Abstract
Psoriasis and atopic dermatitis fall within the category of cutaneous immune-mediated inflammatory diseases (IMIDs). The prevalence of IMIDs is increasing in industrialized societies, influenced by both environmental changes and a genetic predisposition. However, the exact immune factors driving these chronic, progressive diseases are not fully understood. By using multi-omics techniques in cutaneous IMIDs, it is expected to advance the understanding of skin biology, uncover the underlying mechanisms of skin conditions, and potentially devise precise and personalized approaches to diagnosis and treatment. We provide a narrative review of the current knowledge in genomics, epigenomics, and proteomics of atopic dermatitis and psoriasis. A literature search was performed for articles published until 30 November 2023. Although there is still much to uncover, recent evidence has already provided valuable insights, such as proteomic profiles that permit differentiating psoriasis from mycosis fungoides and β-defensin 2 correlation to PASI and its drop due to secukinumab first injection, among others.
1. Introduction
Psoriasis and atopic dermatitis are cutaneous immune-mediated inflammatory diseases (IMIDs) and, as such, belong to a spectrum of inflammatory conditions with two pathophysiologic poles: autoinflammatory and autoimmune. Autoinflammatory diseases are caused by the activation of the innate immune system, which leads to systemic inflammation [1,2]. On the other hand, autoimmune diseases are caused by the activation of the adaptive immune system, with high levels of autoantibodies and self-reactive lymphocytes, leading to inflammation and derangement of local tissues. Some diseases can be classified as either (predominantly) autoimmune (e.g., pemphigus vulgaris) or autoinflammatory diseases (e.g., familial Mediterranean fever), but psoriasis and atopic dermatitis are considered to be mixed-pattern diseases [2,3].
The incidence of IMIDs is growing in industrial societies; environmental changes combined with a genetic background trigger the development of IMIDs [4,5,6]. Recent evidence underpins the presence of shared common pathways among IMIDs [2,6,7], but knowledge of the immune factors that drive these chronic progressive diseases is still incomplete. Treatments of IMIDs are effective in providing clinical benefits to patients, but long-term disease control is still largely unmet [6].
Multi-omics, in the context of dermatology, refers to the use of various omics technologies to comprehensively study and analyze the skin and its related conditions. “Omics” fields encompass various biological data types, and in dermatology, this may include genomics (study of genes and DNA), transcriptomics (study of gene expression), proteomics (study of proteins), metabolomics (study of small molecules and metabolites), and microbiomics (study of the skin microbiome) [8,9,10]. The goal is to advance our knowledge of skin biology, better understand the underlying mechanisms of skin conditions, and potentially develop more precise and personalized approaches to diagnosis and treatment. For example, it can help identify specific biomarkers associated with skin diseases or responses to therapies, ultimately leading to more effective and tailored treatments for dermatological conditions. Due to the considerable length of the manuscript, we found it necessary to constrain the scope of our review. Hereby, we review the current knowledge of genomics, epigenomics, and proteomics in psoriasis and atopic dermatitis. Nevertheless, intriguing facts are also being unveiled by other omics technologies. For example, the combination of proteomics, single-cell transcriptomics, and spatial transcriptomics by Mitamura et al. uncovered the cellular crosstalk between the immune cells involved in skin lesions of atopic dermatitis [11]. Also, metabolomics has enabled the identification of urine metabolites associated with IMIDs, which could be valuable for their diagnosis [12].
2. Material and Methods
An electronic literature search was conducted on the Medline/PubMed database until November 2023, using Medical Subject Headings terms and pertinent medical terminology. The search criteria encompassed the following terms: ‘psoriasis’, ‘atopic dermatitis’, ‘genetic testing’, ‘genomics’, ‘genetics’, ‘epigenomics’, ‘epigenetics’, and ‘proteomics’. We considered original genome studies, reviews, systematic reviews, and meta-analyses that were specifically relevant to the genetic, epigenetic, or proteomic investigation of atopic dermatitis and psoriasis. Manuscripts written in the English language were eligible for inclusion, while letters to the editor, editorials, expert opinions, conference proceedings, studies exclusively involving patients with psoriatic arthritis, those focused on specific psoriasis treatments, or those centered on transcriptomics or proteomics analysis were excluded. The process of selecting publications was performed by two independent researchers (LR and LP), and any disparities were resolved through consensus.
3. Current Knowledge in Pathogenesis of Atopic Dermatitis and Psoriasis
Atopic dermatitis (AD) is the most common chronic cutaneous IMID, with a prevalence of 10% among adults and 20% in children [13]. The underlying pathophysiological mechanisms in AD are complex, encompassing a pronounced genetic susceptibility, epidermal dysfunction, and inflammation driven by T-cells [14,15,16,17,18]. Both the innate and adaptive immune systems contribute to its etiopathogenesis [14,16]. AD was initially considered a purely Th2-mediated inflammatory disease since most patients show high counts of eosinophils and high levels of immunoglobulin E (IgE). However, the immunological pathway of AD is complex, with predominant activation of Th2/Th22 and variable activation of Th17/Th1 lymphocytes; a biphasic switch from Th2 to Th1 responses has been reported in both acute and chronic cutaneous lesions of patients with AD [14,15,16]. The main cytokines related to AD etiopathogenesis are IL-4 and IL-13 [19,20]. They play a crucial role in the differentiation of Th2 cells and the production of IgE. Acute AD skin shows increased expression of Th2 cytokines, like IL-4, IL-5, and IL-13 [20]. In addition, IL-4 and IL-13 cause a disruption in epidermal barrier integrity by decreasing the number of main terminal differentiation proteins, such as filaggrin, loricrin, and involucrin. Levels of IL-13 correlate with the disease severity of AD [21,22,23].
Psoriasis is a chronic skin IMID with a prevalence of 1–3% in Western populations. The pathogenesis of psoriasis is characterized by an interaction of keratinocytes with the innate and adaptive immune systems [24,25,26]. Multiple theories have been put forth attempting to explain the pathogenesis of psoriasis [26,27,28]. The most accepted contemplates an initiation phase followed by a chronic inflammatory phase that is sustained by a feed-forward mechanism based on cytokine-mediated keratinocyte activation and proliferation [29,30]. Genetically predisposed patients become exposed to a trigger (trauma, infection, drugs) that will induce keratinocytes to undergo apoptosis or necrosis, releasing nucleic acids (DNA or RNA) that may induce a type I interferon-mediated autoinflammatory response, and potential autoantigens such as cathelicidin (LL37), ADAMTSL-5, and phospholipase A2 (PLA2G4D). LL37 interacts and forms complexes with self-DNA or -RNA that will trigger the innate immune system via Toll-like receptors (TLR) [31,32]. The activation of TLR7 and TLR8 leads to the production of interferon (IFN)-α and IFN-β by keratinocytes and plasmacytoid dendritic cells (pDCs), and interleukin (IL)-6 and tumor necrosis factor (TNF) by myeloid dendritic cells (mDCs). IL-6 induces differentiation of CD4+ naive T-cells into T helper (Th)-17 cells [33,34], whereas type I IFNs and TNF promote the secretion of IL-12 and IL-23 by mDCs [35,36]. IL-12 and IL-23 are clue cytokines in the immune chain reaction causing psoriasis. CD4+ naïve T-cells differentiate into Th1 cells upon exposure to IL-12, along with tumor growth factor (TGF)-β and IL-6 [37,38,39,40,41]. IL-23 leads to Th17 cell development and activates αβT-cells [37,38,39,40,41]. Th1-activated cells exhibit a distinctive cytokine secretion profile, including IFN-γ and TNF. Meanwhile, Th17-activated cells release IL-17, IL-22, and TNF [42]. Together, these cytokines induce keratinocyte proliferation, differentiation, and inflammatory activation, although IL-17A constitutes the main effector cytokine driving psoriasis pathogenesis and is essential for the development and maintenance of psoriasis plaques [42]. Inflammatory cytokines such as IL-1, IL-6, and TNF, chemokines (including CXCL1, CXCL2, and CXCL3), and AMPs (including S100A7/8/9, human-defensin 2, and LL-37) are produced by stimulated keratinocytes, which attract and activate immune cells and exacerbate psoriatic inflammation [40,43,44]. S100A8/A9 is overexpressed in keratinocytes and innate immune cells, and their transcripts are significantly overexpressed in psoriasis lesions compared to non-lesional psoriasis or atopic dermatitis (AD) skin [45]. Furthermore, psoriasis treatment has been shown to reduce S100A8/A9 levels. Christmann et al. identified the induction of S100-alarmins in an imiquimod-induced murine model of psoriasis-like skin inflammation, which was associated with increased expression of IL-1α, IL-6, IL-17A, or TNF [46]. However, recent evidence has observed that lower epidermal levels of S100A9 in mice lead to more severe psoriasis skin lesions [47].
The maintenance phase of psoriatic inflammation is driven by this positive feedback loop between keratinocytes and T lymphocytes, enhancing further epidermal hyperplasia and a sustained inflammatory response.
Psoriasis encompasses multiple clinical variants, such as pustular psoriasis, guttate psoriasis, and nail psoriasis, among others. Guttate psoriasis was previously thought to be closer to contact dermatitis than psoriasis; however, novel insights by using gene expression profiling and gene set enrichment scores have been observed that are more similar to chronic psoriasis [48]. Regarding pustular psoriasis, in contrast to psoriasis vulgaris, where the IL-23/17 axis plays a pivotal role, pustular psoriasis shows hyperactivation of innate immunity, prominently involving the IL-36 axis [49]. Analysis of gene expression in skin biopsy specimens obtained from individuals affected with either generalized pustular psoriasis (GPP) or plaque psoriasis has unveiled discernible patterns. Specifically, GPP lesions manifest increased expression of IL-1 and IL-36, coupled with diminished levels of IL-17A and interferon-c, when juxtaposed with lesions characteristic of plaque psoriasis [49]. The pathomechanisms of ungual psoriasis remain elusive; however, a variant in IL1RN has been identified in patients with nail psoriasis. IL1RN functions to regulate the proinflammatory activity of IL-1A. The latter has been demonstrated to induce nail changes, suggesting a potential association with nail involvement in patients affected by psoriasis [50,51].
The clinical manifestations and course of both psoriasis and AD are highly variable; most patients achieve satisfactory responses with currently available treatments, but clinical and therapeutic challenges can be vexing in some cases.
4. Omics of Atopic Dermatitis and Psoriasis
4.1. Genomics
4.1.1. Atopic Dermatitis
AD has a strong genetic background, with 70% to 80% heritability [5,52]. The development of AD is significantly influenced by genetic factors, and a family history of AD is the strongest risk factor for this disease [13].
Genetic linkage studies, genome-wide association studies (GWAS), candidate gene techniques, and, more recently, next-generation sequencing (NGS) technologies have all been used to identify the genetic risk factors underlying AD and psoriasis. Genetic linkage studies correlate the inheritance of genetic markers with the existence of clinical features. The first genome-wide linkage study for AD in 2000 identified a major locus of susceptibility on 3q21 [53]. Additional genome-wide linkage studies on AD were performed in the following years, leading to the identification of numerous other loci, including 1q21, 3p, 17q, 18q, and 11.13q. But these loci were frequently excessively wide, necessitating labor-intensive fine mapping [54]. The most recent and largest genome-wide association meta-analysis of AD—including 65,107 AD cases and 1,021,287 controls—was published in October 2023. The authors identified a total of 91 associated loci [55]. Among them, 81 loci were initially identified in individuals of European ancestry, but further studies showed that most of them were replicated in data from other population ancestries. Eight loci were detected in at least one of the populations: European, Latino, and African ancestry, and the remaining 2 loci could be specific to individuals of East Asian ancestry (pending further studies). As stated, most loci identified were shared among ancestries of different populations. However, some loci shared between European, Latino, and Japanese ancestries did not show evidence of association in individuals of African ancestry. Given the distinct AD phenotypes identified among European, Asian, and African individuals, it is tempting to hypothesize that variations in genetic associations at certain loci may be a factor accounting for these clinical phenotypes [55].
The candidate gene technique used to uncover loci linked to the onset and severity of AD has been employed in more than 30 studies. In this method, variation in genes suspected of being involved in disease pathogenesis is compared between affected and healthy individuals [56].
Progress in research has led to the identification of more than 70 genes associated with AD; they can be classified into five groups: genes leading to cutaneous barrier dysfunction, genes associated with altered innate responses, genes associated with acquired immune responses, genes associated with stress responses of the keratinocytes, and genes involved in the metabolism of vitamin D [57,58], as detailed in Table 1.

Table 1.
Genomics of AD.
One of the most well-known genes linked to both disease susceptibility and disease severity is FLG, the gene encoding for filaggrin, the main protein responsible for the maintenance of cutaneous epithelium integrity [58]. Null mutations in the FLG gene result in a deficiency in the epidermal barrier [59,70,71,72]. Patients with heterozygous mutations have an 8-fold increased risk of acquiring AD, while carriers with two mutant alleles are almost always affected by it [58,73].
Apart from FLG, two other key genes have been identified: OLOV1 (Ovo Like Transcriptional Repressor 1) and IL-13. The former is a transcriptional factor that regulates filaggrin expression [60]; the latter mediates Th2 cell responses [57,58], which predominate in AD. The expression of cutaneous barrier proteins such as filaggrin, loricrin, involucrin, cell adhesion proteins, desmosine, and claudins is decreased by Th2 responses with secretion of IL-4 and IL-13 [57,58]. Additionally, Th2 responses disrupt the homeostasis of other epithelia, which results in an unbalanced immune response that is ultimately linked to systemic inflammation, airway hyperresponsiveness, and food allergies [57,74,75,76]. IL-4 gene polymorphisms have also been related to an increased susceptibility to AD [66]. Indeed, IL-4 and IL-13 are successfully targeted by novel biologics used in AD treatment [77].
IL-10 is an anti-inflammatory cytokine that contributes to the modulation of acquired immune and anti-inflammatory responses [67,78]. Polymorphisms in the promoter region of the IL-10 gene can affect the function of IL-10. Among the several polymorphisms identified in the meta-analysis by Zhao et al. two stand out: IL-10-819G/A and IL-10-1082G/A. The former seems to be associated with an increased risk of AD in Caucasian populations, whereas the latter would be associated with an increased risk of AD in Asian populations [67]. However, according to another meta-analysis performed in the same year, available evidence did not support a strong genetic relationship between the IL-10-1082G/A polymorphism and AD risk [78].
IL-6 plays an important role in host defense mechanisms and can act as both a pro-inflammatory cytokine by stimulating the production of IL-4 and IL-5—predominant Th2 cytokines in acute AD—and as an anti-inflammatory cytokine by inhibiting IL-1 and TNF and activating IL-10 and IL1Rα. Single nucleotide polymorphisms (SNPs) in the IL6 gene have been associated with an increased inflammatory response, as in AD [68]. In addition, SNPs in IL6R have also been associated with an increased risk of AD [69].
Alterations in genes codifying proteins that participate in the integrity of the epidermis have also been related to AD. For example, the COL5A3 (Collagen type V Alpha 3 Chain) and the Matrix metalloproteinase (MMP)-9 genes [61]. COL5A3 codifies for type V collagen, which is found in the dermoepidermal junction and in the extracellular matrix associated with type I collagen. The role of MMP9 in tissue remodeling and collagen deposition is well known; it also participates in inflammation by facilitating cellular traffic, including neutrophils and eosinophils. MMP9 would mediate the degradation of type V collagen, leading to eczema susceptibility [61]. COL29A1 gene, which encodes for collagen expressed in the skin, lung, and gastrointestinal tract, has also been associated with AD by genetic linkage. In the outer layers of the epidermis, a lack of collagen XXIX expression would derange keratinocyte cohesion and the integrity and function of the epidermis [62].
Other genes that have been implicated in the pathogenesis of AD are SPINK5 (Serine Peptidase Inhibitor Kazal Type 5), which produces a serine protease inhibitor that contributes to maintaining the skin barrier [63,79], signal transducer and activator of transcription (STAT), thymic stromal lymphopoietin (TLSP), interferon regulatory factor 2 (IRF2), TLR 2, high-affinity IgE receptor (FcεRI α), vitamin D receptor (VDR) [57], ACTL9 (Actin Like 9), KIF3A (Kinesin Family member 3A) [64], LCE1D (Late Cornified Envelope 1D), SPRR3 (Small Proline Rich Protein 3), and S100A3 (S100 Calcium Binding Protein A3) [65].
4.1.2. Psoriasis
Psoriasis is a complex disease characterized by multifactorial inheritance with a strong genetic background and greater than 60% heritability [80,81]. Epidemiological studies revealed a higher prevalence of psoriasis vulgaris among first- and second-degree relatives of affected individuals compared to the general population [82]. Approximately 30% of individuals with psoriasis vulgaris have a first-degree relative affected by the condition. In cases where both parents and a sibling are affected, there is a 50% likelihood of another child developing psoriasis vulgaris. However, if the sibling is affected without the parents’ showing symptoms, the risk decreases to 8%. Moreover, the risk of psoriasis vulgaris is notably elevated in monozygotic twins, being two to three times greater than in dizygotic twins [82].
A minimum of 12 distinct loci suspected to contain genes related to psoriasis susceptibility (PSORS) were identified by genetic linkage studies [83]. However, most of these findings could not be replicated, underscoring the limitations of genetic linkage [80]. PSORS1 was the first locus associated with psoriasis susceptibility; it is the locus with the greatest genetic effect and represents 35–50% of psoriasis heritability explained by known loci [80]. PSORS1 contains HLA-Cw6, the major psoriasis susceptibility allele, located at 6p21.3 [84], as well as the candidate gene corneodesmosin (CDSN), which codifies for a desmosomal protein involved in keratinocyte cohesion and desquamation [85]. PSORS2 and PSORS4 loci have also been validated. PSORS2 contains the CARD14 (caspase recruitment domain family member 14) gene, which encodes for a nuclear factor kB (NF-kB) activator; some of its variations have been linked to common and rare variants of psoriasis [86,87]. Lastly, PSORS4 contains LCE genes, which codify stratum corneum proteins that mediate in terminal epidermal differentiation [88].
Genetic linkage studies have been replaced by newer and more potent analytical techniques that permit a thorough examination of genetic risk variation at the genome-wide level (GWAS, NGS). Genome-wide association studies (GWAS) employ highly optimized microarrays capable of efficiently and robustly genotyping millions of genetic markers throughout the genome [80]. By leveraging substantial sample sizes, GWAS facilitates the detection of even minor distinctions in allele frequencies between individuals with the disease and those without and provides a significantly more potent approach compared to linkage analysis. However, GWAS only permits uncovering statistical relationships; in order to identify potential causal susceptibility alleles, genotyping arrays with dense coverage in regions of interest have been used. To enhance the information obtained by GWAS, additional SNPs not included in the GWAS have been genotyped [80]. These studies allowed the identification of additional susceptibility loci and uncovered genes involved in innate immunity, such as DDX58 and CARD14 [89], and genes related to type I interferon signaling (IFIH1 and TYK2) [90]. Nevertheless, nowadays the prevailing practice consists of conducting genome-wide imputation through freely available computational resources. Imputation plays a crucial role in enabling extensive meta-analyses in psoriasis by amalgamating data produced by diverse GWAS platforms [80]. In fact, the largest genetic study of psoriasis susceptibility undertaken to date, a GWAS meta-analysis of 18 international case-control studies, has identified genetic variants at 109 distinct loci, 45 of which had not been previously reported [91]. They include susceptibility variants located within loci that encode therapeutic targets such as IL17RA and AHR, along with deleterious coding variants that suggest potential novel drug targets, including those found in STAP2, CPVL, and POU2F3 [91].
Genes that have been linked to psoriasis participate in pathways of the epidermal barrier, the innate immune system, and the adaptative immune system, as detailed in Table 2 and discussed as follows.

Table 2.
Genomics in psoriasis.
Structural and functional alterations of keratinocytes participate in the pathogenesis of psoriasis, especially as regards its onset [102,106]. A higher risk of psoriasis has been associated with alterations in LCE genes, especially LCE3B and LCE3C, genes coding for defensins (DEFB4), and the GJB2 gene [92,93,94].
The innate immune system plays a critical role in psoriasis pathogenesis. Its stimulation is necessary for the subsequent activation of the adaptive immune system [83]. Gene candidates for innate immunity pathways in psoriasis have been found through numerous single nucleotide polymorphisms (SNPs) [83]. Transcription of these innate immune genes may reduce the threshold necessary to activate the pathogenic adaptive immune response [83,105]. NF-kB plays a critical role in mediating the transcription of several genes that participate in the innate response [107,108]. Some of them stand out: C-REL [95], which is also related to keratinocyte growth; TRAF3-interacting protein 2 (TRAF3IP2) [96], which mediates the interaction of the innate and adaptive immune systems; and genes encoding CARD proteins [86]. The interferon pathway and antiviral response genes, such as Interferon Induced with Helicase C Domain 1 (IFIHI) and RNA sensor RIG-I (known as DDX58), have also been associated with psoriasis [109]. Alterations in the downregulation of the NF-kB pathway, due to mutations in negative regulators, have also been related to psoriasis susceptibility. These mutations reduce the ability to control inflammation and are as important as mutations inducing overactive immune responses [110]. Some of the genes implicated are TNF-α-inducible protein 3 (TNFAIP3), TNFAIP3 interacting protein 1 (TNIP1), the NF-kB inhibitor α (NFKBIA), and the zinc finger DHHC-type containing 23 (ZC3H12C) [89,92,97].
The major histocompatibility complex (MHC) accounts for one-third of the total genetic influence on psoriasis, highlighting the important pathogenetic role of the antigen presentation pathway in psoriasis [102]. PSORS1 risk variant contains HLA-Cw06. This is present in 20–50% of the patients with psoriasis, compared to 16% of individuals without the disease. Psoriasis can be considered an MHC-1-opathy, in which self-immunogenic peptides are presented by the psoriasis-linked MHC-1 alleles to effector cells, leading to the inflammatory response [98].
Interactions between MHC class I risk alleles and variations in endoplasmic reticulum aminopeptidase 1 (ERAP1) have been identified by GWAS. ERAP1 is an aminopeptidase that prepares peptides for effective antigen presentation. These interactions have been linked to psoriasis risk [99]. Finally, other loci have also been associated with a higher risk of psoriasis [111]. For example, the MHC class I polypeptide-related sequence A (MICA), is found near the HLA-B locus in the MHC region. MICA acts as a ligand for an activating receptor in natural killer cells, NKT cells, and T-cells [83].
The Th1 signaling pathway carries out a crucial role in the pathogenesis of psoriasis, whose lesions have increased numbers of T-cells producing interferon (IFN)-γ [102,112]. IL-12 drives Th1 polarization of T-cells and secretion of IFN-γ, TNF, and IL-2. Multiple genetic psoriasis susceptibility loci affect different parts of the Th1 signaling pathway, including IL-12B (coding for the p40 subunit of IL-12) [100], tyrosine kinase 2 (TYK2) [101], ZC3H12C (transcript involved in macrophage activation) [89], STAT5A, STAT5B, and ILF3 (interleukin enhancer-binding factor 3) [102].
The IL-23/IL-17 pathway plays a major pathogenetic role in psoriasis since Th17 cytokines are the main effectors of keratinocyte alterations and inflammation in this disease [112,113]. Multiple SNPs associated with psoriasis risk have been identified in genomic regions corresponding to both subunits of IL-23 (p19 and p40) and IL23R [100,114]. In addition, the activation of IL-23R leads to downstream signaling through multiple molecules such as TYK2, JAK2, and STAT3 [103,104]. Mutations in genes coding for these molecules have also been shown to be associated with psoriasis [103,104]. Other genetic variants related to IL-23 and psoriasis risk include suppressor of cytokine signaling 1 (SOCS1) and ETS Proto-Oncogen 1 (ETS1) [102]. On the other hand, no association has been yet established between the IL17A gene and psoriasis, but an SNP in the IL17R gene is related to disease susceptibility [115]. IL22 has been associated with psoriasis risk quantitatively by GWAS: a higher number of IL22 gene copies leads to a higher risk of nail psoriasis [103].
The balance between Th17 and regulatory T-cells (Treg) is also altered in psoriasis, where Treg develop expression of RORγt and become functionally impaired; this can be partially explained by polymorphisms in genes such as TNF, IL12RB2, and IL12B [116,117,118,119].
The pathogenesis and genetic background of plaque psoriasis differ from that of generalized pustular psoriasis, an autoinflammatory disorder with special implications for neutrophils and IL-36 [120]. Loss-of-function mutations in IL36RN (coding for the antagonist of the IL-36 receptor) result in DITRA (Deficiency of The Interleukin-36-Receptor Antagonist), a monogenic autoinflammatory disorder with earlier onset and more severe presentation of pustular psoriasis [106,120]. Activating mutations of CARD14 (caspase recruitment domain family member 14), which codes for an inductor of NF-kB signaling expressed in keratinocytes, have also been observed in plaque psoriasis, psoriatic arthritis, and generalized (as well as localized) pustular psoriasis [120]. Other genes with mutations identified in patients with generalized pustular psoriasis include the adaptor-related protein complex 1 subunit sigma 3 (AP1S3) [121], mutations in Myeloperoxidase (MPO) genes [122], Serpin Family A Member 1 (SERPINA1) and SERPINA3, TNIP1, and IL1RN [120].
4.1.3. Mendelian Randomization
Mendelian randomization (MR) is a statistical technique that utilizes genetic variants to evaluate the causal relationship between a specific exposure and the occurrence of a particular disease and has been increasingly used in the study of AD and psoriasis [13]. The increasing availability of GWAS has contributed to the growing prominence of MR in exploring potential causal associations within observational studies. Genetic variants are used as surrogates for the targeted risk factor, and MR mitigates the potential for confounding and reverse causation, thereby yielding a more reliable estimate of causality.
Mendelian Randomization in AD
Multiple studies facing MR in AD have been published lately, including a systematic review in which 30 MR studies were included [123]. This systematic review points body mass index, gut microbial flora, the IL-18 signaling pathway, and gastroesophageal reflux disease as causal factors for AD. On the other hand, AD would be causal for several medical conditions, including health failure, rheumatoid arthritis, and conjunctivitis. Prior studies also disclosed that AD would carry a greater risk of type 2 diabetes and alopecia areata [124,125], whereas no association between AD and higher cancer risk has been found [126].
Mendelian Randomization in Psoriasis
Extensive Mendelian randomization approaches have been performed to identify a genetic relationship between psoriasis and several comorbid diseases. A systematic review, in which 27 MR studies were included, was recently published [127]. Exposures like smoking, obesity, cardiovascular disease, and Crohn’s disease were identified as causal factors for psoriasis and psoriatic arthritis. Whereas, psoriasis was found to be causally associated with cardiovascular complications (myocardial infarction, large artery stroke), a mild risk for lung cancer, COVID-19, and Parkinson disease.
Chalitsios et al. aimed to determine different clusters of co-existing long-term conditions (LTC) in patients with psoriasis [128]. By employing latent class analysis, the cross-sectional data of individuals diagnosed with psoriasis from the UK Biobank underwent examination to delineate specific co-morbidity profiles associated with psoriasis. Linkage disequilibrium score regression was subsequently used to calculate the genetic correlation existing between psoriasis and LTC. They identified 5 different clusters of psoriatic patients [128]. Patients included in clusters 1, 2 and 4 were associated with metabolic syndrome traits, cardiovascular diseases, airway and pulmonary diseases, and depression. Psoriatic patients included in cluster 3 were more prompt to suffer from psoriatic arthritis and rheumatoid arthritis, along with depression and hypertension. Finally, patients in cluster 5 were “relatively healthy”, showing a low probability of developing multimorbidity.
4.1.4. Ferroptosis, Pyroptosis, and Cuproptosis
Cell death plays a critical role in embryonic development, cell fate determination, and the maintenance of immune homeostasis. It is categorized into necroptosis, apoptosis, pyroptosis, cuproptosis, ferroptosis, and necrosis. In this chapter, we will focus on cuproptosis, ferroptosis, and pyroptosis [129].
Ferroptosis, a form of iron-dependent cell death, results in a toxic accumulation of reactive oxygen species. The disruption of iron homeostasis and the oxidation of phospholipids can instigate the occurrence of ferroptosis. Due to its distinctive mechanism, ferroptosis may play a role in determining cellular outcomes, the advancement of inflammatory processes, and various pathological conditions [130].
Pyroptosis, a recently identified form of programmed cell death, is orchestrated by pyroptotic caspases [129]. This modality is characterized by the prompt rupture of the plasma membrane, leading to the release of inflammatory intracellular contents. Recognition of pathogen-associated molecular patterns (PAMPs), damage-associated molecular patterns (DAMPs), and lipopolysaccharide (LPS) by specific inflammasomes and caspases induces the activation of pyroptosis pathways. There are two pyroptosis pathways: canonical and non-canonical. The former is caused by PAMPs and DAMPs, which permit the formation of inflammasomes. Inflammasomes cleave procaspase-1 into caspase-1; activated capsase-1 cleaves gasdermin D (GSDMD) and the maturation of IL-1B and IL-18. Whereas the non-canonical pathway, lipopolysaccharide can activate caspase-4/5/11 to induce pyroptosis by cleavage of GSDMD. Increasing evidence suggests the profound involvement of pyroptosis in infectious diseases, hematologic disorders, and tumorigenesis. Furthermore, activated inflammasomes triggered by both PAMPs and DAMPs are implicated in the initiation of chronic inflammation and autoimmune diseases.
Cuproptosis represents a novel type of cellular death associated with mitochondrial metabolism and facilitated through protein lipoylation [131]. The occurrence of cell death induced by copper ionophores depends predominantly upon the intracellular accumulation of copper. Research findings indicate that FDX1 and protein fatty acylation play pivotal roles as regulators in the context of cuproptosis.
Ferroptosis, Pyroptosis, and Cuproptosis in AD
Pyroptosis has been related to AD pathogenesis [132]. Research has focused on the identification of pyroptosis-related genes (PRGs). Recent studies have identified an increased expression of inflammasome PRGs, such as NLR Family Pyrin Domain Containing 3 (NLRP3) and Absent In Melanoma-2 (AIM2) in AD, which have been related to epidermal inflammation [133,134]. Also, genes of gasdermin C and GSDMD were found to be highly expressed in lesional skin-derived keratinocytes [135,136]. TNF, which plays a role in the pathogenesis of allergic inflammation in AD, forms a complex with caspase-8 and GSDMC. Pyroptosis mediated by the complex TNF-Caspase8-GSDMC could trigger keratinocyte inflammation and death [136]. All these findings suggest that PRGs may play a pivotal role in AD pathogenesis, and ultimately, the identification of pyroptosis-related biological markers (PRBMs) could provide a novel perspective on the diagnosis and treatment of AD [129].
To our knowledge, no ferroptosis or cuproptosis alterations related to AD pathogenesis have been identified yet.
Ferroptisis, Pyroptosis and Cuproptosis in Psoriasis
Ferroptosis is an iron-dependent, lipid peroxidation-mediated cell death pattern that is intricately linked to inflammation within psoriatic lesions [137]. The extent of the impact of ferroptosis on psoriasis remains largely undetermined, but keratinocytes isolated from psoriatic lesions exhibit irregularities in lipid metabolism and expression. At the individual cell level, there is pronounced activation of lipid oxidation and peroxidation in psoriasis lesional keratinocytes [138].
Ferroptosis can be activated by the generation of reactive oxygen species (ROS) [137]. ROS induces GPX4 (glutathione peroxidase 4) deletion. In normal conditions, GPX4 inhibits ferroptosis [139]. The GPX4 deletion decreases keratinocyte adhesion and increases intracellular lipid peroxidation [140]. GPX4 expression is decreased in psoriatic skin lesions compared to healthy skin. Furthermore, augmentation in the cellular import of iron indicates the activation of ferroptosis in psoriasis lesions [141]. Other genes that are related to ferroptosis and regulate the immune microenvironment in psoriasis are PEBP1, PRKAA2, and ACSF2 [138]. PEBP1 mediates ferroptosis vulnerability due to GPX4 deletion, and PRKAA2 induces ferroptosis by inhibiting the transcription of SLC7A11 [142]. Finally, a positive correlation between lipid oxidation and the Th22/Th17 pathway at a single-cell level has been demonstrated: Fer-1 (a specific inhibitor of ferroptosis) decreases inflammation in mice with imiquimod-induced psoriasis, with significantly decreased expression of TNF, IL-6, IL-1α, IL-1β, IL-17, IL-22, and IL-23 [139].
Pyroptosis plays a crucial role in the pathogenesis of disorders with an aberrant Th17 immune response, like psoriasis [143]. Keratinocytes apparently engage cell death by pyroptosis, paradoxically accelerating cell proliferation in the pathogenic environment of psoriasis. In addition, GSDMD-mediated pyroptosis would contribute to the inflammatory environment by allowing the release of DAMPs and inflammatory cytokines. Effective inhibition of pyroptosis with topical application of disulfiram 2% and 5% has been reported to reduce psoriasis-like inflammation in a murine model [143].
Cuproptosis and elevated copper levels in serum may be involved in the pathogenesis of psoriasis by mechanisms largely unknown [131]. The accumulation of intracellular copper leads to cell death, and the cuproptosis-related genes MTF1, ATP7B, and SLC31A1 are increasingly expressed in patients with psoriasis compared to patients without psoriasis [131].
4.2. Epigenomics
Epigenetics consists of the examination of reversible and inheritable alterations in gene expression that occur without changes in the DNA sequence [144]. Epigenetic modification of gene expression occurs either at the transcriptional or post-transcriptional level. The former is mediated by modifications in the chromatin structure that lead to the activation of transcription factors for specific genes without impacting the underlying DNA sequence (Figure 1). Epigenetic mechanisms at the post-transcriptional level are driven by non-coding RNAs (ncRNAs) that alter the structural properties of RNA, DNA, and proteins, modifying gene expression.
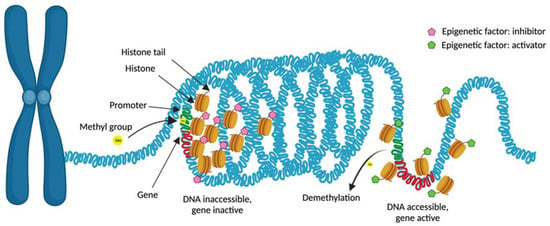
Figure 1.
Epigenetic mechanisms at the transcriptional level. Condensed chromatin (heterochromatin) limits gene transcription, whereas open chromatin (euchromatin) allows transcription factor activation and DNA transcription. Methylation of CpG islands in the promoter region of a gene inhibits its expression. Regarding histone modifications, multiple mechanisms can induce modifications in chromatin. Some of them will result in relaxed chromatin, allowing DNA transcription (activating modifications), while others will induce chromatin compaction, hence limiting gene transcription. Created with Biorender.com (accessed on 16 December 2023).
Epigenetic mechanisms are sensible to external stimuli, representing a link between genetics and the environment. Therefore, epigenetic mechanisms may have the potential to modify the susceptibility to diseases where environmental factors contribute to their etiopathogenesis, such as AD and psoriasis. Nonetheless, the impact of epigenetics is most important during embryogenesis, when pluripotency cells are highly common. The stabilized epigenome in differentiated cells is less susceptible to modification by environmental factors [145]. Epigenetic changes in patients with AD and psoriasis involve genes that regulate immune response and inflammatory processes and those that code for structural proteins of the epidermis [56,146,147,148,149].
The epigenome characteristics associated with any disease can be described according to the epigenetic mechanisms involved, which can alter gene transcription (namely, DNA methylation and histone modifications) or gene translation via non-coding RNAs, including microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA) [56,144,150].
4.2.1. DNA Methylation
DNA methylation is among the most prevalent epigenetic mechanisms that regulate gene expression. This process targets specific DNA sections known as CpG sites, where cytosine is followed by guanine. Around 70% of the human genome’s proximal promoter regions are made up of these main sequences [58,150,151]. DNA methylation consists of adding a methyl group to the C5 position of CpG dinucleotides (cytosine becomes 5-methylcytosine) by DNA methyltransferases (DNMTs); this allows methylcytosine-binding proteins (MBPs) to attach and induce chromatin compaction [152]. When DNA methylation occurs in the promoter region of a gene, this leads to suppression of gene expression since transcriptional factors cannot bind (Figure 1). Conversely, loss of DNA methylation permits gene expression due to chromatin decompaction [152]. Cytosine demethylation is catalyzed by ten-eleven translocation (TET) enzymes (Figure 2).
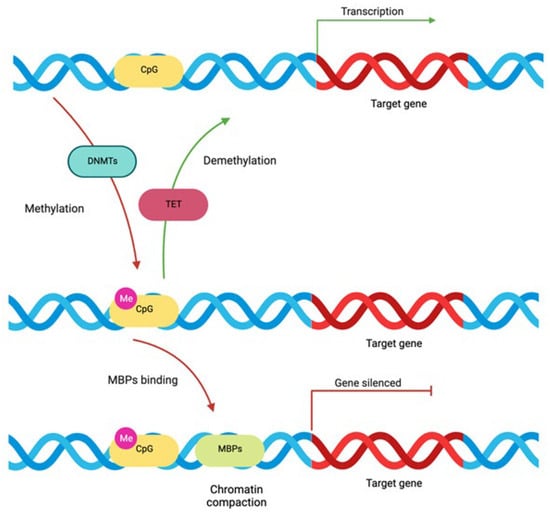
Figure 2.
DNA methylation process: Approximately 70% of the proximal promoter regions of the human genome are made up of CpG islands, where cytosine is followed by guanine. Methyltransferases (DNMTs) catalyze the addition of a methyl group to the C5 position of CpG dinucleotides (cytosine becomes 5-methylcytosine), allowing methylcytosine-binding proteins (MBPs) to attach and induce chromatin compaction. Therefore, DNA methylation leads to suppression of gene expression. Conversely, loss of DNA methylation by ten-eleven translocation (TET) enzymes induces chromatin decompaction and gene expression. Created with Biorender.com (accessed on 16 December 2023).
Several alterations in DNA methylation have been identified in AD and psoriasis, with potential etiopathogenetic implications.
DNA Methylation in AD
The DNA methylation profile observed in the skin of patients with AD is different from that of healthy individuals [56]. The keratinocytes obtained from AD lesions showed demethylation of the thymic stromal lymphopoietin gene (TSLP) promoter, which led to overexpression of the corresponding Th2-biasing alarmin in the epidermis [153]. The promoter of FCER1G, which codes for the high-affinity IgE receptor FcεRI, was also shown to be demethylated in the monocytes of AD patients, leading to the overexpression of this receptor on the surface of these cells [154].
AD pathogenesis in many patients is primarily driven by mutations that result in decreased filaggrin expression. According to research by Ziyab et al., heterozygotic carriers of the null mutations R501X, 2282del4, and S3247X may exhibit excessive methylation of the filaggrin gene (FLG). This interacts with epigenetic regulation to increase the risk of AD in the carriers [155]. Variants of another gene implicated in skin barrier function, kinesin family member 3A (KIF3A), are associated with an increased risk of AD and present alterations in DNA methylation [156,157].
Epigenetic changes play a fundamental role in the differentiation process of subpopulations of T lymphocytes [158,159]. Activated Th2 lymphocytes produce IL-4, IL-5, and IL-13 cytokines. Their epigenetic activation is related to the demethylation of IL-13 and IL-4 gene promoters and the methylation of the IFG gene promoter. On the other hand, activation of Th1 lymphocytes can be caused by methylation of CpG islands of the IL-4, IL-13, and IL17A gene promoters, methylation of the RORC gene transcription factor, and demethylation of IFG and TBX21 [158,159,160]. Treg deficiency, which is one of the hallmarks of AD pathogenesis, is associated with methylation of the FOXP3 promoter and demethylation of RORC [161,162,163].
DNA Methylation in Psoriasis
DNA methylation in psoriasis occurs in immune-active sites other than the skin. Moreover, this is associated with disease severity, the distribution of lesions, and the different tissue types involved [164]. Epigenome-wide methylation studies have shown differentially methylated genes and pathways in psoriatic lesions compared to skin from healthy controls or even to non-lesional skin of patients with psoriasis [165,166,167], even though the methylation pattern of the latter is closer to that of skin in healthy individuals than to psoriatic skin lesions [164]. In recent studies comparing psoriasis-involved skin to adjacent non-lesional skin, differences in CpG methylation were found at the promoters of known PSORS genes, such as S100A9, PTPN22, SELENBP1, CARD14, and KAZN [167]. In addition, DNA methylation differences can also be related to the histological features of psoriasis lesions. Patients with Munro’s microabscess present a characteristic DNA methylation profile with the involvement of genes related to the chemotaxis of neutrophils [167]. S100A8 (involved in epidermal differentiation), CYP2S1 (metabolism of retinoic acid), and EIF2C2 (with a role in RNA processing) are also differentially methylated in psoriatic lesions compared with non-lesional skin and with the normal skin of healthy controls [165]. The MGRN1 gene codes for E3 ubiquitin-protein ligase, which is involved in the degradation of misfolded proteins. Its methylation pattern is different between psoriatic scales and psoriatic skin lesions [168].
Tissue-resident memory T-cells in the skin have been related to the local relapse of psoriasis. DNA methylation alterations could be involved in this local memory since they do not revert completely to lesional skin after treatment. Indeed, distinct methylation patterns have been observed between never-lesional skin and resolved psoriasis lesions [169]. Methylation differences in Wnt and cadherin pathway genes have also been shown between never-lesional psoriatic skin and healthy skin in volunteers, suggesting that uninvolved skin could be a pre-psoriatic condition with an underlying vulnerability to disease [170].
Variations in DNA methylation have been found in peripheral blood mononuclear cells (PBMCs) from patients with psoriasis. The expression of methyl-CpG binding domain protein 2 (MBD2) and methyl-CpG binding protein 2 (MeCP2), two crucial regulators of DNA methylation, is markedly downregulated in PBMCs from psoriatic patients, whereas the methyltransferase DNMT1 is overexpressed [171]. Likewise, mesenchymal stem cells from psoriasis patients have been found to exhibit differential methylation of the promoters of genes involved in cell interaction, signal transduction, nucleotide degradation, skin differentiation, and cell motility [172].
4.2.2. Histone Modification
Histones are highly conserved proteins that are found in cell nuclei as structural components of chromatin and are involved in the control of gene expression. The accessibility of DNA sequences can be altered by post-transcriptional changes of histones, hence modifying the transcription of these DNA sequences [167]. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are the enzymes that maintain the general equilibrium between histone acetylation and deacetylation.
A wide spectrum of histone modifications with potential pathogenic implications have been described in psoriatic and AD skin. The mechanisms that influence chromatin compression due to post-translational modifications of histones are acetylation, phosphorylation, methylation, ubiquitination, and SUMOylation (Figure 3) [58].
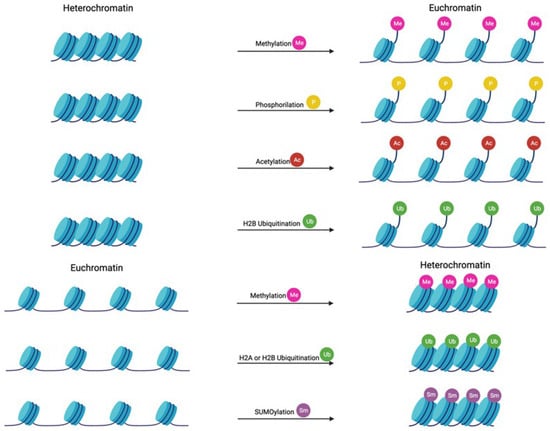
Figure 3.
Mechanisms involved in histone modification Nucleosomes are the basic repeating units that package DNA into the nucleus. DNA is wrapped around octamers of histone proteins (two H2A, two H2AB, two H3, and two H4), stabilized by an H1 histone. Histone acetylation and phosphorylation result in chromatin relaxation, allowing DNA transcription. On the other hand, histone SUMOylation and H2A ubiquitination cause chromatin compaction. Histone methylation and H2B ubiquitination may induce both relaxation or compaction of chromatin. Created with Biorender.com (accessed on 16 December 2023).
While histone deacetylation loosens the chromatin structure and activates gene transcription, acetylation causes chromatin to become more tightly packed and prevents transcription (Figure 1). On the other hand, methylation of histones will cause activation or suppression of the transcription, depending on the site of methylation and the number of methyl groups introduced.
Histone Modification in AD
Histone alterations related to AD pathogenesis can involve processes related to epidermal dysfunction, epidermal differentiation, and immune dysfunction. Regarding epidermal dysfunction, trimethylation of histone H3 at its Lys 27 residue by the histone transmethylase EZH2 prevents premature recruitment of activator protein 1 (AP1) to activate transcription of several structural genes required for epidermal differentiation [173,174]. Also, mixed-lineage leukemia (MLL), along with the multisubunit (WDR5, RbBP5, ASH2L, and DPY30) complex, catalyzes the trimethylation of H3K4, regulating the expression of genes related to differentiation in human skin [175]. Finally, histone hyperacetylation promoted by 5-azacytidine (5AC) and sodium butyrate (NaB) leads to keratinocyte differentiation through increased expression of Sprr1/2 and involucrin in human keratinocytes [176].
Decreased expression of proteins involved in tight junctions, such as transmembrane proteins and intracellular cytoplasmic proteins, has been observed in AD. Steelant et al. identified a new pathway associated with histone deacetylase (HDAC) activation and epidermal barrier dysfunction. In addition, they demonstrated that the application of HDAC inhibitors restored barrier integrity and reduced hyperresponsiveness in asthmatic mice [177].
Regarding immune dysfunction and histone modifications, Harb et al. demonstrated that placental histone hyperacetylation was linked to a low incidence of food allergen sensitization in children [178]. They noticed H3 acetylation of the SH2B3 gene, H3 acetylation of the IFNG gene, and H4 acetylation of the HDAC4 gene. The latter boosted HDAC4 expression in the placenta. HDAC4 possesses deacetylase activity for cytoplasmic nonhistone proteins, like STAT1. According to the authors, the higher placental expression of HDAC4 would lead to genome-wide deacetylation of immune genes and/or transcription factors involved in Th1 tilting. Thus, would enhance early immune polarization, aiding newborns to avoid IgE sensitization [178]. There is abundant additional evidence that histone modification and DNA methylation participate in the differentiation process of T lymphocytes [158,159,160].
Histone Modification in Psoriasis
Several histone modifications have been described with potential implications for psoriasis pathogenesis. For example, reduced levels of methylated H3K27 have been correlated with Th17 differentiation [179]. Other alterations reported are methylation at cystine 9 of histone 3 (H3K9), which can modify IL23 expression in keratinocytes, and H3K4 methylation in PBMCs from psoriasis patients [180,181]. H3K27 and H3K4 methylation of cystines 4 and 27 of histone 3 could influence the response to treatment since their levels have been found to differ between responders and non-responders to biological treatment [181].
Dysregulation of HATs and HDACs has been found in lesional psoriasis skin compared to healthy skin [167]. Elevated levels of HDAC-1 in psoriatic skin might lead to VEGF overexpression, endothelial cell proliferation, and keratinocyte survival [182]. High levels of H3K9 and H3K27 acetylation have been found in the IL17A promoter region in the immune cells of patients with psoriasis; they would promote Th17 differentiation and psoriasis development [183].
Acetylated histones are recognized by BET proteins that mediate the transcription of proinflammatory and immunoregulatory genes. Inhibition of BET proteins reduces the expression of important pro-inflammatory factors (IL-22, IL-17, and RORC) and could represent a potential new treatment for psoriasis [184].
4.2.3. Non-Coding RNA
Non-coding RNAs (ncRNAs) are not translated into proteins; instead, they interact with RNA, DNA, and proteins to change their structural properties and affect gene expression (Figure 4) [185]. Non-coding RNAs can be classified as short ncRNAs, intermediate ncRNAs, and long ncRNAs (lncRNAs), depending on their size. Indeed, short ncRNA can be divided into short interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), and microRNAs (miRNAs).
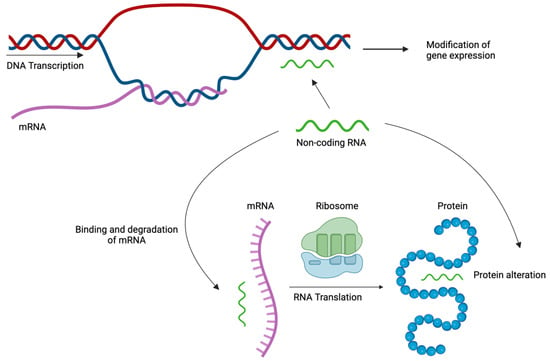
Figure 4.
Epigenetic mechanisms at translational level: Non-coding RNAs interact with DNA, mRNA or proteins altering their structure and affecting gene expression, mRNA translation, and protein function. Created with Biorender.com (accessed on 16 December 2023).
Micro-RNAs (mi-RNAs) are a class of small, single-stranded, non-coding molecules approximately 19–25 base pairs in length and are essential for controlling post-transcriptional gene expression in eukaryotes. Relative preservation of these RNA molecules across time attests to their biological importance. They have been linked to multiple cellular processes such as morphogenesis, proliferation, differentiation, regulation of cellular metabolism, signal transduction, and apoptosis [151,161,186,187,188].
MicroRNAs bind to the targeted messenger RNA (mRNA) strand, resulting in mRNA instability. Unstable mRNA is degraded in the cytoplasm; thus, translation of the corresponding gene is stopped, and the function of the protein is inhibited [186]. Mi-RNAs can also alter DNA expression by modulating DNA methylation, targeting enzymes necessary for DNA methylation, or through histone modifications [185,189]. This epigenetic mechanism is thought to control 1–3% of the human genome, corresponding to at least 30% of the protein-coding genes. Different miRNA species may regulate the same gene, while the expression of different genes may be influenced by the same class of miRNA [186].
Non-Coding RNA in AD
Multiple alterations in the expression of specific miRNAs have been observed in the skin and serum of AD patients. They control the expression of genes that mediate Th2 polarization, the activity of Treg, multiple inflammatory processes, the formation of tight junctions, the proliferation and death of epidermal keratinocytes, and the synthesis of cytokines and chemokines [188,190,191,192,193]. Increased levels of 10 miRNA species and decreased levels of 34 have been found in AD skin lesions [191]. Major miRNA alterations related to AD pathogenesis are summarized in Table 3.

Table 3.
Epigenomics in AD.
The activation of TLR receptors by bacterial lipopolysaccharides and the NFkB-associated activation of the relevant intracellular pathways are two examples of IFN-inducing activities suppressed by miR-146a. Additionally, the TRAF6, IRAK1, IRAK2, and RELB genes whose transcripts control the activation of this pathway, are directly inhibited by miR-146. As previously mentioned, NFkB is a transcription factor that mediates the expression of several genes coding proinflammatory cytokines (IL1B, TNFA, CARD10, IRAK1, CCL5, and CXCL). If the expression of these genes is inhibited, there is a reduction in the effectiveness of the innate immune response, which is typically seen in AD [186,190,192].
T helper type 17 (Th17) and Treg cell differentiation depend on miR-155, one of the most upregulated miRNAs in AD patients [191]. In addition, miR-155 blocks the expression of CTLA-4 (cytotoxic T-lymphocyte-associated protein 4), an immune checkpoint receptor that inhibits T-cell responses. Therefore, miR-155 increased expression in the AD skin causes a loss of Treg-dependent regulatory mechanisms, leading to increased effector T-cell proliferation and sustained inflammation [150,191,195,197,198]. Additionally, overexpression of miR-155 in AD is positively correlated with AD severity [199]. A positive correlation between miR-155 expression and Th17 cell percentage (which is increased in AD patients) has also been demonstrated [199].
The miRNA molecules have also been related to the Th2 bias. The Let-7 a-d family acts as an inhibitor of IL-13 and CCR7 synthesis. A decreased expression of Let-7 a-d has been observed in atopic skin, promoting the overproduction of IL-13 and contributing to the Th2 bias characteristic of AD [192]. Furthermore, miR-375 induces TSLP production and promotes Th2 responses in the skin by blocking KLF5 expression [186]. Previous reports have described down-regulation of miR-375 in AD [58,196], but Simpson et al. and Behesti et al. observed a positive correlation between miR-375 levels in breast milk and the risk of AD development in newborn infants [187,200].
Elevated serum levels of mir-151a have been found in AD patients; mir-151a inhibits the expression of the IL12RB2-subunit of the IL-12 receptor in human T helper cells, leading to an increase of the Th2 cells [194,201].
Finally, loss of hsa-miR-26a-5a is linked to increased expression of the enzyme hyaluronan 3 synthase (HAS3) in AD skin; HAS3 is involved in the synthesis of hyaluronic acid, a crucial component of the extracellular matrix [202].
The miRNA molecules have also been studied as possible therapeutic targets. The chemokine CCL22 influences the recruitment of Th2 lymphocytes expressing cutaneous lymphocyte antigen (CLA) into the skin. Dietary treatment with Salmonella typhimurium expressing CCL22-targeting miRNA prevented the expression of CCL22 in vivo, resulting in differences in the severity of AD lesions, pruritus, serum levels of IL-4, and serum and skin tissue levels of CCL22 [203].
Non-Coding RNA in Psoriasis
Short non-coding RNAs have been strongly associated with the pathogenesis of psoriasis, especially micro-RNAs, which are able to interfere in the immune response and modify skin inflammation as well as keratinocyte proliferation and differentiation [204,205,206]. They are detailed in Table 4.

Table 4.
Epigenomics in psoriasis.
One of the most downregulated micro-RNAs in psoriasis lesions is miRNA125b, which has been related to inhibition of keratinocyte proliferation and promotion of keratinocyte differentiation [204,205].
Lipocalin 2 (LCN2) has been implicated in the inflammatory process, playing an important role in cell survival and apoptosis. LCN2 is targeted by miR-383, reducing the expression of JAK3/STAT3 and leading to immune suppression. However, psoriasis lesions have shown downregulation of miR-383, therefore a proinflammatory status [211]. Another micro-RNA downregulated in psoriasis is miR-214-3p, which induces keratinocyte proliferation [212]. Lastly, miR-125a-5p is also downregulated in psoriasis lesions; the methyltransferase EZH2 catalyzes the methylation of histone H3 at lysine 27, repressing transcription of miR-125a-5pn [231] and eventually leading to increased expression of IL-17A-induced cytokines and cell proliferation [213].
Other micro-RNAs are upregulated in psoriasis. MiR-203 is overexpressed in psoriasis lesions, inducing keratinocyte proliferation via inhibition of LXR-α or PPAR-γ [232]. Overexpression of miR-378a (also upregulated in psoriasis) would be induced by IL-17A via the NF-kB pathway [207]. MiR-31 is activated by NF-kB to promote keratinocyte hyperproliferation in psoriatic lesions [208]. High levels of miR-210 have been found in psoriasis lesions and animal models; it stimulates Th17 and Th1 cell differentiation but inhibits Th2 differentiation by suppressing STAT6 and LYN expression [209]. Other micro-RNAs upregulated in psoriasis are miR-200c and miR-155. These micro-RNAs might become potential targets for psoriasis [210,233].
Long non-coding RNAs (>200 nucleotides) have also been associated with psoriasis pathogenesis. LncRNAs modulate gene expression by interacting with transcription factors and proteins that alter the chromatin structure or by acting as competitive endogenous RNAs that limit the accessibility of miRNAs to inhibit their target genes [185,189]. LncRNA-RP6-65G23.1 is significantly upregulated and promotes keratinocyte proliferation and suppression of apoptosis by altering the expression of Bcl-xl, Bcl2, and the p-ERK1/2 p-AKT signaling pathway [214]. MIR31HG is another lncRNA upregulated in psoriasis lesions; it seems to be activated by NF-kB and would participate in keratinocyte proliferation [215]. Several lncRNAs are upregulated in psoriasis, such as MSX2P1 [216], XIST [217], FABP5P3 [218], KLDHC7B-DT [219], or SPRR2C [220], whereas MEG3 [221], GAS5 [222], PRINS [223] or NEAT1 [224] are downregulated in psoriasis.
Finally, circular RNAs (circRNAs) are intermediate single-stranded ncRNAs that show continuous circular structure and covalent coupling between the 5′ and 3′ ends, which makes them unique [189]. CircRNAs participate in the modulation of gene expression or translation of regulatory proteins through their involvement in the degeneration of microRNAs and RNA-binding proteins [234]. Some circRNAs are upregulated in psoriasis, such as circOAS3 [225], circEIF5 [226], circ_0060531 [227], hsa_circ_0003738 [228], or hsa_circ_0061012 [229], whereas circRAB3B is downregulated [230]. The latter acts as an inductor of PTEN, suppressing keratinocyte hyperproliferation. CircRAB3B levels in psoriasis lesions are diminished compared to healthy skin.
4.3. Proteomics
Proteomics encompasses the examination of protein structure, function, interactions, and post-translational modifications at various phases of the complete set of proteins produced by organisms (proteome). It is widely acknowledged that the regulation of protein expression can operate independently of gene expression; thus, proteomics studies are essential in elucidating the underlying mechanisms responsible for the pathogenesis of diseases [13]. Moreover, proteomic studies allow the identification and characterization of proteins implicated in disease mechanisms without any previous knowledge about these proteins within the disease paradigm [235].
Powerful methods of proteomics allow for extensive examination of proteins potentially implicated in disease pathogenesis [13].
4.3.1. Proteomics in AD
Initial studies of proteomics in AD identified a limited number of proteins potentially involved in AD, such as ALDH1, NCC27, S100/A11, cyclinA2, caldesmon 1 isoform 5, nucleophosmin 1, esterase D, chloride intracellular channel 4, vinculin, and filamin A proteins [236,237,238,239,240,241,242]. S100/A11 induces human beta-defensin (HBD)-3 and FLG expression, and it was found to be significantly downregulated in the presence of IL-4 and IL-13 [239].
Proteomic research with mass spectrometry on tape-strips derived from AD skin has led to the identification of several proteins with pathogenetic implications [243,244,245,246]. Broccardo et al. demonstrated that e-fabp and alpha-enolase were distinctively expressed in AD samples, with increased expression of triosephosphate isomerase and serpin B3, and downregulation of keratin type I cytoskeletal 10, keratin type II cytoskeletal 1, caspase 14 precursor, and dermcidin precursor in comparison to control samples [244]. Additional tape-strip studies found decreased levels of proteins involved in natural moisturizing factor and barrier function compared to non-lesional AD skin; dry skin would predispose to skin infections [243]. Other tape-strip studies have identified 252 and 35 differentially expressed proteins in AD lesional versus healthy skin, respectively [246,247].
Broad proteomic studies with OLINK and SOMAscan have identified dysregulation of several crucial immune and skin barrier AD markers, providing further insight into AD pathogenesis. Proteins associated with general inflammation (MMP12), dendritic cells (CD40, TNF), macrophages (CD163), T-cell activation/migration (IL-2RA, CCL19), Th2 (IL-1RL1, IL-13), Th1 (CXCL9/10/11), Th17/Th22 (IL-12B, IL-17RA), and innate immunity (IL-6, IL-8) are upregulated, whereas proteins related to barrier function are downregulated (FLG, LOR) [248,249,250,251,252,253,254,255]. Although most of these proteins had already been identified by transcriptomic studies, proteomics allowed validation; hence, they could be used as disease severity indicators and even as therapeutic targets [256,257].
Proteomic studies in the skin and/or blood have also been performed in the pediatric AD population [257]. Brunner et al. demonstrated proteomic signs of systemic inflammation in pediatric patients with a 6-month course of moderate-to-severe AD, such as Selectin E (SELE), MMP3, MMP9, MMP10, and urokinase receptor [249]. Cardiovascular risk mediators seen in adult patients were also found in early-onset AD children, with some differences [248,249]: adults with AD exhibited increased expression of a wider array of Th2-associated mediators such as TARC/CCL17, CCL13, and IL-13, as well as the Th17 lymphokine CCL20. Additionally, Th1 markers (CXCL9, CXCL10, and IFN-γ) were upregulated in adults and absent in blood from pediatric AD patients. Therefore, Th1 activation is a marker of AD chronicity and a suitable target to prevent the progression from pediatric AD to chronic AD [249,258].
AD proteomics have been compared to those of other inflammatory diseases. Brunner et al. compared the proteomic blood signatures of moderate-to-severe AD and psoriasis patients. Ten proteins were increased in both diseases compared to controls, including markers of Th1 (IFN-γ, CXCL9, TNF-β/lymphotoxin) and Th17 (CCL20, IL-17C) immune responses, IL-16, BLMH, IL-20, CDCP1, and IL2RA. However, significant differences in their proteomic blood profiles among the two diseases were observed. Markers of systemic inflammation are stronger in AD compared to psoriasis [248,249,257]. Concretely, levels of Th1 (CXCL10, CXCL11), Th2 (IL-13, CCL13, CCL17/TARC, CCL11, IL-10), Th17/Th22 (S100A12) and Th1/Th17/Th22 (IL-12/IL-23p40) associated proteins, and proteins involved in the development and activation of T-cells (CD40L, IL-7, CCL25, IL2RB, IL15RA, CD6, RANKL, TNFRSF9/CD137) and dendritic cells (CD40, FLT3 ligand), mediators associated with atherosclerosis (CX3CL1/fractalkine, CCL8, M-CSF, CXCL5, CCL4, and HGF), factors involved in tissue remodeling (MMP-12, MMP-1, and MMP-10), angiogenesis (VEGF-A), TGF-α, NF-κB activator IKBKG/NEMO, molecules facilitating the chemotaxis of T-cells, B-cells, eosinophils (CCL28), and neutrophils (CXCL5, CXCL6), as well as the soluble cytokine receptors IL-10RB and IL-18R1, were observed to be higher in individuals with AD, compared to psoriasis [248].
Finally, proteomics studies have been used to evaluate the responses of AD to treatment. A tape-strip proteomic study of 353 inflammatory proteins showed a significant decrease of 136 after dupilumab treatment, which suppressed the Th2 pathway without significant modulation of Th1 [252].
4.3.2. Proteomics in Psoriasis
Proteomics studies have provided enhanced insight into the pathogenesis of psoriasis, since pronounced alterations in gene expression may not necessarily lead to substantial modifications at the protein level; conversely, marked shifts in protein expression may not reflect significant alterations in the transcriptome [235]. One of the initial proteomics studies included skin samples taken from individuals with acute guttate psoriasis, chronic plaque psoriasis, and nickel-induced contact eczema; the proteome profile of acute guttate psoriasis closely resembled that of contact eczema, suggesting that the length of disease progression might influence the proteomic signature [259]. Another study showed that cytoskeletal proteins, actin-binding proteins/peptides, and calcium-binding components would participate in the altered protease activity seen in psoriasis [260].
Comparison of psoriasis and healthy skin proteome patterns has led to the identification of several proteins with different expressions in psoriasis [261]. Some of these proteins (SFN protein, peroxiredoxin 2, among others) are involved in cell proliferation, regulatory/balancing system, and inflammatory response. Schonthaler et al. demonstrated higher expression of S100A8, S100A9, and complement C3 in psoriatic epidermis compared to healthy skin [262]. Furthermore, the deletion of the gene coding for S100A9 led to an attenuation of psoriasis-like disease in a murine model [262]. Swindell et al. identified 748 proteins differentially expressed in psoriasis as compared to healthy skin [263]; some of them were categorized as psoriasis-specific, which showed a disproportionate induction by IL-17A in cultured keratinocytes compared to increased but non-specific proteins. Recent evidence has brought to light the importance of peptidase inhibitor 3 (PI3), which is highly expressed in keratinocytes from psoriatic lesions. PI3 is significantly correlated with local gene expression and with disease severity. Hence, PI3 may be a psoriasis-specific biomarker for disease severity and hyper-keratinization [264].
In 2016, Bottoni et al. observed structural protein alterations in the saliva of patients affected by psoriasis, which were similar to those observed in diabetic patients; in both cases, they differed from normal saliva [265].
Utilizing antibody-based methodologies and protein array technology, elevated concentrations of chemoattractants for neutrophils, Th1 cells, monocytes, and dendritic cells were detected in the stratum corneum of individuals with psoriasis; on the other hand, no upregulation of cytokines associated with Th2 cells, which are not implicated in the pathophysiology of psoriasis, was identified [266].
Decreased levels of proteins related to vitamin D regulation and lipid metabolism were observed by Gegotek et al., but their precise role in psoriasis pathogenesis remains unclear [267].
In 2018, Chularojanamontri et al. attempted to define normal skin autoantigens by immunoproteomics in sera from patients with psoriasis and healthy volunteers [268]. By using a healthy skin sample, no significant differences among psoriatic and healthy sera were observed in the patterns of IgG- and IgM-immunoreactivity to both epidermal and dermal autoantigens. These findings suggest that the autoantigens originating from the epidermis and dermis in psoriatic skin, along with the associated humoral immune response, are probably a consequence of downstream processes. In contrast, those linked to upstream mechanisms may likely result from cell-mediated immune mechanisms [268].
Regarding systemic inflammation in patients affected by psoriasis, Brunnet et al. [248] observed significant increases in Th17-associated proteins, proteins involved in coagulation and angiogenesis (t-PA), endothelial activation (IL-6), lipid metabolism (LDL-R), cardiovascular disease risk proteins (galectin-3), mediators of cell adhesion, and T-cell activation (TREML2), among others.
The discovery of biomarkers is one of the major goals of proteomics in psoriasis. A study by Cowen et al. found differences in serum proteome patterns between patients with psoriasis, tumor-staged mycosis fungoides, and healthy participants [269]. Likewise, Williamson et al. identified increased levels of profilin 1, a potential biomarker for psoriasis, in plasma from psoriasis patients [270]. Matsuura et al. identified consistently increased fibrinogen α chain-derived and filaggrin-derived peptide levels in patients with psoriasis vulgaris and psoriatic arthritis [271]. Another protein that holds promise as a potential biomarker is kynureninase, with elevated levels in sera from individuals with psoriasis but no alterations in patients with AD or contact dermatitis [250]. Méhul et al. demonstrated distinct lesional proteomic profiles between psoriasis and cutaneous T-cell lymphoma (CTCL) [272]: psoriasis lesions exhibited differential expression of IL36G and IL37 in contrast to healthy skin, whereas CTCL lesions did not; conversely, CCL27 was differentially expressed in CTCL when compared to healthy skin, with no variations observed in psoriasis lesions. Lastly, desmoplakin, complement C3, polymeric immunoglobulin receptor, and cytokeratin 17 demonstrated significant associations with the Psoriasis Area and Severity Index (PASI) score, indicating their potential as novel biomarkers for disease severity assessment [273].
Proteomics have also been studied regarding the response of psoriasis to treatments. Kolbinger et al. examined the proteomic profile of psoriasis skin and sera before and after secukinumab (anti-IL-17A monoclonal antibody) treatment [274]. Among several proteins examined, β-defensin 2 was found to be correlated with PASI and IL-17 levels. Following the administration of a single dose (300 mg) of secukinumab, the aberrantly regulated antimicrobial peptides, proinflammatory cytokines, and neutrophil chemoattractants reverted to their normal levels. Additionally, a noteworthy decrease in β-defensin 2 was noted. Hence, β-defensin 2 holds potential as a biomarker for IL-17A-mediated psoriatic pathology and as a predictor of response to anti-IL-17A treatment [274].
Proteomics have also been used to identify hepatic and renal toxicity due to systemic therapies for psoriasis. Multiple proteins were identified in urine samples as possible biomarkers of methotrexate-induced hepatic toxicity [275]; however, the reliability of these proteins was not validated with liver biopsy or other alternative methods, such as Fibroscan®, magnetic resonance elastography, or FibroTest®. Cyclosporine-induced proteomic alterations in human kidney cell lines, which were partially resolved with the administration of N-acetylcysteine, remain to be elucidated in psoriatic patients [276].
5. Conclusions
This narrative review illustrates the complex nature of AD and psoriasis genetics, epigenetics, and proteomics, involving a wide array of genes and proteins contributing to innate and adaptative immune systems as well as to skin barrier integrity. Rapid advancements in genetic research technology have not only deepened our understanding of the pathophysiology of psoriasis but have also led to the development of new diagnostic biomarkers and novel medications. Psoriasis and AD represent complex diseases with distinct phenotypes; most studies in psoriasis have been focused on plaque psoriasis, and further research is required addressing topographic or morphological variants, such as pustular psoriasis.
6. Future Directions
Multi-omics has furthered our knowledge of the pathogenesis of multiple IMIDs and led to the identification of novel disease biomarkers and suitable treatment targets. The goal of personalized medicine, namely to choose treatments taking into account genetic factors and proteomic profiles, seems closer. The molecular underpinnings of psoriasis and AD can be analyzed using a systems biology approach based on microarray data analysis. Various bioinformatics tools can be used to highlight differentially expressed genes, enrich gene ontology, and conduct network analysis to identify critical regulatory pathways. Ultimately, these studies may provide novel therapeutic targets, opening new horizons in medical treatment [277]. Furthermore, response to treatment can be predicted after the first injection of a monoclonal antibody like secukinumab [274]. Multiomic studies may further our understanding of cellular and transcriptomic alterations associated with optimal response to treatments, remission, and potential modification of disease course in psoriasis and other diseases [278,279]. Recently available tools such as mind.px (https://minderahealth.com/psoriasis/) (accessed on 27 December 2023) might contribute to predicting the response of psoriasis to biological treatments and potentially improve the efficiency of treatment.
Author Contributions
All authors declare to have contributed to the conception and design of the article, the writing of the draft, the critical review of the intellectual content. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
L. Puig has perceived consultancy/speaker’s honoraria from and/or participated in clinical trials sponsored by Abbvie, Almirall, Amgen, Biogen, Bristol Myers Squibb, Boehringer Ingelheim, Celgene, Fresenius-Kabi, Gebro, Janssen, JS BIOCAD, Leo-Pharma, Lilly, Novartis, Pfizer, Samsung-Bioepis, and UCB. L. Rusiñol declares no conflicts of interest for the related topic.
References
- Surace, A.E.A.; Hedrich, C.M. The Role of Epigenetics in Autoimmune/Inflammatory Disease. Front. Immunol. 2019, 10, 1525. [Google Scholar] [CrossRef]
- Dorochow, E.; Köhm, M.; Hahnefeld, L.; Gurke, R. Metabolic Profiling in Rheumatoid Arthritis, Psoriatic Arthritis, and Psoriasis: Elucidating Pathogenesis, Improving Diagnosis, and Monitoring Disease Activity. J. Pers. Med. 2022, 12, 924. [Google Scholar] [CrossRef]
- van Kempen, T.S.; Wenink, M.H.; Leijten, E.F.A.; Radstake, T.R.D.J.; Boes, M. Perception of self: Distinguishing autoimmunity from autoinflammation. Nat. Rev. Rheumatol. 2015, 11, 483–492. [Google Scholar] [CrossRef]
- Anchang, C.G.; Xu, C.; Raimondo, M.G.; Atreya, R.; Maier, A.; Schett, G.; Zaburdaev, V.; Rauber, S.; Ramming, A. The Potential of OMICs Technologies for the Treatment of Immune-Mediated Inflammatory Diseases. Int. J. Mol. Sci. 2021, 22, 7506. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, S.F.; Ulrik, C.S.; Kyvik, K.O.; Hjelmborg, J.V.B.; Skadhauge, L.R.; Steffensen, I.; Backer, V. Importance of genetic factors in the etiology of atopic dermatitis: A twin study. Allergy Asthma Proc. 2007, 28, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Vetrano, S.; Bouma, G.; Benschop, R.J.; Birngruber, T.; Costanzo, A.; D’Haens, G.R.A.M.; Frasca, L.; Hillenbrand, R.; Iversen, L.; Johansen, C.; et al. ImmUniverse Consortium: Multi-omics integrative approach in personalized medicine for immune-mediated inflammatory diseases. Front. Immunol. 2022, 13, 1002629. [Google Scholar] [CrossRef]
- Radulska, A.; Pelikant-Małecka, I.; Jendernalik, K.; Dobrucki, I.T.; Kalinowski, L. Proteomic and Metabolomic Changes in Psoriasis Preclinical and Clinical Aspects. Int. J. Mol. Sci. 2023, 24, 9507. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.; Farlik, M.; Sheffield, N.C. Multi-Omics of Single Cells: Strategies and Applications. Trends Biotechnol. 2016, 34, 605–608. [Google Scholar] [CrossRef]
- Vilanova, C.; Porcar, M. Are multi-omics enough? Nat. Microbiol. 2016, 1, 16101. [Google Scholar] [CrossRef]
- Bersanelli, M.; Mosca, E.; Remondini, D.; Giampieri, E.; Sala, C.; Castellani, G.; Milanesi, L. Methods for the integration of multi-omics data: Mathematical aspects. BMC Bioinform. 2016, 17 (Suppl. S2), 15. [Google Scholar] [CrossRef]
- Mitamura, Y.; Reiger, M.; Kim, J.; Xiao, Y.; Zhakparov, D.; Tan, G.; Rückert, B.; Rinaldi, A.O.; Baerenfaller, K.; Akdis, M.; et al. Spatial transcriptomics combined with single-cell RNA-sequencing unravels the complex inflammatory cell network in atopic dermatitis. Allergy 2023, 78, 2215–2231. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Julià, A.; Vinaixa, M.; Domènech, E.; Fernández-Nebro, A.; Cañete, J.D.; Ferrándiz, C.; Tornero, J.; Gisbert, J.P.; Nos, P.; et al. Urine metabolome profiling of immune-mediated inflammatory diseases. BMC Med. 2016, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Carrascosa-Carrillo, J.M.; Aterido, A.; Li, T.; Guillén, Y.; Martinez, S.; Marsal, S.; Julià, A. Toward Precision Medicine in Atopic Dermatitis Using Molecular-Based Approaches. Actas Dermosifiliogr. 2023, 115, T66–T75. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Luo, L.; Zhu, J.; Li, C. Multi-Omics Research Strategies for Psoriasis and Atopic Dermatitis. Int. J. Mol. Sci. 2023, 24, 8018. [Google Scholar] [CrossRef] [PubMed]
- Sroka-Tomaszewska, J.; Trzeciak, M. Molecular Mechanisms of Atopic Dermatitis Pathogenesis. Int. J. Mol. Sci. 2021, 22, 4130. [Google Scholar] [CrossRef] [PubMed]
- David Boothe, W.; Tarbox, J.A.; Tarbox, M.B. Atopic Dermatitis: Pathophysiology. Adv. Exp. Med. Biol. 2017, 1027, 21–37. [Google Scholar] [PubMed]
- Weidinger, S.; Beck, L.A.; Bieber, T.; Kabashima, K.; Irvine, A.D. Atopic dermatitis. Nat. Rev. Dis. Primers 2018, 4, 1. [Google Scholar] [CrossRef]
- Langan, S.M.; Irvine, A.D.; Weidinger, S. Atopic dermatitis. Lancet 2020, 396, 345–360. [Google Scholar] [CrossRef]
- Çetinarslan, T.; Kümper, L.; Fölster-Holst, R. The immunological and structural epidermal barrier dysfunction and skin microbiome in atopic dermatitis-an update. Front. Mol. Biosci. 2023, 10, 1159404. [Google Scholar] [CrossRef]
- Oyoshi, M.K.; He, R.; Kumar, L.; Yoon, J.; Geha, R.S. Cellular and molecular mechanisms in atopic dermatitis. Adv. Immunol. 2009, 102, 135–226. [Google Scholar]
- Tsoi, L.C.; Rodriguez, E.; Degenhardt, F.; Baurecht, H.; Wehkamp, U.; Volks, N.; Szymczak, S.; Swindell, W.R.; Sarkar, M.K.; Raja, K.; et al. Atopic Dermatitis Is an IL-13–Dominant Disease with Greater Molecular Heterogeneity Compared to Psoriasis. J. Investig. Dermatol. 2019, 139, 1480–1489. [Google Scholar] [CrossRef] [PubMed]
- Szegedi, K.; Lutter, R.; Res, P.C.; Bos, J.D.; Luiten, R.M.; Kezic, S.; Middelkamp-Hup, M.A. Cytokine profiles in interstitial fluid from chronic atopic dermatitis skin. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 2136–2144. [Google Scholar] [CrossRef]
- Guttman-Yassky, E.; Suárez-Fariñas, M.; Chiricozzi, A.; Nograles, K.E.; Shemer, A.; Fuentes-Duculan, J.; Cardinale, I.; Lin, P.; Bergman, R.; Bowcock, A.M.; et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J. Allergy Clin. Immunol. 2009, 124, 1235–1244.e58. [Google Scholar] [CrossRef] [PubMed]
- Rendon, A.; Schäkel, K. Psoriasis pathogenesis and treatment. Int. J. Mol. Sci. 2019, 20, 1475. [Google Scholar] [CrossRef]
- Lowes, M.A.; Suárez-Fariñas, M.; Krueger, J.G. Immunology of psoriasis. Annu. Rev. Immunol. 2014, 32, 227–255. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Lai, Y. Crosstalk between keratinocytes and immune cells in inflammatory skin diseases. Explor. Immunol. 2021, 1, 418–431. [Google Scholar] [CrossRef]
- Boehncke, W.-H.; Schön, M.P. Psoriasis. Lancet 2015, 386, 983–994. [Google Scholar] [CrossRef] [PubMed]
- Nestle, F.O.; Kaplan, D.H.; Barker, J. Psoriasis. N. Engl. J. Med. 2009, 361, 496–509. [Google Scholar] [CrossRef]
- Ben Abdallah, H.; Johansen, C.; Iversen, L. Key Signaling Pathways in Psoriasis: Recent Insights from Antipsoriatic Therapeutics. Psoriasis Targets Ther. 2021, 11, 83–97. [Google Scholar] [CrossRef]
- Hawkes, J.E.; Chan, T.C.; Krueger, J.G. Psoriasis pathogenesis and the development of novel targeted immune therapies. J. Allergy Clin. Immunol. 2017, 140, 645–653. [Google Scholar] [CrossRef]
- Ganguly, D.; Chamilos, G.; Lande, R.; Gregorio, J.; Meller, S.; Facchinetti, V.; Homey, B.; Barrat, F.J.; Zal, T.; Gilliet, M. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J. Exp. Med. 2009, 206, 1983–1994. [Google Scholar] [CrossRef] [PubMed]
- Lande, R.; Gregorio, J.; Facchinetti, V.; Chatterjee, B.; Wang, Y.-H.; Homey, B.; Cao, W.; Wang, Y.-H.; Su, B.; Nestle, F.O.; et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 2007, 449, 564–569. [Google Scholar] [CrossRef]
- Park, H.; Li, Z.; Yang, X.O.; Chang, S.H.; Nurieva, R.; Wang, Y.-H.; Wang, Y.; Hood, L.; Zhu, Z.; Tian, Q.; et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005, 6, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.E.; Hatton, R.D.; Mangan, P.R.; Turner, H.; Murphy, T.L.; Murphy, K.M.; Weaver, C.T. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005, 6, 1123–1132. [Google Scholar] [CrossRef]
- Nakajima, A.; Matsuki, T.; Komine, M.; Asahina, A.; Horai, R.; Nakae, S.; Ishigame, H.; Kakuta, S.; Saijo, S.; Iwakura, Y. TNF, but not IL-6 and IL-17, is crucial for the development of T cell-independent psoriasis-like dermatitis in Il1rn-/- mice. J. Immunol. 2010, 185, 1887–1893. [Google Scholar] [CrossRef]
- Lande, R.; Botti, E.; Jandus, C.; Dojcinovic, D.; Fanelli, G.; Conrad, C.; Chamilos, G.; Feldmeyer, L.; Marinari, B.; Chon, S.; et al. The antimicrobial peptide LL37 is a T-cell autoantigen in psoriasis. Nat. Commun. 2014, 5, 5621. [Google Scholar] [CrossRef]
- Aggarwal, S.; Ghilardi, N.; Xie, M.-H.; de Sauvage, F.J.; Gurney, A.L. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 2003, 278, 1910–1914. [Google Scholar] [CrossRef]
- Tait Wojno, E.D.; Hunter, C.A.; Stumhofer, J.S. The Immunobiology of the Interleukin-12 Family: Room for Discovery. Immunity 2019, 50, 851–870. [Google Scholar] [CrossRef]
- Cai, Y.; Shen, X.; Ding, C.; Qi, C.; Li, K.; Li, X.; Jala, V.R.; Zhang, H.; Wang, T.; Zheng, J.; et al. Pivotal role of dermal IL-17-producing γδ T cells in skin inflammation. Immunity 2011, 35, 596–610. [Google Scholar] [CrossRef] [PubMed]
- Bielecki, P.; Riesenfeld, S.J.; Hütter, J.-C.; Torlai Triglia, E.; Kowalczyk, M.S.; Ricardo-Gonzalez, R.R.; Lian, M.; Amezcua Vesely, M.C.; Kroehling, L.; Xu, H.; et al. Skin-resident innate lymphoid cells converge on a pathogenic effector state. Nature 2021, 592, 128–132. [Google Scholar] [CrossRef]
- Matos, T.R.; O’Malley, J.T.; Lowry, E.L.; Hamm, D.; Kirsch, I.R.; Robins, H.S.; Kupper, T.S.; Krueger, J.G.; Clark, R.A. Clinically resolved psoriatic lesions contain psoriasis-specific IL-17-producing αβ T cell clones. J. Clin. Investig. 2017, 127, 4031–4041. [Google Scholar] [CrossRef] [PubMed]
- Blauvelt, A.; Chiricozzi, A. The Immunologic Role of IL-17 in Psoriasis and Psoriatic Arthritis Pathogenesis. Clin. Rev. Allergy Immunol. 2018, 55, 379–390. [Google Scholar] [CrossRef] [PubMed]
- McGeachy, M.J.; Cua, D.J.; Gaffen, S.L. The IL-17 Family of Cytokines in Health and Disease. Immunity 2019, 50, 892–906. [Google Scholar] [CrossRef]
- Chiricozzi, A.; Guttman-Yassky, E.; Suárez-Fariñas, M.; Nograles, K.E.; Tian, S.; Cardinale, I.; Chimenti, S.; Krueger, J.G. Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J. Investig. Dermatol. 2011, 131, 677–687. [Google Scholar] [CrossRef]
- Matsunaga, Y.; Hashimoto, Y.; Ishiko, A. Stratum corneum levels of calprotectin proteins S100A8/A9 correlate with disease activity in psoriasis patients. J. Dermatol. 2021, 48, 1518–1525. [Google Scholar] [CrossRef]
- Christmann, C.; Zenker, S.; Martens, L.; Hübner, J.; Loser, K.; Vogl, T.; Roth, J. Interleukin 17 Promotes Expression of Alarmins S100A8 and S100A9 During the Inflammatory Response of Keratinocytes. Front. Immunol. 2021, 11, 599947. [Google Scholar] [CrossRef] [PubMed]
- Mellor, L.F.; Gago-Lopez, N.; Bakiri, L.; Schmidt, F.N.; Busse, B.; Rauber, S.; Jimenez, M.; Megías, D.; Oterino-Sogo, S.; Sanchez-Prieto, R.; et al. Keratinocyte-derived S100A9 modulates neutrophil infiltration and affects psoriasis-like skin and joint disease. Ann. Rheum. Dis. 2022, 81, 1400–1408. [Google Scholar] [CrossRef]
- Duffin, K.C.; Hwang, S.T.; Krueger, J.G. Advances and Controversies in Our Understanding of Guttate and Plaque Psoriasis. J. Rheumatol. 2023, 50, 4–7. [Google Scholar] [CrossRef]
- Sugiura, K. Role of Interleukin 36 in Generalised Pustular Psoriasis and Beyond. Dermatol. Ther. 2022, 12, 315–328. [Google Scholar] [CrossRef]
- Julià, A.; Tortosa, R.; Hernanz, J.M.; Cañete, J.D.; Fonseca, E.; Ferrándiz, C.; Unamuno, P.; Puig, L.; Fernández-Sueiro, J.L.; Sanmartí, R.; et al. Risk variants for psoriasis vulgaris in a large case-control collection and association with clinical subphenotypes. Hum. Mol. Genet. 2012, 21, 4549–4557. [Google Scholar] [CrossRef]
- Canal-García, E.; Bosch-Amate, X.; Belinchón, I.; Puig, L. [Translated article] Nail Psoriasis. Actas Dermosifiliogr. 2022, 113, T481–T490. [Google Scholar] [CrossRef]
- Stensen, L.; Thomsen, S.F.; Backer, V. Change in prevalence of atopic dermatitis between 1986 and 2001 among children. Allergy Asthma Proc. 2008, 29, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-A.; Wahn, U.; Kehrt, R.; Tarani, L.; Businco, L.; Gustafsson, D.; Andersson, F.; Oranje, A.P.; Wolkertstorfer, A.; Berg, A.V.; et al. A major susceptibility locus for atopic dermatitis maps to chromosome 3q21. Nat. Genet. 2000, 26, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Elias, M.S.; Bradley, M. Genetics in Atopic Dermatitis: Historical Perspective and Future Prospects. Acta Derm. Venereol. 2020, 100, adv00163. [Google Scholar] [CrossRef]
- Budu-Aggrey, A.; Kilanowski, A.; Sobczyk, M.K.; Shringarpure, S.S.; Mitchell, R.; Reis, K.; Reigo, A.; Mägi, R.; Nelis, M.; Tanaka, N.; et al. European and multi-ancestry genome-wide association meta-analysis of atopic dermatitis highlights importance of systemic immune regulation. Nat. Commun. 2023, 14, 6172. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Estravís, M.; García-Sánchez, A.; Dávila, I.; Isidoro-García, M.; Sanz, C. Genetics and epigenetics of atopic dermatitis: An updated systematic review. Genes 2020, 11, 442. [Google Scholar] [CrossRef]
- Bratu, D.; Boda, D.; Caruntu, C. Genomic, Epigenomic, Transcriptomic, Proteomic and Metabolomic Approaches in Atopic Dermatitis. Curr. Issues Mol. Biol. 2023, 45, 5215–5231. [Google Scholar] [CrossRef]
- Nedoszytko, B.; Reszka, E.; Gutowska-Owsiak, D.; Trzeciak, M.; Lange, M.; Jarczak, J.; Niedoszytko, M.; Jablonska, E.; Romantowski, J.; Strapagiel, D.; et al. Genetic and Epigenetic Aspects of Atopic Dermatitis. Int. J. Mol. Sci. 2020, 21, 6484. [Google Scholar] [CrossRef]
- Irvine, A.D.; Mclean, W.H.I.; Leung, D.Y.M. Mechanisms of Disease Filaggrin Mutations Associated with Skin and Allergic Diseases. N. Engl. J. Med. 2011, 365, 1315–1327. [Google Scholar] [CrossRef]
- Tsuji, G.; Hashimoto-Hachiya, A.; Kiyomatsu-Oda, M.; Takemura, M.; Ohno, F.; Ito, T.; Morino-Koga, S.; Mitoma, C.; Nakahara, T.; Uchi, H.; et al. Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis. 2017, 8, e2931. [Google Scholar] [CrossRef]
- Margaritte-Jeannin, P.; Babron, M.C.; Laprise, C.; Lavielle, N.; Sarnowski, C.; Brossard, M.; Moffatt, M.; Gagné-Ouellet, V.; Etcheto, A.; Lathrop, M.; et al. The COL5A3 and MMP9 genes interact in eczema susceptibility. Clin. Exp. Allergy 2018, 48, 297–305. [Google Scholar] [CrossRef]
- Söderhäll, C.; Marenholz, I.; Kerscher, T.; Rüschendorf, F.; Esparza-Gordillo, J.; Worm, M.; Gruber, C.; Mayr, G.; Albrecht, M.; Rohde, K.; et al. Variants in a novel epidermal collagen gene (COL29A1) are associated with atopic dermatitis. PLoS Biol. 2007, 5, 1952–1961. [Google Scholar] [CrossRef] [PubMed]
- Kabesch, M.; Carr, D.; Weilandw, S.K.; Von Mutius, E. Association between polymorphisms in serine protease inhibitor, kazal type 5 and asthma phenotypes in a large German population sample. Clin. Exp. Allergy 2004, 34, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Paternoster, L.; Standl, M.; Chen, C.M.; Ramasamy, A.; Bøpnnelykke, K.; Duijts, L.; Ferreira, M.A.; Alves, A.C.; Thyssen, J.P.; Albrecht, E.; et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat. Genet. 2011, 44, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Marenholz, I.; Rivera, V.A.G.; Esparza-Gordillo, J.; Bauerfeind, A.; Lee-Kirsch, M.A.; Ciechanowicz, A.; Kurek, M.; Piskackova, T.; MacEk, M.; Lee, Y.A. Association screening in the epidermal differentiation complex (EDC) identifies an SPRR3 repeat number variant as a risk factor for eczema. J. Investig. Dermatol. 2011, 131, 1644–1649. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Liu, Y.; Xue, R.; Chen, L.; Chen, H.; Shao, L.; Wang, J.; Zhang, X. Interleukin 4 -590C/T (rs2243250) Polymorphism Is Associated with Increased Risk of Atopic Dermatitis: Meta-Analysis of Case-Control Studies. Dermatitis 2017, 28, 144–151. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, Z.Y.; Li, L.F. Association between the IL-10-1082G/A.; IL-10-592A/C.; and IL-10-819G/A Polymorphisms and Atopic Dermatitis Susceptibility: A Meta-Analysis. Genet. Test Mol. Biomark. 2019, 23, 332–341. [Google Scholar] [CrossRef]
- Gharagozlou, M.; Farhadi, E.; Khaledi, M.; Behniafard, N.; Sotoudeh, S.; Salari, R.; Darabi, B.; Fathi, S.M.; Mahmoudi, M.; Aghamohammadi, A.; et al. Association Between the Interleukin 6 Genotype at Position-174 and Atopic Dermatitis. J. Investig. Allergol. Clin. Immunol. 2013, 23, 89–93. [Google Scholar]
- Esparza-Gordillo, J.; Schaarschmidt, H.; Liang, L.; Cookson, W.; Bauerfeind, A.; Lee-Kirsch, M.A.; Nemat, K.; Henderson, J.; Paternoster, L.; Harper, J.I.; et al. A functional IL-6 receptor (IL6R) variant is a risk factor for persistent atopic dermatitis. J. Allergy Clin. Immunol. 2013, 132, 371–377. [Google Scholar] [CrossRef]
- Palmer, C.N.A.; Irvine, A.D.; Terron-Kwiatkowski, A.; Zhao, Y.; Liao, H.; Lee, S.P.; Goudie, D.R.; Sandilands, A.; Campbell, L.E.; Smith, F.J.D.; et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006, 38, 441–446. [Google Scholar] [CrossRef]
- Rodríguez, E.; Baurecht, H.; Herberich, E.; Wagenpfeil, S.; Brown, S.J.; Cordell, H.J.; Irvine, A.D.; Weidinger, S. Meta-analysis of filaggrin polymorphisms in eczema and asthma: Robust risk factors in atopic disease. J. Allergy Clin. Immunol. 2009, 123, 1361–1370.e7. [Google Scholar] [CrossRef] [PubMed]
- Elhaji, Y.; Sasseville, D.; Pratt, M.; Asai, Y.; Matheson, K.; McLean, W.H.I.; Hull, P.R. Filaggrin gene loss-of-function mutations constitute a factor in patients with multiple contact allergies. Contact Dermat. 2019, 80, 354–358. [Google Scholar] [CrossRef] [PubMed]
- McLean, W.H.I. Filaggrin failure—From ichthyosis vulgaris to atopic eczema and beyond. Br. J. Dermatol. 2016, 175, 4–7. [Google Scholar] [CrossRef]
- Kim, J.; Kim, B.E.; Leung, D.Y.M. Pathophysiology of atopic dermatitis: Clinical implications. Allergy Asthma Proc. 2019, 40, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Boguniewicz, M. Biologic Therapy for Atopic Dermatitis: Moving Beyond the Practice Parameter and Guidelines. J. Allergy Clin. Immunol. Pract. 2017, 5, 1477–1487. [Google Scholar] [CrossRef]
- Howell, M.D.; Kim, B.E.; Gao, P.; Grant, A.V.; Boguniewicz, M.; DeBenedetto, A.; Schneider, L.; Beck, L.A.; Barnes, K.C.; Leung, D.Y.M. Cytokine modulation of atopic dermatitis filaggrin skin expression. J. Allergy Clin. Immunol. 2009, 124, R7–R12. [Google Scholar] [CrossRef]
- Adam, D.N.; Gooderham, M.J.; Beecker, J.R.; Hong, C.H.; Jack, C.S.; Jain, V.; Lansang, P.; Lynde, C.W.; Papp, K.A.; Prajapati, V.H.; et al. Expert consensus on the systemic treatment of atopic dermatitis in special populations. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 1135–1148. [Google Scholar] [CrossRef]
- Qi, Y.; Kong, J.; He, J. Genetic relationship between IL-10 gene polymorphisms and the risk of clinical atopic dermatitis. BMC Med. Genet. 2019, 20, 83. [Google Scholar] [CrossRef]
- Yoon, N.Y.; Wang, H.Y.; Jun, M.; Jung, M.; Kim, D.H.; Lee, N.R.; Hong, K.W.; Seo, S.J.; Choi, E.; Lee, J.; et al. Simultaneous detection of barrier- and immune-related gene variations in patients with atopic dermatitis by reverse blot hybridization assay. Clin. Exp. Dermatol. 2018, 43, 430–436. [Google Scholar] [CrossRef]
- Dand, N.; Mahil, S.K.; Capon, F.; Smith, C.H.; Simpson, M.A.; Barker, J.N. Psoriasis and genetics. Acta Derm. Venereol. 2020, 100, 54–64. [Google Scholar] [CrossRef]
- Huang, Y.H.; Chiou, M.J.; Yang, S.F.; Kuo, C.F. The effect of paternal psoriasis on neonatal outcomes: A nationwide population-based study. Front. Immunol. 2023, 14, 1172274. [Google Scholar] [CrossRef]
- Griffiths, C.E.; Barker, J.N. Pathogenesis and clinical features of psoriasis. Lancet 2007, 370, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Harden, J.L.; Krueger, J.G.; Bowcock, A.M. The immunogenetics of Psoriasis: A comprehensive review. J. Autoimmun. 2015, 64, 66–73. [Google Scholar] [CrossRef]
- Veal, C.D.; Capon, F.; Allen, M.H.; Heath, E.K.; Evans, J.C.; Jones, A.; Patel, S.; Burden, D.; Tillman, D.; Barker, J.N.W.N.; et al. Family-based analysis using a dense single-nucleotide polymorphism–based map defines genetic variation at PSORS1, the major psoriasis-susceptibility locus. Am. J. Hum. Genet. 2002, 71, 554–564. [Google Scholar] [CrossRef]
- Jonca, N.; Leclerc, E.A.; Caubet, C.; Simon, M.; Guerrin, M.; Serre, G. Corneodesmosomes and corneodesmosin: From the stratum corneum cohesion to the pathophysiology of genodermatoses. Eur. J. Dermatol. 2011, 21 (Suppl. S2), 35–42. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.T.; Cao, L.; Roberson, E.D.O.; Pierson, K.C.; Yang, C.-F.; Joyce, C.E.; Ryan, C.; Duan, S.; Helms, C.A.; Liu, Y.; et al. PSORS2 is due to mutations in CARD14. Am. J. Hum. Genet. 2012, 90, 784–795. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.T.; Cao, L.; Roberson, E.D.O.; Duan, S.; Helms, C.A.; Nair, R.P.; Duffin, K.C.; Stuart, P.E.; Goldgar, D.; Hayashi, G.; et al. Rare and common variants in CARD14, encoding an epidermal regulator of NF-kappaB, in psoriasis. Am. J. Hum. Genet. 2012, 90, 796–808. [Google Scholar] [CrossRef]
- Zhang, X.-J.; Huang, W.; Yang, S.; Sun, L.-D.; Zhang, F.-Y.; Zhu, Q.-X.; Zhang, F.-R.; Zhang, C.; Du, W.-H.; Pu, X.-M.; et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 2009, 41, 205–210. [Google Scholar] [CrossRef]
- Tsoi, L.C.; Spain, S.L.; Knight, J.; Ellinghaus, E.; Stuart, P.E.; Capon, F.; Ding, J.; Li, Y.; Tejasvi, T.; Gudjonsson, J.E.; et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012, 44, 1341–1348. [Google Scholar] [CrossRef]
- Dand, N.; Mucha, S.; Tsoi, L.C.; Mahil, S.K.; Stuart, P.E.; Arnold, A.; Baurecht, H.; Burden, A.D.; Callis Duffin, K.; Chandran, V.; et al. Exome-wide association study reveals novel psoriasis susceptibility locus at TNFSF15 and rare protective alleles in genes contributing to type I IFN signalling. Hum. Mol. Genet. 2017, 26, 4301–4313. [Google Scholar] [CrossRef] [PubMed]
- Dand, N.; Stuart, P.E.; Bowes, J.; Ellinghaus, D.; Nititham, J.; Saklatvala, J.R.; Teder-Laving, M.; Thomas, L.F.; Traks, T.; Uebe, S.; et al. GWAS meta-analysis of psoriasis identifies new susceptibility alleles impacting disease mechanisms and therapeutic targets. medRxiv 2023. [Google Scholar]
- Sun, L.D.; Cheng, H.; Wang, Z.X.; Zhang, A.P.; Wang, P.G.; Xu, J.H.; Zhu, Q.X.; Zhou, H.S.; ELinghaus, E.; Zhang, F.R.; et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat. Genet. 2010, 42, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Hollox, E.J.; Huffmeier, U.; Zeeuwen, P.L.J.M.; Palla, R.; Lascorz, J.; Rodijk-Olthuis, D.; Van De Kerkhof, P.C.M.; Traupe, H.; De Jongh, G.; Heijer, M.D.; et al. Psoriasis is associated with increased β-defensin genomic copy number. Nat. Genet. 2008, 40, 23–25. [Google Scholar] [CrossRef]
- Li, M.; Wu, Y.; Chen, G.; Yang, Y.; Zhou, D.; Zhang, Z.; Zhang, D.; Chen, Y.; Lu, Z.; He, L.; et al. Deletion of the late cornified envelope genes LCE3C and LCE3B is associated with psoriasis in a Chinese population. J. Investig. Dermatol. 2011, 131, 1639–1643. [Google Scholar] [CrossRef]
- Stuart, P.E.; Nair, R.P.; ELinghaus, E.; Ding, J.; Tejasvi, T.; GudjonSon, J.E.; Li, Y.; Weidinger, S.; Eberlein, B.; Gieger, C.; et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat. Genet. 2010, 42, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Hüffmeier, U.; Uebe, S.; Ekici, A.B.; Bowes, J.; Giardina, E.; Korendowych, E.; Juneblad, K.; Apel, M.; McManus, R.; Ho, P.; et al. CoMon variants at TRAF3IP2 are associated with susceptibility to psoriatic arthritis and psoriasis. Nat. Genet. 2010, 42, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Shared Principles in NF-κB Signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef]
- Kuiper, J.J.; Prinz, J.C.; Stratikos, E.; Kuśnierczyk, P.; Arakawa, A.; Springer, S.; Mintoff, D.; Padjen, I.; Shumnalieva, R.; Vural, S.; et al. EULAR study group on “MHC-I-opathy”: Identifying disease-overarching mechanisms across disciplines and borders. Ann. Rheum. Dis. 2023, 82, 887–896. [Google Scholar] [CrossRef]
- Arakawa, A.; Reeves, E.; Vollmer, S.; Arakawa, Y.; He, M.; Galinski, A.; Stöhr, J.; Dornmair, K.; James, E.; Prinz, J.C. ERAP1 Controls the Autoimmune Response against Melanocytes in Psoriasis by Generating the Melanocyte Autoantigen and Regulating Its Amount for HLA-C*06:02 Presentation. J. Immunol. 2021, 207, 2235–2244. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.P.; Ruether, A.; Stuart, P.E.; Jenisch, S.; Tejasvi, T.; Hiremagalore, R.; Schreiber, S.; Kabelitz, D.; Lim, H.W.; Voorhees, J.J.; et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J. Investig. Dermatol. 2008, 128, 1653–1661. [Google Scholar] [CrossRef]
- Strange, A.; Capon, F.; Spencer, C.C.A.; Knight, J.; Weale, M.E.; Allen, M.H.; Barton, A.; Band, G.; Bellenguez, C.; Bergboer, J.G.M.; et al. A genome-wide asociation study identifies new psoriasis susceptibility loci and an interaction betwEn HLA-C and ERAP1. Nat. Genet. 2010, 42, 985–990. [Google Scholar]
- Babaie, F.; Omraninava, M.; Gorabi, A.M.; Khosrojerdi, A.; Aslani, S.; Yazdchi, A.; Torkamandi, S.; Mikaeili, H.; Sathyapalan, T.; Sahebkar, A. Etiopathogenesis of Psoriasis from Genetic Perspective: An updated Review. Curr. Genom. 2022, 23, 163–174. [Google Scholar]
- Ellinghaus, D.; Ellinghaus, E.; Nair, R.P.; Stuart, P.E.; Esko, T.; Metspalu, A.; Debrus, S.; Raelson, J.V.; Tejasvi, T.; Belouchi, M.; et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am. J. Hum. Genet. 2012, 90, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Q.; Yang, Z.; Wang, W.; Li, B.; Bai, M.; Wu, J.; Ge, H.; Dong, Z.; Shen, J.; Tang, H.; et al. Genetic Study on Small Insertions and Deletions in Psoriasis Reveals a Role in Complex Human Diseases. J. Investig. Dermatol. 2019, 139, 2302–2312.e14. [Google Scholar] [CrossRef]
- Mateu-Arrom, L.; Puig, L. Genetic and Epigenetic Mechanisms of Psoriasis. Genes 2023, 14, 1619. [Google Scholar] [CrossRef] [PubMed]
- Ray-Jones, H.; Eyre, S.; Barton, A.; Warren, R.B. One SNP at a Time: Moving beyond GWAS in Psoriasis. J. Investig. Dermatol. 2016, 136, 567–573. [Google Scholar] [CrossRef]
- Goldminz, A.M.; Au, S.C.; Kim, N.; Gottlieb, A.B.; Lizzul, P.F. NF-κB: An essential transcription factor in psoriasis. J. Dermatol. Sci. 2013, 69, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Hoesel, B.; Schmid, J.A. The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 2013, 12, 86. [Google Scholar] [CrossRef]
- Prens, E.P.; Kant, M.; Van Dijk, G.; Van Der Wel, L.I.; Mourits, S.; Van Der Fits, L. IFN-α enhances poly-IC responses in human keratinocytes by inducing expression of cytosolic innate RNA receptors: Relevance for psoriasis. J. Investig. Dermatol. 2008, 128, 932–938. [Google Scholar] [CrossRef]
- Gibson, F.; Hanly, A.; Grbic, N.; Grunberg, N.; Wu, M.; Collard, M.; Alani, R.M. Epigenetic Dysregulation in Autoimmune and Inflammatory Skin Diseases. Clin. Rev. Allergy Immunol. 2022, 63, 447–471. [Google Scholar] [CrossRef]
- Feng, B.J.; Sun, L.D.; Soltani-Arabshahi, R.; Bowcock, A.M.; Nair, R.P.; Stuart, P.; Elder, J.T.; Schrodi, S.J.; Begovich, A.B.; Abecasis, G.R.; et al. Multiple loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet. 2009, 5, e1000606. [Google Scholar] [CrossRef]
- Kagami, S.; Rizzo, H.L.; Lee, J.J.; Koguchi, Y.; Blauvelt, A. Circulating Th17, Th22, and Th1 cells are increased in psoriasis. J. Investig. Dermatol. 2010, 130, 1373–1383. [Google Scholar] [CrossRef]
- McGeachy, M.J.; Chen, Y.; Tato, C.M.; Laurence, A.; Joyce-Shaikh, B.; Blumenschein, W.M.; McClanahan, T.K.; O’Shea, J.J.; Cua, D.J. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 2009, 10, 314–324. [Google Scholar] [CrossRef]
- Cargill, M.; Schrodi, S.J.; Chang, M.; Garcia, V.E.; Brandon, R.; Callis, K.P.; Matsunami, N.; Ardlie, K.G.; Civello, D.; Catanese, J.J.; et al. k Present affiliation: Oklahoma Medical Research Foundation, Oklahoma City. Am. J. Hum. Genet. 2007, 80, 273–290. [Google Scholar] [CrossRef]
- Ammar, M.; Bouchlaka-Souissi, C.; Zaraa, I.; Helms, C.; Doss, N.; Bouazizi, F.; Dhaoui, R.; Ossman, A.B.; Ammar-el Gaied, A.B.; Mokni, M. Family-based association study in Tunisian familial psoriasis. Int. J. Dermatol. 2012, 51, 1329–1334. [Google Scholar] [CrossRef]
- Purzycka-Bohdan, D.; Nedoszytko, B.; Sobalska-Kwapis, M.; Zabłotna, M.; Żmijewski, M.A.; Wierzbicka, J.; Gleń, J.; Strapagiel, D.; Szczerkowska-Dobosz, A.; Nowicki, R.J. Assessment of the Potential Role of Selected Single Nucleotide Polymorphisms (SNPs) of Genes Related to the Functioning of Regulatory T Cells in the Pathogenesis of Psoriasis. Int. J. Mol. Sci. 2023, 24, 6061. [Google Scholar] [CrossRef]
- Kim, J.; Moreno, A.; Krueger, J.G. The imbalance between Type 17 T-cells and regulatory immune cell subsets in psoriasis vulgaris. Front. Immunol. 2022, 13, 1005115. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, Z.; Zhao, Z.; Yu, Y.; Fan, H.; Xu, X.; Bu, X.; Gu, J. IL-21 induces an imbalance of Th17/treg cells in moderate-to-severe plaque psoriasis patients. Front. Immunol. 2019, 10, 1865. [Google Scholar] [CrossRef]
- Zhong, L.; Luo, N.; Zhong, X.; Xu, T.; Hao, P. The immunoregulatory effects of natural products on psoriasis via its action on Th17 cells versus regulatory T cells balance. Int. Immunopharmacol. 2022, 110, 109032. [Google Scholar] [CrossRef]
- Young, K.Z.; Sarkar, M.K.; Gudjonsson, J.E. Pathophysiology of generalized pustular psoriasis. Exp. Dermatol. 2023, 32, 1194–1203. [Google Scholar] [CrossRef]
- Mahil, S.K.; Twelves, S.; Farkas, K.; Setta-Kaffetzi, N.; Burden, A.D.; Gach, J.E.; Irvine, A.D.; Képíró, L.; Mockenhaupt, M.; Oon, H.H.; et al. AP1S3 Mutations Cause Skin Autoinflammation by Disrupting Keratinocyte Autophagy and Up-Regulating IL-36 Production. J. Investig. Dermatol. 2016, 136, 2251–2259. [Google Scholar] [CrossRef]
- Onitsuka, M.; Farooq, M.; Iqbal, M.N.; Yasuno, S.; Shimomura, Y. A homozygous loss-of-function variant in the MPO gene is associated with generalized pustular psoriasis. J. Dermatol. 2023, 50, 664–671. [Google Scholar] [CrossRef]
- Elhage, K.G.; Kranyak, A.; Jin, J.Q.; Haran, K.; Spencer, R.K.; Smith, P.; Davis, M.S.; Hakimi, M.; Bhutani, T.; Liao, W. Mendelian randomization studies in atopic dermatitis: A systematic review. J. Investig. Dermatol. 2023. [Google Scholar] [CrossRef]
- Zhou, W.; Cai, J.; Li, Z.; Lin, Y. Association of atopic dermatitis with autoimmune diseases: A bidirectional and multivariable two-sample mendelian randomization study. Front. Immunol. 2023, 14, 1132719. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, W. Genome-Wide Integration of Genetic and Genomic Studies of Atopic Dermatitis: Insights into Genetic Architecture and Pathogenesis. J. Investig. Dermatol. 2022, 142, 2958–2967.e8. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, L.; Wang, Y.; Wang, X.; Lewis, S.J.; Wang, J. Atopic dermatitis and risk of 14 site-specific cancers: A Mendelian randomization study. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 2490–2497. [Google Scholar] [CrossRef]
- Jin, J.Q.; Elhage, K.G.; Spencer, R.K.; Davis, M.S.; Hakimi, M.; Bhutani, T.; Liao, W. Mendelian Randomization Studies in Psoriasis and Psoriatic Arthritis: A Systematic Review. J. Investig. Dermatol. 2023, 143, 762–776.e3. [Google Scholar] [CrossRef]
- Chalitsios, C.V.; Meena, D.; Manou, M.; Papagiannopoulos, C.; Markozannes, G.; Gill, D.; Su, B.; Tsilidis, K.K.; Evangelou, E.; Tzoulaki, I. Multiple long-term conditions in people with psoriasis: A latent class and bidirectional Mendelian randomisation analysis. Br. J. Dermatol. 2023, ljad410. [Google Scholar] [CrossRef]
- Zhang, D.; Li, Y.; Du, C.; Sang, L.; Liu, L.; Li, Y.; Wang, F.; Fan, W.; Tang, P.; Zhang, S.; et al. Evidence of pyroptosis and ferroptosis extensively involved in autoimmune diseases at the single-cell transcriptome level. J. Transl. Med. 2022, 20, 363. [Google Scholar] [CrossRef]
- Zhou, Q.; Yang, L.; Li, T.; Wang, K.; Huang, X.; Shi, J.; Wang, Y. Mechanisms and inhibitors of ferroptosis in psoriasis. Front. Mol. Biosci. 2022, 9, 1019447. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zhu, J.; Chen, J.; Jia, S.; Nie, S. Significance of cuproptosis- related genes in the diagnosis and classification of psoriasis. Front. Mol. Biosci. 2023, 10, 1115091. [Google Scholar] [CrossRef]
- Wu, W.; Chen, G.; Zhang, Z.; He, M.; Li, H.; Yan, F. Construction and verification of atopic dermatitis diagnostic model based on pyroptosis related biological markers using machine learning methods. BMC Med. Genom. 2023, 16, 138. [Google Scholar] [CrossRef]
- Zheng, J.; Yao, L.; Zhou, Y.; Gu, X.; Wang, C.; Bao, K.; Sun, Y.; Hong, M. A novel function of NLRP3 independent of inflammasome as a key transcription factor of IL-33 in epithelial cells of atopic dermatitis. Cell Death Dis. 2021, 12, 871. [Google Scholar] [CrossRef] [PubMed]
- de Koning, H.D.; Bergboer, J.G.M.; van den Bogaard, E.H.; van Vlijmen-Willems, I.M.J.J.; Rodijk-Olthuis, D.; Simon, A.; Zeeuwen, P.L.J.M.; Schalkwijk, J. Strong induction of AIM2 expression in human epidermis in acute and chronic inflammatory skin conditions. Exp. Dermatol. 2012, 21, 961–964. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zhao, R.; Xia, W.; Chang, C.-W.; You, Y.; Hsu, J.-M.; Nie, L.; Chen, Y.; Wang, Y.-C.; Liu, C.; et al. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat. Cell Biol. 2020, 22, 1264–1275. [Google Scholar] [CrossRef] [PubMed]
- Ong, P.Y. New insights in the pathogenesis of atopic dermatitis. Pediatr. Res. 2014, 75, 171–175. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef]
- Wu, M.N.; Zhou, D.M.; Jiang, C.Y.; Chen, W.W.; Chen, J.C.; Zou, Y.M.; Han, T.; Zhou, L.J.M. Genetic analysis of potential biomarkers and therapeutic targets in ferroptosis from psoriasis. Front. Immunol. 2023, 13, 1104462. [Google Scholar] [CrossRef]
- Shou, Y.; Yang, L.; Yang, Y.; Xu, J. Inhibition of keratinocyte ferroptosis suppresses psoriatic inflammation. Cell Death Dis. 2021, 12, 1009. [Google Scholar] [CrossRef]
- Sengupta, A.; Lichti, U.F.; Carlson, B.A.; Cataisson, C.; Ryscavage, A.O.; Mikulec, C.; Conrad, M.; Fischer, S.M.; Hatfield, D.L.; Yuspa, S.H. Targeted disruption of glutathione peroxidase 4 in mouse skin epithelial cells impairs postnatal hair follicle morphogenesis that is partially rescued through inhibition of COX-2. J. Investig. Dermatol. 2013, 133, 1731–1741. [Google Scholar] [CrossRef]
- Arbiser, J.L.; Bonner, M.Y.; Ward, N.; Elsey, J.; Rao, S. Selenium unmasks protective iron armor: A possible defense against cutaneous inflammation and cancer. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2518–2527. [Google Scholar] [CrossRef]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.-J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Lian, N.; Chen, Y.; Chen, S.; Zhang, Y.; Chen, H.; Yang, Y.; Gu, H.; Chen, Q.; Li, M.; Chen, X. Gasdermin D-mediated keratinocyte pyroptosis as a key step in psoriasis pathogenesis. Cell Death Dis. 2023, 14, 595. [Google Scholar] [CrossRef] [PubMed]
- Dopytalska, K.; Ciechanowicz, P.; Wiszniewski, K.; Szymańska, E.; Walecka, I. The role of epigenetic factors in psoriasis. Int. J. Mol. Sci. 2021, 22, 9294. [Google Scholar] [CrossRef] [PubMed]
- Frischknecht, L.; Vecellio, M.; Selmi, C. The role of epigenetics and immunological imbalance in the etiopathogenesis of psoriasis and psoriatic arthritis. Ther. Adv. Musculoskelet. Dis. 2019, 11, 1759720X19886505. [Google Scholar] [CrossRef] [PubMed]
- Bonamonte, D.; Filoni, A.; Vestita, M.; Romita, P.; Foti, C.; Angelini, G. The Role of the Environmental Risk Factors in the Pathogenesis and Clinical Outcome of Atopic Dermatitis. BioMed Res. Int. 2019, 2019, 2450605. [Google Scholar] [CrossRef]
- Løset, M.; Brown, S.J.; Saunes, M.; Hveem, K. Genetics of Atopic Dermatitis: From DNA Sequence to Clinical Relevance. Dermatology 2019, 235, 355–364. [Google Scholar] [CrossRef]
- Stemmler, S.; Hoffjan, S. Trying to understand the genetics of atopic dermatitis. Mol. Cell. Probes 2016, 30, 374–385. [Google Scholar] [CrossRef]
- Nguyen, C.M.; Liao, W. Genomic imprinting in psoriasis and atopic dermatitis: A review. J. Dermatol. Sci. 2015, 80, 89–93. [Google Scholar] [CrossRef]
- Bin, L.; Leung, D.Y.M. Genetic and epigenetic studies of atopic dermatitis. Allergy Asthma Clin. Immunol. 2016, 12, 52. [Google Scholar] [CrossRef]
- Botchkarev, V.A.; Gdula, M.R.; Mardaryev, A.N.; Sharov, A.A.; Fessing, M.Y. Epigenetic regulation of gene expression in keratinocytes. J. Investig. Dermatol. 2012, 132, 2505–2521. [Google Scholar] [CrossRef]
- Alagia, A.; Gullerova, M. The Methylation Game: Epigenetic and Epitranscriptomic Dynamics of 5-Methylcytosine. Front. Cell Dev. Biol. 2022, 10, 915685. [Google Scholar] [CrossRef]
- Luo, Y.; Zhou, B.; Zhao, M.; Tang, J.; Lu, Q. Promoter demethylation contributes to TSLP overexpression in skin lesions of patients with atopic dermatitis. Clin. Exp. Dermatol. 2014, 39, 48–53. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, P.; Zhao, M.; Liang, G.; Yin, H.; Zhang, G.; Wen, H.; Lu, Q. Demethylation of the FCER1G promoter leads to FcεRI overexpression on monocytes of patients with atopic dermatitis. Allergy Eur. J. Allergy Clin. Immunol. 2012, 67, 424–430. [Google Scholar] [CrossRef]
- Ziyab, A.H.; Karmaus, W.; Holloway, J.W.; Zhang, H.; Ewart, S.; Arshad, S.H. DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. J. Eur. Acad. Dermatol. Venereol. 2013, 27, e420–e423. [Google Scholar] [CrossRef]
- Stevens, M.L.; Zhang, Z.; Johansson, E.; Ray, S.; Jagpal, A.; Ruff, B.P.; Kothari, A.; He, H.; Martin, L.J.; Ji, H.; et al. Disease-associated KIF3A variants alter gene methylation and expression impacting skin barrier and atopic dermatitis risk. Nat. Commun. 2020, 11, 4092. [Google Scholar] [CrossRef]
- Johansson, E.; Biagini Myers, J.M.; Martin, L.J.; He, H.; Pilipenko, V.; Mersha, T.; Weirauch, M.; Salomonis, N.; Ryan, P.; LeMasters, G.K.; et al. KIF3A genetic variation is associated with pediatric asthma in the presence of eczema independent of allergic rhinitis. J. Allergy Clin. Immunol. 2017, 140, 595–598.e5. [Google Scholar] [CrossRef]
- Potaczek, D.P.; Harb, H.; Michel, S.; Alhamwe, B.A.; Renz, H.; Tost, J. Epigenetics and allergy: From basic mechanisms to clinical applications. Epigenomics 2017, 9, 539–571. [Google Scholar] [CrossRef]
- Lee, D.U.; Agarwal, S.; Rao, A. Th2 Lineage Commitment and Efficient IL-4 Production Involves Extended Demethylation of the IL-4 Gene. Immunity 2002, 16, 649–660. [Google Scholar] [CrossRef]
- Jones, B.; Chen, J. Inhibition of IFN-γ transcription by site-specific methylation during T helper cell development. EMBO J. 2006, 25, 2443–2452. [Google Scholar] [CrossRef]
- Harb, H.; Renz, H. Update on epigenetics in allergic disease. J. Allergy Clin. Immunol. 2015, 135, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Kehrmann, J.; Tatura, R.; Zeschnigk, M.; Probst-Kepper, M.; Geffers, R.; Steinmann, J.; Buer, J. Impact of 5-aza-2′-deoxycytidine and epigallocatechin-3-gallate for induction of human regulatory T cells. Immunology 2014, 142, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Nedoszytko, B.; Sokołowska-Wojdyło, M.; Renke, J.; Lange, M.; Trzonkowski, P.; Sobjanek, M.; Szczerkowska-Dobosz, A.; Niedoszytko, M.; Górska, A.; Romantowski, J.; et al. The role of regulatory T cells and genes involved in their differentiation in pathogenesis of selected inflammatory and neoplastic skin diseases. Part III: Polymorphisms of genes involved in Tregs’ activation and function. Postęp. Dermatol. Alergol. 2017, 34, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Qu, K.; Kuai, L.; Ru, Y.; Huang, K.; Yan, X.; Xing, M. Epigenetics in psoriasis: Perspective of DNA methylation. Mol. Genet. Genom. 2021, 296, 1027–1040. [Google Scholar] [CrossRef] [PubMed]
- Chandra, A.; Senapati, S.; Roy, S.; Chatterjee, G.; Chatterjee, R. Epigenome-wide DNA methylation regulates cardinal pathological features of psoriasis. Clin. Epigenet. 2018, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Wang, W.; Shen, C.; Li, H.; Zuo, X.; Zheng, X.; Yue, M.; Zhang, C.; Yu, L.; Chen, M.; et al. Epigenome-Wide Association Analysis Identified Nine Skin DNA Methylation Loci for Psoriasis. J. Investig. Dermatol. 2016, 136, 779–787. [Google Scholar] [CrossRef]
- Gao, L.; Lu, Q. The critical importance of epigenetics in autoimmune-related skin diseases. Front. Med. 2023, 17, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Nobeyama, Y.; Umezawa, Y.; Nakagawa, H. Less-invasive analysis of DNA methylation using psoriatic scales. J. Dermatol. Sci. 2016, 83, 70–73. [Google Scholar] [CrossRef]
- Ghaffarinia, A.; Ayaydin, F.; Póliska, S.; Manczinger, M.; Bolla, B.S.; Flink, L.B.; Balogh, F.; Veréb, Z.; Bozó, R.; Szabó, K.; et al. Psoriatic Resolved Skin Epidermal Keratinocytes Retain Disease-Residual Transcriptomic and Epigenomic Profiles. Int. J. Mol. Sci. 2023, 24, 4556. [Google Scholar] [CrossRef]
- Verma, D.; Ekman, A.K.; Bivik Eding, C.; Enerbäck, C. Genome-Wide DNA Methylation Profiling Identifies Differential Methylation in Uninvolved Psoriatic Epidermis. J. Investig. Dermatol. 2018, 138, 1088–1093. [Google Scholar] [CrossRef]
- Zhang, P.; Su, Y.; Chen, H.; Zhao, M.; Lu, Q. Abnormal DNA methylation in skin lesions and PBMCs of patients with psoriasis vulgaris. J. Dermatol. Sci. 2010, 60, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Yin, G.; An, P.; Wang, C.; Liu, R.; Yang, Y.; Yan, X.; Li, J.; Li, X.; Zhang, K. DNA methylation of dermal MSCs in psoriasis: Identification of epigenetically dysregulated genes. J. Dermatol. Sci. 2013, 72, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zeng, J.; Lu, J. Critical role of epigenetic modification in the pathogenesis of atopic dermatitis. Indian J. Dermatol. Venereol. Leprol. 2023, 89, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Ezhkova, E.; Pasolli, H.A.; Parker, J.S.; Stokes, N.; Su, I.; Hannon, G.; Tarakhovsky, A.; Fuchs, E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 2009, 136, 1122–1135. [Google Scholar] [CrossRef] [PubMed]
- Hopkin, A.S.; Gordon, W.; Klein, R.H.; Espitia, F.; Daily, K.; Zeller, M.; Baldi, P.; Andersen, B. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 2012, 8, e1002829. [Google Scholar] [CrossRef]
- Elder, J.T.; Zhao, X. Evidence for local control of gene expression in the epidermal differentiation complex. Exp. Dermatol. 2002, 11, 406–412. [Google Scholar] [CrossRef]
- Steelant, B.; Wawrzyniak, P.; Martens, K.; Jonckheere, A.C.; Pugin, B.; Schrijvers, R.; Bullens, D.M.; Vanoirbeek, J.A.; Krawczyk, K.; Dreher, A.; et al. Blocking histone deacetylase activity as a novel target for epithelial barrier defects in patients with allergic rhinitis. J. Allergy Clin. Immunol. 2019, 144, 1242–1253.e7. [Google Scholar] [CrossRef]
- Harb, H.; Alashkar Alhamwe, B.; Acevedo, N.; Frumento, P.; Johansson, C.; Eick, L.; Papadogiannakis, N.; Alm, J.; Renz, H.; Potaczek, D.P.; et al. Epigenetic Modifications in Placenta are Associated with the Child’s Sensitization to Allergens. BioMed Res. Int. 2019, 2019, 1315257. [Google Scholar] [CrossRef]
- Liu, Z.; Cao, W.; Xu, L.; Chen, X.; Zhan, Y.; Yang, Q.; Liu, S.; Chen, P.; Jiang, Y.; Sun, X.; et al. The histone H3 lysine-27 demethylase Jmjd3 plays a critical role in specific regulation of Th17 cell differentiation. J. Mol. Cell Biol. 2015, 7, 505–516. [Google Scholar] [CrossRef]
- Li, H.; Yao, Q.; Mariscal, A.G.; Wu, X.; Hülse, J.; Pedersen, E.; Helin, K.; Waisman, A.; Vinkel, C.; Thomsen, S.F.; et al. Epigenetic control of IL-23 expression in keratinocytes is important for chronic skin inflammation. Nat. Commun. 2018, 9, 1420. [Google Scholar] [CrossRef]
- Ovejero-Benito, M.C.; Reolid, A.; Sánchez-Jiménez, P.; Saiz-Rodríguez, M.; Muñoz-Aceituno, E.; Llamas-Velasco, M.; Martín-Vilchez, S.; Cabaleiro, T.; Román, M.; Ochoa, D.; et al. Histone modifications associated with biological drug response in moderate-to-severe psoriasis. Exp. Dermatol. 2018, 27, 1361–1371. [Google Scholar] [CrossRef]
- Tovar-Castillo, L.E.; Cancino-Díaz, J.C.; García-Vázquez, F.; Cancino-Gómez, F.G.; León-Dorantes, G.; Blancas-González, F.; Jiménez-Zamudio, L.; García-Latorre, E.; Cancino-Díaz, M.E. Under-expression of VHL and over-expression of HDAC-1, HIF-1alpha, LL-37, and IAP-2 in affected skin biopsies of patients with psoriasis. Int. J. Dermatol. 2007, 46, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Cao, G.; Sun, G.; Zhu, L.; Tian, Y.; Song, Y.; Guo, C.; Wang, X.; Zhong, J.; Zhou, W.; et al. GLS1-mediated glutaminolysis unbridled by MALT1 protease promotes psoriasis pathogenesis. J. Clin. Investig. 2020, 130, 5180–5196. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, A.; Al-Harbi, N.O.; Al-Harbi, M.M.; El-Sherbeeny, A.M.; Ahmad, S.F.; Siddiqui, N.; Ansari, M.A.; Zoheir, K.M.A.; Attia, S.M.; Al-Hosaini, K.A.; et al. Imiquimod-induced psoriasis-like skin inflammation is suppressed by BET bromodomain inhibitor in mice through RORC/IL-17A pathway modulation. Pharmacol. Res. 2015, 99, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Tsoi, L.C.; Gudjonsson, J.E. Dysregulated epigenetic modifications in psoriasis. Exp. Dermatol. 2021, 30, 1156–1166. [Google Scholar] [CrossRef]
- Rebane, A.; Akdis, C.A. MicroRNAs: Essential players in the regulation of inflammation. J. Allergy Clin. Immunol. 2013, 132, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.R.; Brede, G.; Johansen, J.; Johnsen, R.; Storrø, O.; Sætrom, P.; Øien, T. Human breast milk miRNA; maternal probiotic supplementation and atopic dermatitis in offspring. PLoS ONE 2015, 10, e0143496. [Google Scholar] [CrossRef]
- Sonkoly, E.; Ståhle, M.; Pivarcsi, A. MicroRNAs and immunity: Novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 2008, 18, 131–140. [Google Scholar] [CrossRef]
- Ghosh, D.; Ganguly, T.; Chatterjee, R. Emerging roles of non-coding RNAs in psoriasis pathogenesis. Funct. Integr. Genom. 2023, 23, 129. [Google Scholar] [CrossRef]
- Rebane, A.; Runnel, T.; Aab, A.; Maslovskaja, J.; Rückert, B.; Zimmermann, M.; Plaas, M.; Kärner, J.; Treis, A.; Pihlap, M.; et al. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J. Allergy Clin. Immunol. 2014, 134, 836–847.e11. [Google Scholar] [CrossRef]
- Sonkoly, E.; Janson, P.; Majuri, M.L.; Savinko, T.; Fyhrquist, N.; Eidsmo, L.; Xu, N.; Meisgen, F.; Wei, T.; Bradley, M.; et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J. Allergy Clin. Immunol. 2010, 126, 581–589.e20. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Rothenberg, M.E. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J. Allergy Clin. Immunol. 2013, 132, 3–13. [Google Scholar] [CrossRef]
- Lu, T.X.; Hartner, J.; Lim, E.-J.; Fabry, V.; Mingler, M.K.; Cole, E.T.; Orkin, S.H.; Aronow, B.J.; Rothenberg, M.E. MicroRNA-21 Limits In Vivo Immune Response-Mediated Activation of the IL-12/IFN-γ Pathway, Th1 Polarization, and the Severity of Delayed-Type Hypersensitivity. J. Immunol. 2011, 187, 3362–3373. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, M.; Li, L.; Zhang, L.; Chan, M.T.V.; Wu, W.K.K. MicroRNAs in atopic dermatitis: A systematic review. J. Cell. Mol. Med. 2020, 24, 5966–5972. [Google Scholar] [CrossRef]
- Yao, R.; Ma, Y.L.; Liang, W.; Li, H.H.; Ma, Z.J.; Yu, X.; Liao, Y.H. MicroRNA-155 Modulates Treg and Th17 Cells Differentiation and Th17 Cell Function by Targeting SOCS1. PLoS ONE 2012, 7, e46082. [Google Scholar] [CrossRef] [PubMed]
- Specjalski, K.; Jassem, E. MicroRNAs: Potential Biomarkers and Targets of Therapy in Allergic Diseases? Arch. Immunol. Ther. Exp. 2019, 67, 213–223. [Google Scholar] [CrossRef]
- Liang, Y.; Chang, C.; Lu, Q. The Genetics and Epigenetics of Atopic Dermatitis—Filaggrin and Other Polymorphisms. Clin. Rev. Allergy Immunol. 2016, 51, 315–328. [Google Scholar] [CrossRef]
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk: An exosomal microRNA transmitter promoting thymic regulatory T cell maturation preventing the development of atopy? J. Transl. Med. 2014, 12, 43. [Google Scholar] [CrossRef]
- Ma, L.; Xue, H.B.; Wang, F.; Shu, C.M.; Zhang, J.H. MicroRNA-155 may be involved in the pathogenesis of atopic dermatitis by modulating the differentiation and function of T helper type 17 (Th17) cells. Clin. Exp. Immunol. 2015, 181, 142–149. [Google Scholar] [CrossRef]
- Beheshti, R.; Halstead, S.; McKeone, D.; Hicks, S.D. Understanding immunological origins of atopic dermatitis through multi-omic analysis. Pediatr. Allergy Immunol. 2022, 33, e13817. [Google Scholar] [CrossRef]
- Chen, X.F.; Zhang, L.J.; Zhang, J.; Dou, X.; Shao, Y.; Jia, X.J.; Zhang, W.; Yu, B. MiR-151a is involved in the pathogenesis of atopic dermatitis by regulating interleukin-12 receptor β2. Exp. Dermatol. 2018, 27, 427–432. [Google Scholar] [CrossRef]
- Li, H.M.; Xiao, Y.J.; Min, Z.S.; Tan, C. Identification and interaction analysis of key genes and microRNAs in atopic dermatitis by bioinformatics analysis. Clin. Exp. Dermatol. 2019, 44, 257–264. [Google Scholar] [CrossRef]
- Yoon, W.S.; Lee, S.S.; Chae, Y.S.; Park, Y.K. Therapeutic effects of recombinant Salmonella typhimurium harboring CCL22 miRNA on atopic dermatitis-like skin in mice. Exp. Mol. Med. 2011, 43, 63–70. [Google Scholar] [CrossRef]
- Wei, T.; Folkersen, L.; Biskup, E.; Xu, N.; Manfe, V.; Niazi, O.; Gniadecki, R. Ubiquitin-specific peptidase 2 as a potential link between microRNA-125b and psoriasis. Br. J. Dermatol. 2017, 176, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Huang, Y.; Zhu, X.; Lin, X.; Luo, D. miR-125b-mediated regulation of cell proliferation through the Jagged-1/Notch signaling pathway by inhibiting BRD4 expression in psoriasis. Mol. Med. Rep. 2019, 19, 5227–5236. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Shi, R.; Ma, R.; Tang, X.; Gong, Y.; Yu, Z.; Shi, Y. The role of microRNA in psoriasis: A review. Exp. Dermatol. 2023, 32, 1598–1612. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Pasquali, L.; Gao, C.; Srivastava, A.; Khera, N.; Freisenhausen, J.C.; Luo, L.; Rosén, E.; van Lierop, A.; Homey, B.; et al. miR-378a regulates keratinocyte responsiveness to interleukin-17A in psoriasis*. Br. J. Dermatol. 2022, 187, 211–222. [Google Scholar] [CrossRef]
- Yan, S.; Xu, Z.; Lou, F.; Zhang, L.; Ke, F.; Bai, J.; Liu, Z.; Liu, J.; Wang, H.; Zhu, H.; et al. NF-κB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat. Commun. 2015, 6, 7652. [Google Scholar] [CrossRef]
- Wu, R.; Zeng, J.; Yuan, J.; Deng, X.; Huang, Y.; Chen, L.; Zhang, P.; Feng, H.; Liu, Z.; Wang, Z.; et al. MicroRNA-210 overexpression promotes psoriasis-like inflammation by inducing Th1 and Th17 cell differentiation. J. Clin. Investig. 2018, 128, 2551–2568. [Google Scholar] [CrossRef]
- Magenta, A.; D’Agostino, M.; Sileno, S.; Di Vito, L.; Uras, C.; Abeni, D.; Martino, F.; Barillà, F.; Madonna, S.; Albanesi, C.; et al. The Oxidative Stress-Induced miR-200c Is Upregulated in Psoriasis and Correlates with Disease Severity and Determinants of Cardiovascular Risk. Oxidative Med. Cell. Longev. 2019, 2019, 8061901. [Google Scholar] [CrossRef]
- Wang, H.; Xu, Y.; Jin, M.; Li, H.; Li, S. miR-383 reduces keratinocyte proliferation and induces the apoptosis in psoriasis via disruption of LCN2-dependent JAK/STAT pathway activation. Int. Immunopharmacol. 2021, 96, 107587. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, F.; Tian, Q.; Dong, J.; Chen, L.; Hu, R. Involvement of miR-214-3p/FOXM1 Axis During the Progression of Psoriasis. Inflammation 2022, 45, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Liu, Z.; Wang, B. EZH2 is involved in psoriasis progression by impairing miR-125a-5p inhibition of SFMBT1 and leading to inhibition of the TGFβ/SMAD pathway. Ther. Adv. Chronic Dis. 2021, 12, 2040622320987348. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Wang, G.; Wang, M.; Chen, C.; Zhang, M.; Liu, M.; Shao, Y.; Zheng, Y. LncRNA RP6-65G23.1 accelerates proliferation and inhibits apoptosis via p-ERK1/2/p-AKT signaling pathway on keratinocytes. J. Cell Biochem. 2020, 121, 4580–4589. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Chen, F.; Hua, M.; Guo, J.; Nong, Y.; Tang, Q.; Zhong, F.; Qin, L. Knockdown of lncRNA MIR31HG inhibits cell proliferation in human HaCaT keratinocytes. Biol. Res. 2018, 51, 30. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Li, R.; Zhao, X.; Yan, J.; Sun, Q. Up-regulated lncRNA-MSX2P1 promotes the growth of IL-22-stimulated keratinocytes by inhibiting miR-6731-5p and activating S100A7. Exp. Cell Res. 2018, 363, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.; Wu, X.; Xiang, Y. Sinomenine Suppressed Keratinocyte Proliferation and Imiquimod-Induced Psoriasis-Like Dermatitis by Regulating lncRNA XIST. Ski. Pharmacol. Physiol. 2022, 35, 328–342. [Google Scholar] [CrossRef]
- Huang, S.; Zhen, Y.; Yin, X.; Yang, Z.; Li, X.; Wang, R.; Wen, H.; Zhong, H.; Yan, J.; Sun, Q. KMT2C Induced by FABP5P3 Aggravates Keratinocyte Hyperproliferation and Psoriasiform Skin Inflammation by Upregulating the Transcription of PIK3R3. J. Investig. Dermatol. 2023, 143, 37–47.e8. [Google Scholar] [CrossRef]
- Yin, X.; Yang, Z.; Zhu, M.; Chen, C.; Huang, S.; Li, X.; Zhong, H.; Wen, H.; Sun, Q.; Yu, X.; et al. ILF2 Contributes to Hyperproliferation of Keratinocytes and Skin Inflammation in a KLHDC7B-DT-Dependent Manner in Psoriasis. Front. Genet. 2022, 13, 890624. [Google Scholar] [CrossRef]
- Yin, X.; Yang, Z.; Zhu, M.; Chen, C.; Sun, Q. Role of the long non-coding RNA, SPRR2C, based on an in vitro psoriatic keratinocyte cell model. Eur. J. Dermatol. 2022, 32, 171–180. [Google Scholar]
- Jia, H.-Y.; Zhang, K.; Lu, W.-J.; Xu, G.-W.; Zhang, J.-F.; Tang, Z.-L. LncRNA MEG3 influences the proliferation and apoptosis of psoriasis epidermal cells by targeting miR-21/caspase-8. BMC Mol. Cell Biol. 2019, 20, 46. [Google Scholar] [CrossRef]
- Ahmed Shehata, W.; Maraee, A.; Abd El Monem Ellaithy, M.; Tayel, N.; Abo-Ghazala, A.; Mohammed El-Hefnawy, S. Circulating long noncoding RNA growth arrest-specific transcript 5 as a diagnostic marker and indicator of degree of severity in plaque psoriasis. Int. J. Dermatol. 2021, 60, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, H.Y.; Tawfik, N.Z.; Soliman, N.H.; Eldeen, L.A.T. The lncRNA PRINS-miRNA-mRNA Axis Gene Expression Profile as a Circulating Biomarker Panel in Psoriasis. Mol. Diagn. Ther. 2022, 26, 451–465. [Google Scholar] [CrossRef]
- Wang, D.; Cheng, S.; Zou, G.; Ding, X. Paeoniflorin inhibits proliferation and migration of psoriatic keratinocytes via the lncRNA NEAT1/miR-3194-5p/Galectin-7 axis. Anticancer Drugs 2022, 33, e423–e433. [Google Scholar] [CrossRef]
- Yang, Z.; Yin, X.; Chen, C.; Huang, S.; Li, X.; Yan, J.; Sun, Q. CircOAS3 Regulates Keratinocyte Proliferation and Psoriatic Inflammation by Interacting with Hsc70 via the JNK/STAT3/NF-κB Signaling Pathway. Inflammation 2022, 45, 1924–1935. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yang, Z.; Yin, X.; Huang, S.; Yan, J.; Sun, Q. CircEIF5 contributes to hyperproliferation and inflammation of keratinocytes in psoriasis via p-NFκB and p-STAT3 signalling pathway. Exp. Dermatol. 2022, 31, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Luo, J.; Chen, W.; He, Q.; Long, J.; Zhang, B. Circ_0060531 knockdown ameliorates IL-22-induced keratinocyte damage by binding to miR-330-5p to decrease GAB1 expression. Autoimmunity 2022, 55, 243–253. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, C.; Bai, X.; Xiao, C.; Dang, E.; Wang, G. hsa_circ_0003738 Inhibits the Suppressive Function of Tregs by Targeting miR-562/IL-17A and miR-490-5p/IFN-γ Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 21, 1111–1119. [Google Scholar] [CrossRef]
- Qiao, M.; Ding, J.; Yan, J.; Li, R.; Jiao, J.; Sun, Q. Circular RNA Expression Profile and Analysis of Their Potential Function in Psoriasis. Cell Physiol. Biochem. 2018, 50, 15–27. [Google Scholar] [CrossRef]
- Lu, J.; Xu, X.; Li, Y.; Yu, N.; Ding, Y.; Shi, Y. CircRAB3B suppresses proliferation, motility, cell cycle progression and promotes the apoptosis of IL-22-induced keratinocytes depending on the regulation of miR-1228-3p/PTEN axis in psoriasis. Autoimmunity 2021, 54, 303–312. [Google Scholar] [CrossRef]
- Xiong, J.; Tu, Y.; Feng, Z.; Li, D.; Yang, Z.; Huang, Q.; Li, Z.; Cao, Y.; Jie, Z. Epigenetics mechanisms mediate the miR-125a/BRMS1 axis to regulate invasion and metastasis in gastric cancer. OncoTargets Ther. 2019, 12, 7513–7525. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, H.; Wang, C.; Zeng, B.; Tang, X.; Zhang, Y.; Peng, Y.; Luo, M.; Huang, P.; Yang, Z. miR-203 promotes HaCaT cell overproliferation through targeting LXR-α and PPAR-γ. Cell Cycle 2020, 19, 1928–1940. [Google Scholar] [CrossRef]
- Luo, Q.; Zeng, J.; Li, W.; Lin, L.; Zhou, X.; Tian, X.; Liu, W.; Zhang, L.; Zhang, X. Silencing of miR-155 suppresses inflammatory responses in psoriasis through inflammasome NLRP3 regulation. Int. J. Mol. Med. 2018, 42, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-Y.; Kuo, H.-C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 29. [Google Scholar] [CrossRef] [PubMed]
- Chularojanamontri, L.; Charoenpipatsin, N.; Silpa-Archa, N.; Wongpraparut, C.; Thongboonkerd, V. Proteomics in Psoriasis. Int. J. Mol. Sci. 2019, 20, 1141. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-D.; Kim, S.-Y.; Jang, H.-S.; Seo, E.-Y.; Namkung, J.-H.; Park, H.-S.; Cho, S.Y.; Paik, Y.-K.; Yang, J.-M. Towards a proteomic analysis of atopic dermatitis: A two-dimensional-polyacrylamide gel electrophoresis/mass spectrometric analysis of cultured patient-derived fibroblasts. Proteomics 2004, 4, 3446–3455. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-D.; Jang, H.-S.; Kim, S.-Y.; Ko, S.-K.; Lyou, Y.-J.; Lee, D.-Y.; Paik, Y.-K.; Yang, J.-M. Two-dimensional electrophoretic profiling of atopic dermatitis in primary cultured keratinocytes from patients. Proteomics 2006, 6, 1362–1370. [Google Scholar] [CrossRef]
- Park, Y.-D.; Lyou, Y.-J.; Yang, J.-M. Detection of down-regulated acetaldehyde dehydrogenase 1 in atopic dermatitis patients by two-dimensional electrophoresis. Exp. Dermatol. 2007, 16, 130–134. [Google Scholar] [CrossRef]
- Howell, M.D.; Fairchild, H.R.; Kim, B.E.; Bin, L.; Boguniewicz, M.; Redzic, J.S.; Hansen, K.C.; Leung, D.Y.M. Th2 cytokines act on S100/A11 to downregulate keratinocyte differentiation. J. Investig. Dermatol. 2008, 128, 2248–2258. [Google Scholar] [CrossRef]
- Park, Y.-D.; Lyou, Y.-J.; Yang, J.-M. Two-dimensional electrophoresis analyses of atopic dermatitis and the chances to detect new candidate proteins by the variations in immobilized pH gradient strips. J. Dermatol. Sci. 2007, 47, 9–17. [Google Scholar] [CrossRef]
- Yoon, S.W.; Kim, T.Y.; Sung, M.H.; Kim, C.J.; Poo, H. Comparative proteomic analysis of peripheral blood eosinophils from healthy donors and atopic dermatitis patients with eosinophilia. Proteomics 2005, 5, 1987–1995. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.K.; Cho, H.J.; Ryu, S.I.; Hwang, H.-R.; Kim, D.-H.; Ryu, H.Y.; Chung, J.W.; Kim, T.-Y.; Park, B.C.; Bae, K.-H.; et al. Comparative proteomic analysis of peripheral blood mononuclear cells from atopic dermatitis patients and healthy donors. BMB Rep. 2008, 41, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Broccardo, C.J.; Mahaffey, S.; Schwarz, J.; Wruck, L.; David, G.; Schlievert, P.M.; Reisdorph, N.A.; Leung, D.Y.M. Comparative proteomic profiling of patients with atopic dermatitis based on history of eczema herpeticum infection and Staphylococcus aureus colonization. J. Allergy Clin. Immunol. 2011, 127, 186–193.e11. [Google Scholar] [CrossRef] [PubMed]
- Broccardo, C.J.; Mahaffey, S.B.; Strand, M.; Reisdorph, N.A.; Leung, D.Y.M. Peeling off the layers: Skin taping and a novel proteomics approach to study atopic dermatitis. J. Allergy Clin. Immunol. 2009, 124, 1113–1115.e11. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, J.-I.; Kamiya, K.; Yamaguchi, H.; Ikeya, S.; Suzuki, T.; Aoshima, M.; Tatsuno, K.; Fujiyama, T.; Suzuki, M.; Yatagai, T.; et al. Proteome analysis of stratum corneum from atopic dermatitis patients by hybrid quadrupole-orbitrap mass spectrometer. J. Allergy Clin. Immunol. 2014, 134, 957–960.e8. [Google Scholar] [CrossRef] [PubMed]
- Winget, J.M.; Finlay, D.; Mills, K.J.; Huggins, T.; Bascom, C.; Isfort, R.J.; Moritz, R.L. Quantitative Proteomic Analysis of Stratum Corneum Dysfunction in Adult Chronic Atopic Dermatitis. J. Investig. Dermatol. 2016, 136, 1732–1735. [Google Scholar] [CrossRef] [PubMed]
- Morelli, P.; Gaspari, M.; Gabriele, C.; Dastoli, S.; Bennardo, L.; Pavel, A.B.; Patruno, C.; Del Duca, E.; Nisticò, S.P. Proteomic analysis from skin swabs reveals a new set of proteins identifying skin impairment in atopic dermatitis. Exp. Dermatol. 2021, 30, 811–819. [Google Scholar] [CrossRef]
- Brunner, P.M.; Suárez-Fariñas, M.; He, H.; Malik, K.; Wen, H.-C.; Gonzalez, J.; Chan, T.C.-C.; Estrada, Y.; Zheng, X.; Khattri, S.; et al. The atopic dermatitis blood signature is characterized by increases in inflammatory and cardiovascular risk proteins. Sci. Rep. 2017, 7, 8707. [Google Scholar] [CrossRef]
- Brunner, P.M.; He, H.; Pavel, A.B.; Czarnowicki, T.; Lefferdink, R.; Erickson, T.; Canter, T.; Puar, N.; Rangel, S.M.; Malik, K.; et al. The blood proteomic signature of early-onset pediatric atopic dermatitis shows systemic inflammation and is distinct from adult long-standing disease. J. Am. Acad. Dermatol. 2019, 81, 510–519. [Google Scholar] [CrossRef]
- Wang, J.; Suárez-Fariñas, M.; Estrada, Y.; Parker, M.L.; Greenlees, L.; Stephens, G.; Krueger, J.; Guttman-Yassky, E.; Howell, M.D. Identification of unique proteomic signatures in allergic and non-allergic skin disease. Clin. Exp. Allergy 2017, 47, 1456–1467. [Google Scholar] [CrossRef]
- Pavel, A.B.; Zhou, L.; Diaz, A.; Ungar, B.; Dan, J.; He, H.; Estrada, Y.D.; Xu, H.; Fernandes, M.; Renert-Yuval, Y.; et al. The proteomic skin profile of moderate-to-severe atopic dermatitis patients shows an inflammatory signature. J. Am. Acad. Dermatol. 2020, 82, 690–699. [Google Scholar] [CrossRef]
- He, H.; Olesen, C.M.; Pavel, A.B.; Clausen, M.-L.; Wu, J.; Estrada, Y.; Zhang, N.; Agner, T.; Guttman-Yassky, E. Tape-Strip Proteomic Profiling of Atopic Dermatitis on Dupilumab Identifies Minimally Invasive Biomarkers. Front. Immunol. 2020, 11, 1768. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Del Duca, E.; Diaz, A.; Kim, H.J.; Gay-Mimbrera, J.; Zhang, N.; Wu, J.; Beaziz, J.; Estrada, Y.; Krueger, J.G.; et al. Mild atopic dermatitis lacks systemic inflammation and shows reduced nonlesional skin abnormalities. J. Allergy Clin. Immunol. 2021, 147, 1369–1380. [Google Scholar] [CrossRef]
- Rojahn, T.B.; Vorstandlechner, V.; Krausgruber, T.; Bauer, W.M.; Alkon, N.; Bangert, C.; Thaler, F.M.; Sadeghyar, F.; Fortelny, N.; Gernedl, V.; et al. Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis. J. Allergy Clin. Immunol. 2020, 146, 1056–1069. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.; Wang, J.; Yu, L.; Liu, H.; Estrada, Y.; Greenlees, L.; McPhee, R.; Ruzin, A.; Guttman-Yassky, E.; Howell, M.D. Atopic Dermatitis Endotypes Based on Allergen Sensitization, Reactivity to Staphylococcus aureus Antigens, and Underlying Systemic Inflammation. J. Allergy Clin. Immunol. Pract. 2020, 8, 236–247.e3. [Google Scholar] [CrossRef] [PubMed]
- Renert-Yuval, Y.; Thyssen, J.P.; Bissonnette, R.; Bieber, T.; Kabashima, K.; Hijnen, D.; Guttman-Yassky, E. Biomarkers in atopic dermatitis-a review on behalf of the International Eczema Council. J. Allergy Clin. Immunol. 2021, 147, 1174–1190.e1. [Google Scholar] [CrossRef] [PubMed]
- Mikhaylov, D.; Del Duca, E.; Guttman-Yassky, E. Proteomic signatures of inflammatory skin diseases: A focus on atopic dermatitis. Expert Rev. Proteom. 2021, 18, 345–361. [Google Scholar] [CrossRef]
- Guttman-Yassky, E.; Krueger, J.G.; Lebwohl, M.G. Systemic immune mechanisms in atopic dermatitis and psoriasis with implications for treatment. Exp. Dermatol. 2018, 27, 409–417. [Google Scholar] [CrossRef]
- Carlén, L.M.; Sánchez, F.; Bergman, A.-C.; Becker, S.; Hirschberg, D.; Franzén, B.; Coffey, J.; Jörnvall, H.; Auer, G.; Alaiya, A.A.; et al. Proteome analysis of skin distinguishes acute guttate from chronic plaque psoriasis. J. Investig. Dermatol. 2005, 124, 63–69. [Google Scholar] [CrossRef]
- Plavina, T.; Hincapie, M.; Wakshull, E.; Subramanyam, M.; Hancock, W.S. Increased plasma concentrations of cytoskeletal and Ca2+-binding proteins and their peptides in psoriasis patients. Clin. Chem. 2008, 54, 1805–1814. [Google Scholar] [CrossRef]
- Ryu, J.; Park, S.G.; Park, B.C.; Choe, M.; Lee, K.-S.; Cho, J.-W. Proteomic analysis of psoriatic skin tissue for identification of differentially expressed proteins: Up-regulation of GSTP1, SFN and PRDX2 in psoriatic skin. Int. J. Mol. Med. 2011, 28, 785–792. [Google Scholar] [PubMed]
- Schonthaler, H.B.; Guinea-Viniegra, J.; Wculek, S.K.; Ruppen, I.; Ximénez-Embún, P.; Guío-Carrión, A.; Navarro, R.; Hogg, N.; Ashman, K.; Wagner, E.F. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity 2013, 39, 1171–1181. [Google Scholar] [CrossRef] [PubMed]
- Swindell, W.R.; Remmer, H.A.; Sarkar, M.K.; Xing, X.; Barnes, D.H.; Wolterink, L.; Voorhees, J.J.; Nair, R.P.; Johnston, A.; Elder, J.T.; et al. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Leijten, E.; Zhu, Y.; Nordkamp, M.O.; Ye, S.; Pouw, J.; Tao, W.; Balak, D.; Zheng, G.; Radstake, T.; et al. Multi-omics approach identifies PI3 as a biomarker for disease severity and hyper-keratinization in psoriasis. J. Dermatol. Sci. 2023, 111, 101–108. [Google Scholar] [CrossRef]
- Bottoni, U.; Tiriolo, R.; Pullano, S.A.; Dastoli, S.; Amoruso, G.F.; Nisticò, S.P.; Fiorillo, A.S. Infrared Saliva Analysis of Psoriatic and Diabetic Patients: Similarities in Protein Components. IEEE Trans. Biomed. Eng. 2016, 63, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Méhul, B.; Laffet, G.; Séraïdaris, A.; Russo, L.; Fogel, P.; Carlavan, I.; Pernin, C.; Andres, P.; Queille-Roussel, C.; Voegel, J.J. Noninvasive proteome analysis of psoriatic stratum corneum reflects pathophysiological pathways and is useful for drug profiling. Br. J. Dermatol. 2017, 177, 470–488. [Google Scholar] [CrossRef] [PubMed]
- Gęgotek, A.; Domingues, P.; Wroński, A.; Wójcik, P.; Skrzydlewska, E. Proteomic plasma profile of psoriatic patients. J. Pharm. Biomed. Anal. 2018, 155, 185–193. [Google Scholar] [CrossRef]
- Chularojanamontri, L.; Wongpraparut, C.; Silpa-Archa, N.; Peerapen, P.; Boonmark, W.; Kulthanan, K.; Thongboonkerd, V. The humoral immunity to epidermal and dermal antigens in psoriasis: A downstream rather than an upstream event. Clin. Exp. Med. 2018, 18, 453–456. [Google Scholar] [CrossRef]
- Cowen, E.W.; Liu, C.-W.; Steinberg, S.M.; Kang, S.; Vonderheid, E.C.; Kwak, H.S.; Booher, S.; Petricoin, E.F.; Liotta, L.A.; Whiteley, G.; et al. Differentiation of tumour-stage mycosis fungoides, psoriasis vulgaris and normal controls in a pilot study using serum proteomic analysis. Br. J. Dermatol. 2007, 157, 946–953. [Google Scholar] [CrossRef]
- Williamson, J.C.; Scheipers, P.; Schwämmle, V.; Zibert, J.R.; Beck, H.C.; Jensen, O.N. A proteomics approach to the identification of biomarkers for psoriasis utilising keratome biopsy. J. Proteom. 2013, 94, 176–185. [Google Scholar] [CrossRef]
- Matsuura, T.; Sato, M.; Nagai, K.; Sato, T.; Arito, M.; Omoteyama, K.; Suematsu, N.; Okamoto, K.; Kato, T.; Soma, Y.; et al. Serum peptides as putative modulators of inflammation in psoriasis. J. Dermatol. Sci. 2017, 87, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Méhul, B.; Ménigot, C.; Fogel, P.; Seraidaris, A.; Genette, A.; Pascual, T.; Duvic, M.; Voegel, J.J. Proteomic analysis of stratum corneum in Cutaneous T-Cell Lymphomas and psoriasis. Exp. Dermatol. 2019, 28, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Reindl, J.; Pesek, J.; Krüger, T.; Wendler, S.; Nemitz, S.; Muckova, P.; Büchler, R.; Opitz, S.; Krieg, N.; Norgauer, J.; et al. Proteomic biomarkers for psoriasis and psoriasis arthritis. J. Proteom. 2016, 140, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Kolbinger, F.; Loesche, C.; Valentin, M.-A.; Jiang, X.; Cheng, Y.; Jarvis, P.; Peters, T.; Calonder, C.; Bruin, G.; Polus, F.; et al. β-Defensin 2 is a responsive biomarker of IL-17A-driven skin pathology in patients with psoriasis. J. Allergy Clin. Immunol. 2017, 139, 923–932.e8. [Google Scholar] [CrossRef]
- van Swelm, R.P.L.; Laarakkers, C.M.M.; Kooijmans-Otero, M.; de Jong, E.M.G.J.; Masereeuw, R.; Russel, F.G.M. Biomarkers for methotrexate-induced liver injury: Urinary protein profiling of psoriasis patients. Toxicol. Lett. 2013, 221, 219–224. [Google Scholar] [CrossRef]
- Lamoureux, F.; Mestre, E.; Essig, M.; Sauvage, F.L.; Marquet, P.; Gastinel, L.N. Quantitative proteomic analysis of cyclosporine-induced toxicity in a human kidney cell line and comparison with tacrolimus. J. Proteom. 2011, 75, 677–694. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, L.; Du, Q.; Gong, X.; Tian, J. Exploring the molecular mechanism underlying the psoriasis and T2D by using microarray data analysis. Sci. Rep. 2023, 13, 19313. [Google Scholar] [CrossRef]
- Mehta, H.; Mashiko, S.; Angsana, J.; Rubio, M.; Hsieh, Y.C.M.; Maari, C.; Reich, K.; Blauvelt, A.; Bissonnette, R.; Muñoz-Elías, E.J.; et al. Differential Changes in Inflammatory Mononuclear Phagocyte and T-Cell Profiles within Psoriatic Skin during Treatment with Guselkumab vs. Secukinumab. J. Investig. Dermatol. 2021, 141, 1707–1718.e9. [Google Scholar] [CrossRef]
- Blauvelt, A.; Armstrong, A.W.; Langley, R.G.; Gebauer, K.; Thaçi, D.; Bagel, J.; Guenther, L.C.; Paul, C.; Randazzo, B.; Flavin, S.; et al. Efficacy of guselkumab versus secukinumab in subpopulations of patients with moderate-to-severe plaque psoriasis: Results from the ECLIPSE study. J. Dermatol. Treat. 2022, 33, 2317–2324. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).