | −1.13 | 0.02 | 1.52 | down |
| pmb2685 | 2-Deoxyribose 5′-phosphate | 0.83 | 0.02 | 1.50 | up |
| pmb2804 | 13-HPODE | 0.70 | 0.03 | 1.49 | up |
| pmb3023 | Eriodictyol C-hexoside | −1.41 | 0.04 | 1.42 | down |
| pmb3026 | Quercetin O-acetylhexoside | 0.74 | 0.05 | 1.61 | up |
| pmb3028 | Tricin O-sinapic acid | 1.13 | 0.01 | 1.57 | up |
| pme0208 | Amentoflavone | 1.48 | 0.00 | 1.62 | up |
| pme0250 | Azelaic acid | 0.64 | 0.02 | 1.51 | up |
| pme0398 | Chlorogenic acid (3-O-Caffeoylquinic acid) | −1.19 | 0.01 | 1.55 | down |
| pme0413 | Vanillin | 0.82 | 0.05 | 1.42 | up |
| pme0534 | Gluconic acid | 1.16 | 0.00 | 1.60 | up |
| pme0543 | Indole-5-carboxylic acid | 0.64 | 0.03 | 1.50 | up |
| pme1521 | Dihydroquercetin (Taxifolin) | 0.62 | 0.02 | 1.54 | up |
| pme1583 | Eriodictyol | −1.13 | 0.01 | 1.59 | down |
| pme1622 | Kaempferol 3-O-glucoside (Astragalin) | −0.66 | 0.01 | 1.56 | down |
| pme1637 | Coniferyl alcohol | 1.17 | 0.04 | 1.43 | up |
| pme2054 | L-Tryptophan | −1.08 | 0.03 | 1.52 | down |
| pme2901 | 1-O-Caffeoyl quinic acid | −1.59 | 0.01 | 1.60 | down |
| pme2957 | Naringenin chalcone | −0.73 | 0.03 | 1.50 | down |
| pme2984 | Isosakuranetin-7-neohesperidoside (Poncirin) | −1.59 | 0.02 | 1.55 | down |
| pme3123 | Sinapyl alcohol | 1.22 | 0.00 | 1.60 | up |
| pme3191 | Uridine 5′-monophosphate | −0.87 | 0.04 | 1.45 | down |
| pme3210 | Genistein 7-O-Glucoside (Genistin) | 1.39 | 0.03 | 1.58 | up |
| pme3246 | Coniferin | −0.60 | 0.03 | 1.50 | down |
| pme3396 | Fustin | −1.02 | 0.00 | 1.63 | down |
| pme3431 | “Esculin (6,7-Dihydroxycoumarin-6-glucoside)” | −1.01 | 0.02 | 1.51 | down |
| pme3443 | Sinapinaldehyde | 1.48 | 0.00 | 1.58 | up |
| pmf0057 | “4,2′,4′,6′-Tetrahydroxychalcone” | −0.74 | 0.00 | 1.61 | down |
| pmf0232 | Tiliroside | −1.62 | 0.01 | 1.55 | down |
| pmf0348 | “2,6-Dimethyl-7-octene-2,3,6-triol” | 0.65 | 0.02 | 1.53 | up |
| pmf0455 | Peimine | 2.32 | 0.00 | 1.63 | up |
| pmf0559 | Abrine | 1.16 | 0.02 | 1.56 | up |
| Genes | Primer Sequences | |
|---|---|---|
| LOC107262217 (PGLR) | F: GACCTGGACATGGCATTAGTATAG | R: CCATTAGCGGTGTTTGAGAGA |
| LOC8265938 (NAC) | F: CTATAGCCGGAGAACACAGAATAG | R: CAACTCTCTTGTTGGCGTTTG |
| LOC8265956 (HSP7C) | F: ATACCACCATTCCCACAAAGA | R: CTTGCTCTCTCACCCTCATAAA |
| LOC8284441 (CLPB1) | F: CACTGATGCTGCATTGGATTAC | R: TAACTGTGTCACCACCTTCTTC |
| LOC107260773 (SAU50) | F: GCCGAAGAAGAGTTTGGATTTG | R: GTGACGATGTTAGAGAGCGAAA |
| LOC8288420 (PME45) | F: CCCTCATTGGTGGCTTCATC | R: CTTACAGCCTGCAATCCACC |
| LOC8258115 (PER) | F: ATTCTCGCCGCCTTCTACAG | R: GTGGGAGCTTGGCATTGTTG |
| LOC8269707 (Actin) | F: AGGAGTTGGGTGTGTTCATTC | R: ACAAGGACTCCACAGCTTTATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Orlov, Y.L.; Chen, M. Deciphering the Molecular Mechanism of the Intermediate Secondary Growth and Internode Elongation of the Castor Bean (Ricinus communis L.) by the Combined Analysis of the Transcriptome and Metabolome. Int. J. Mol. Sci. 2024, 25, 1053. https://doi.org/10.3390/ijms25021053
Chen Y, Orlov YL, Chen M. Deciphering the Molecular Mechanism of the Intermediate Secondary Growth and Internode Elongation of the Castor Bean (Ricinus communis L.) by the Combined Analysis of the Transcriptome and Metabolome. International Journal of Molecular Sciences. 2024; 25(2):1053. https://doi.org/10.3390/ijms25021053
Chicago/Turabian StyleChen, Yujie, Yuriy L. Orlov, and Ming Chen. 2024. "Deciphering the Molecular Mechanism of the Intermediate Secondary Growth and Internode Elongation of the Castor Bean (Ricinus communis L.) by the Combined Analysis of the Transcriptome and Metabolome" International Journal of Molecular Sciences 25, no. 2: 1053. https://doi.org/10.3390/ijms25021053
APA StyleChen, Y., Orlov, Y. L., & Chen, M. (2024). Deciphering the Molecular Mechanism of the Intermediate Secondary Growth and Internode Elongation of the Castor Bean (Ricinus communis L.) by the Combined Analysis of the Transcriptome and Metabolome. International Journal of Molecular Sciences, 25(2), 1053. https://doi.org/10.3390/ijms25021053








