Challenging the Norm: The Unrecognized Impact of Soluble Guanylyl Cyclase Subunits in Cancer
Abstract
1. Introduction
2. Soluble Guanylyl Cyclase
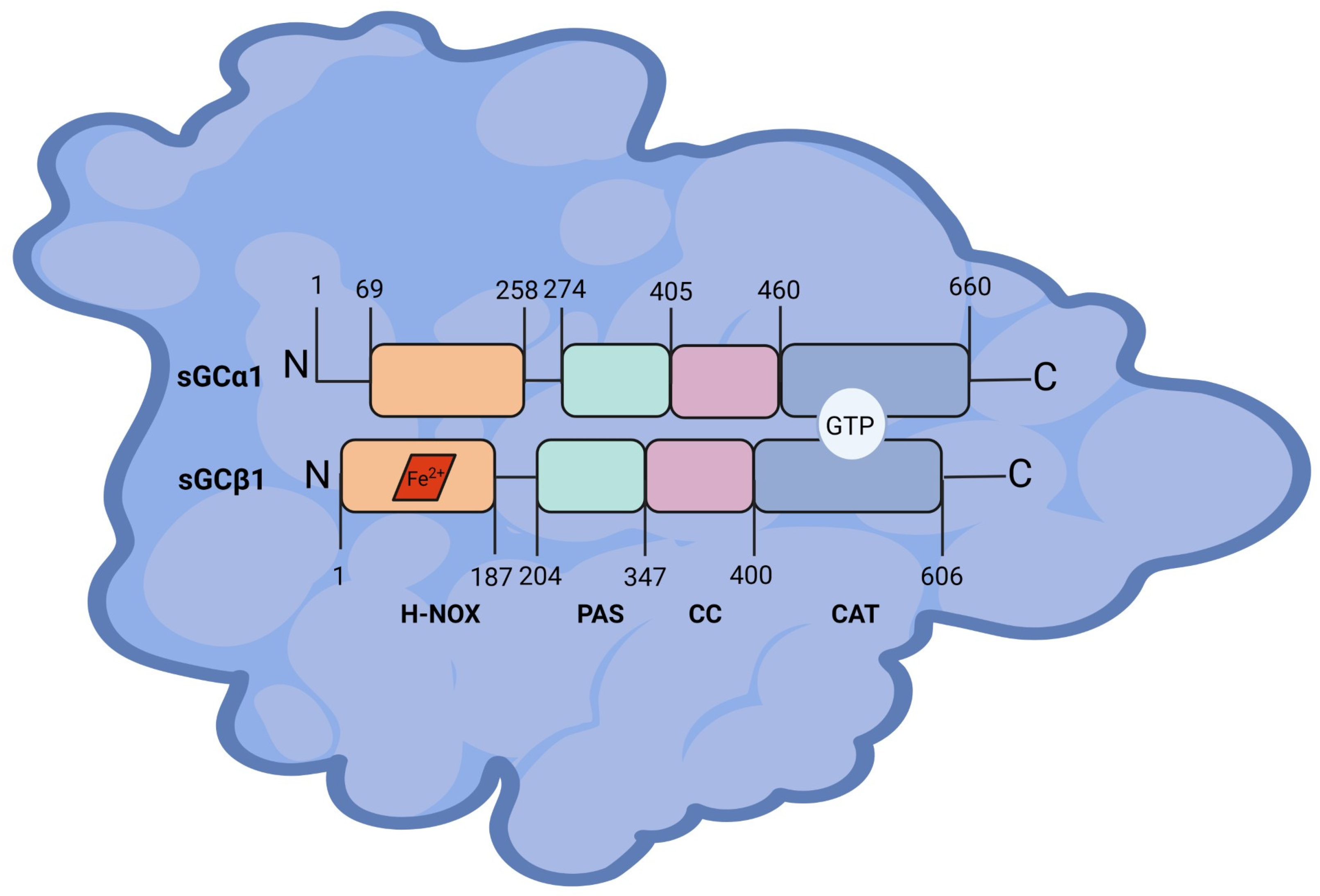
2.1. Structure
2.2. sGC Subunit Gene Localization and Splicing Variants
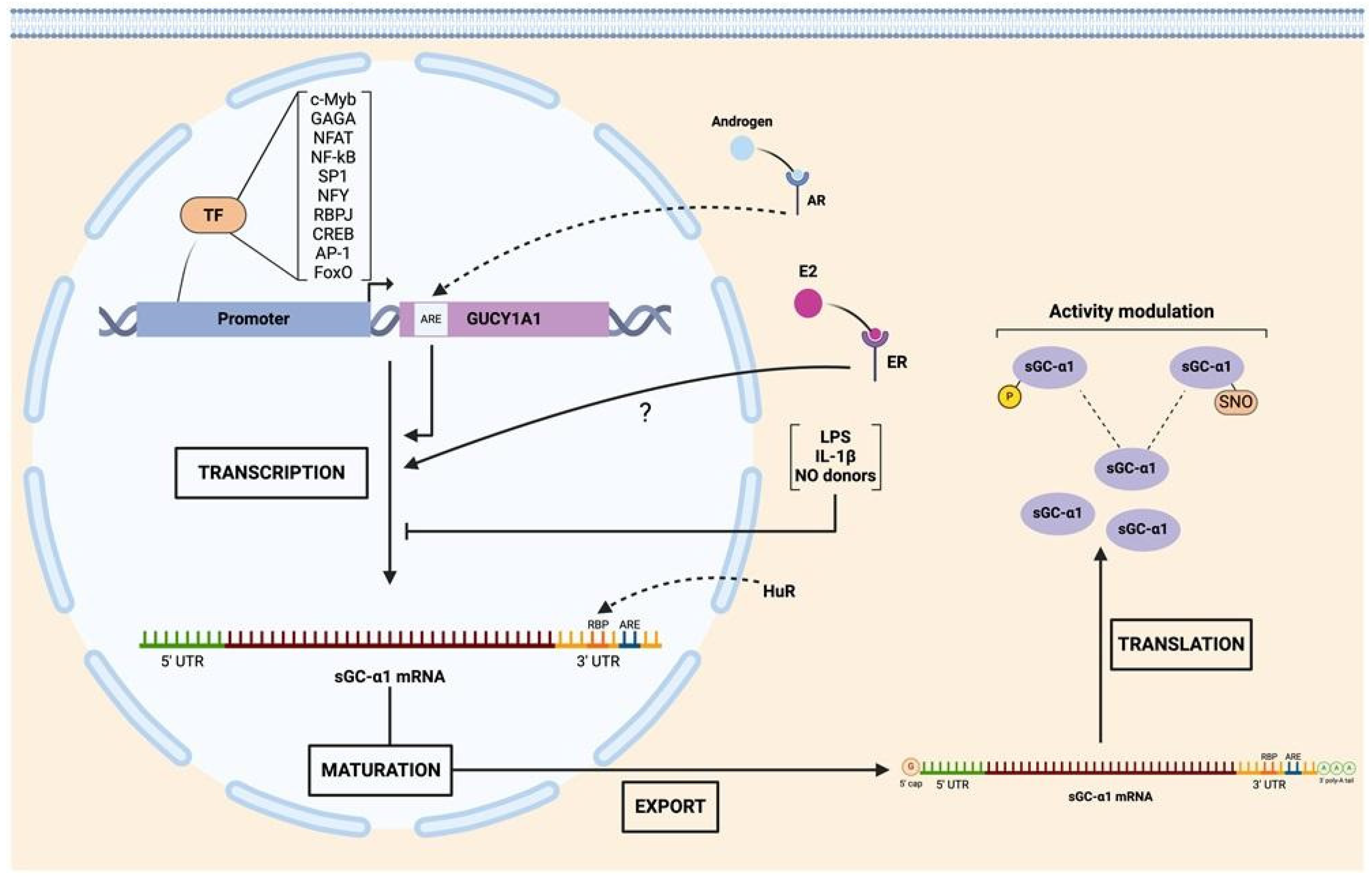
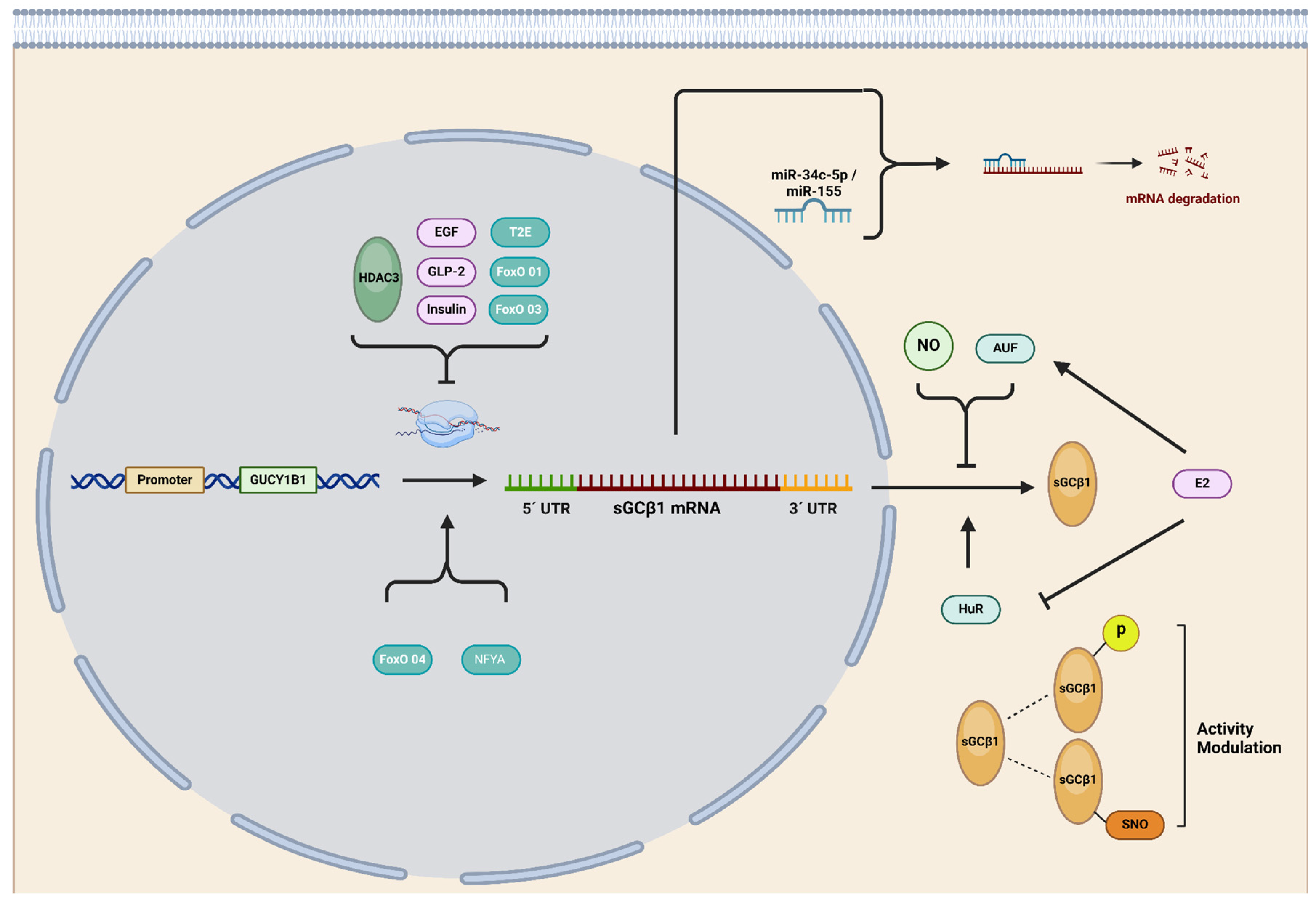
2.3. Transcriptional Control of sGC Subunit Expression
2.4. Post-Transcriptional Regulation of sGC Subunits
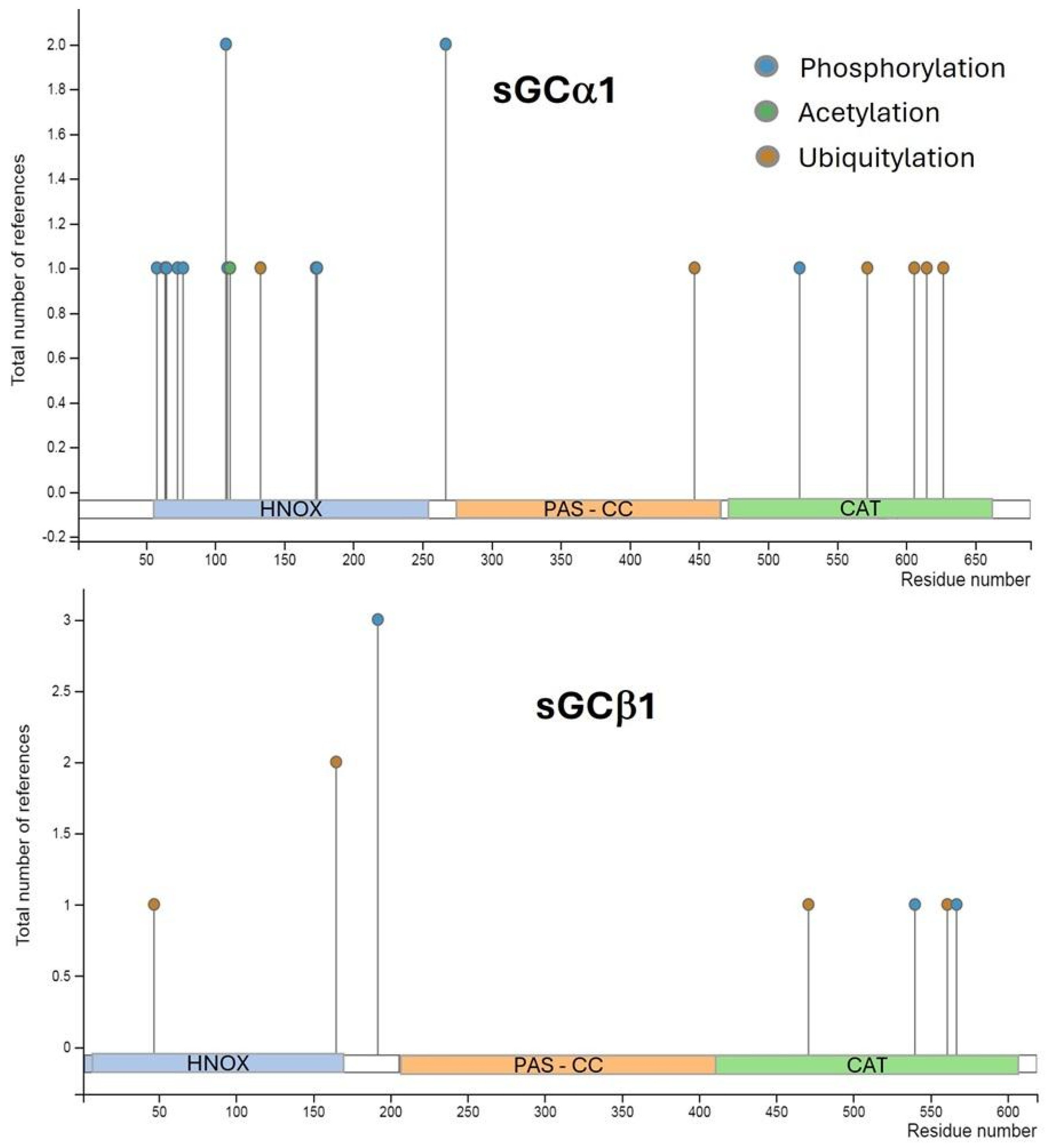
2.5. Post-Translational Regulation of sGC
2.6. Harmony or Discord? Deciphering Interactions between sGCα1 and sGCβ1 Subunits
3. sGC and cGMP in Cancer
4. Unraveling the Role of sGCα and sGCβ1 in Cancer: Insights and Implications
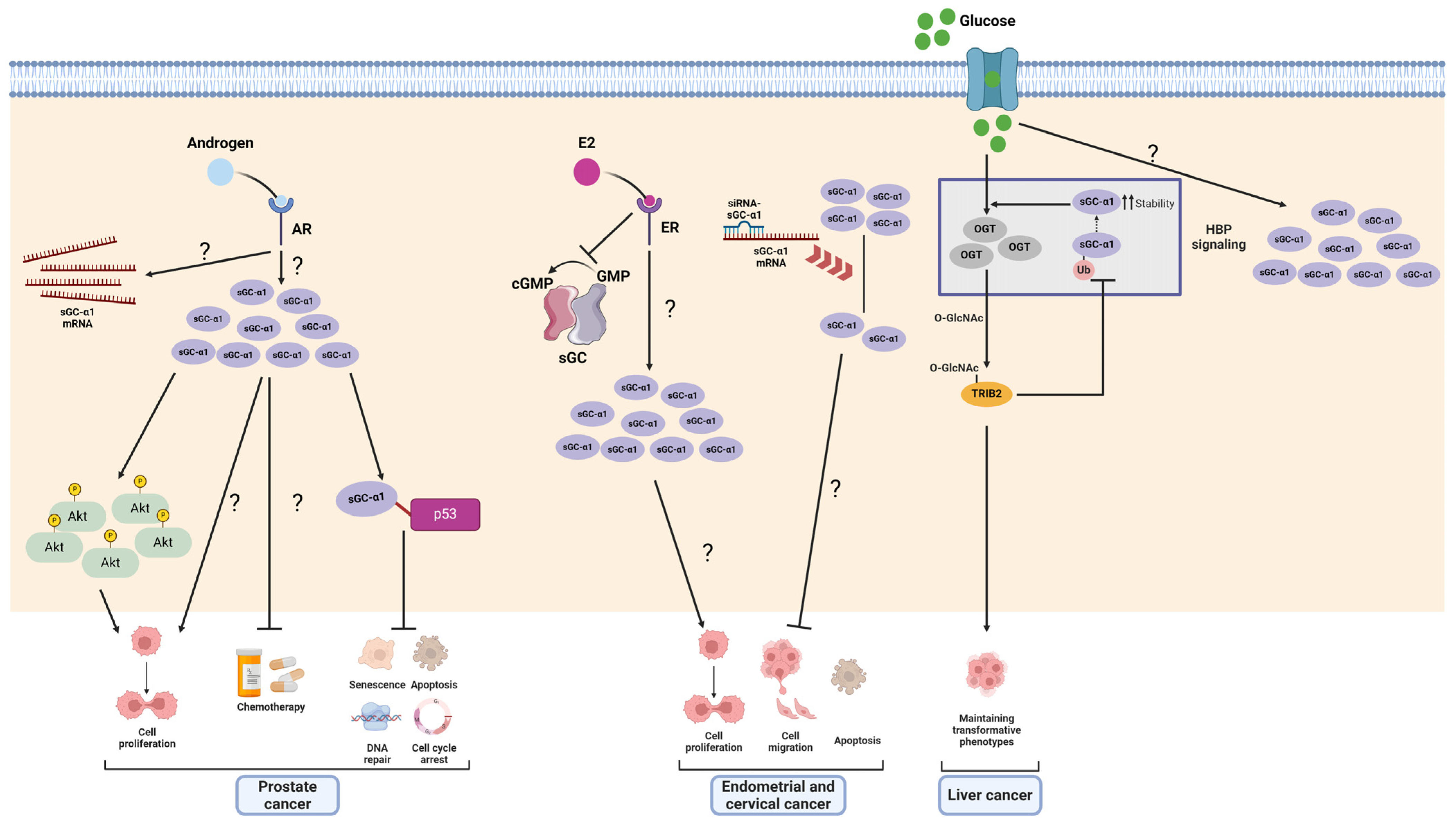
4.1. sGCα1 in Cancer
4.1.1. Prostate Cancer
4.1.2. Breast Cancer
4.1.3. Endometrial and Cervical Cancer
4.1.4. Liver Cancer
4.2. The Emerging Role of sGCα2 as a Potential Therapeutic Target in Cancer
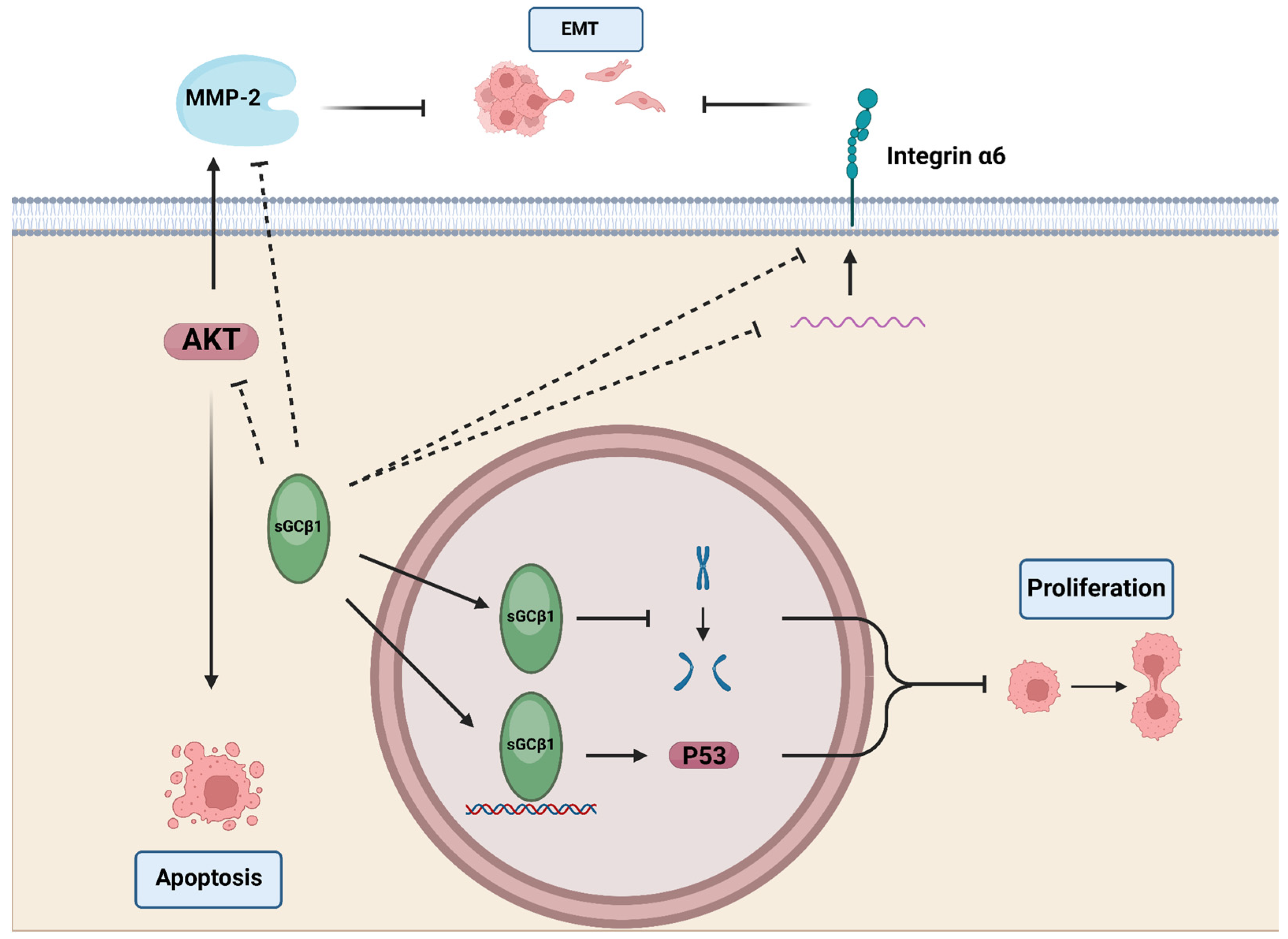
5. sGCβ1 in Cancer
5.1. Astrocytes, Glia, Glioma, and Neuroblastoma
5.2. Breast Cancer
5.3. Endometrial and Cervical Cancer
6. sGCβ2 in Cancer: A Big Question Mark
7. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Radi, R. Oxygen Radicals, Nitric Oxide, and Peroxynitrite: Redox Pathways in Molecular Medicine. Proc. Natl. Acad. Sci. USA 2018, 115, 5839–5848. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Vanhoutte, P.M. Macro- and Microvascular Endothelial Dysfunction in Diabetes. J. Diabetes 2017, 9, 434–449. [Google Scholar] [CrossRef]
- Steinert, J.R.; Amal, H. The Contribution of an Imbalanced Redox Signalling to Neurological and Neurodegenerative Conditions. Free Radic. Biol. Med. 2023, 194, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Ledo, A.; Lourenço, C.F.; Cadenas, E.; Barbosa, R.M.; Laranjinha, J. The Bioactivity of Neuronal-Derived Nitric Oxide in Aging and Neurodegeneration: Switching Signaling to Degeneration. Free Radic. Biol. Med. 2021, 162, 500–513. [Google Scholar] [CrossRef]
- Kamm, A.; Przychodzen, P.; Kuban-Jankowska, A.; Jacewicz, D.; Dabrowska, A.M.; Nussberger, S.; Wozniak, M.; Gorska-Ponikowska, M. Nitric Oxide and Its Derivatives in the Cancer Battlefield. Nitric Oxide Biol. Chem. 2019, 93, 102–114. [Google Scholar] [CrossRef]
- Somasundaram, V.; Basudhar, D.; Bharadwaj, G.; No, J.H.; Ridnour, L.A.; Cheng, R.Y.S.; Fujita, M.; Thomas, D.D.; Anderson, S.K.; McVicar, D.W.; et al. Molecular Mechanisms of Nitric Oxide in Cancer Progression, Signal Transduction, and Metabolism. Antioxid. Redox Signal. 2019, 30, 1124–1143. [Google Scholar] [CrossRef]
- Holotiuk, V.V.; Kryzhanivska, A.Y.; Churpiy, I.K.; Tataryn, B.B.; Ivasiutyn, D.Y. Role of Nitric Oxide in Pathogenesis of Tumor Growth and Its Possible Application in Cancer Treatment. Exp. Oncol. 2019, 41, 210–215. [Google Scholar] [CrossRef] [PubMed]
- López-Sánchez, L.M.; Aranda, E.; Rodríguez-Ariza, A. Nitric Oxide and Tumor Metabolic Reprogramming. Biochem. Pharmacol. 2020, 176, 113769. [Google Scholar] [CrossRef]
- Kim, J.; Thomas, S.N. Opportunities for Nitric Oxide in Potentiating Cancer Immunotherapy. Pharmacol. Rev. 2022, 74, 1146–1175. [Google Scholar] [CrossRef]
- Kimura, H.; Murad, F. Subcellular Localization of Guanylate Cyclase. Life Sci. 1975, 17, 837–843. [Google Scholar] [CrossRef]
- Kimura, H.; Murad, F. Two Forms of Guanylate Cyclase in Mammalian Tissues and Possible Mechanisms for Their Regulation. Metabolism. 1975, 24, 439–445. [Google Scholar] [CrossRef]
- Bian, K.; Murad, F. Nitric Oxide (NO)–Biogeneration, Regulation, and Relevance to Human Diseases. Front. Biosci. J. Virtual Libr. 2003, 8, d264–d278. [Google Scholar] [CrossRef]
- Biel, M.; Michalakis, S. Cyclic Nucleotide-Gated Channels. Handb. Exp. Pharmacol. 2009, 191, 111–136. [Google Scholar] [CrossRef]
- Lincoln, T.M.; Cornwell, T.L. Intracellular Cyclic GMP Receptor Proteins. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1993, 7, 328–338. [Google Scholar] [CrossRef]
- Budworth, J.; Meillerais, S.; Charles, I.; Powell, K. Tissue Distribution of the Human Soluble Guanylate Cyclases. Biochem. Biophys. Res. Commun. 1999, 263, 696–701. [Google Scholar] [CrossRef]
- Kang, Y.; Liu, R.; Wu, J.X.; Chen, L. Structural insights into the mechanism of human soluble guanylate cyclase. Nature 2019, 574, 206–210. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Buechler, W.A.; Nakane, M.; Murad, F. Expression of Soluble Guanylate Cyclase Activity Requires Both Enzyme Subunits. Biochem. Biophys. Res. Commun. 1991, 174, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.; Kim, J.; Yang, L.; Sturley, S.L.; Danziger, R.S. Expression and Purification of Soluble, Active Heterodimeric Guanylyl Cyclase from Baculovirus. Protein Expr. Purif. 1997, 10, 325–330. [Google Scholar] [CrossRef]
- Chhajlani, V.; Frändberg, P.A.; Ahlner, J.; Axelsson, K.L.; Wikberg, J.E. Heterogeneity in Human Soluble Guanylate Cyclase Due to Alternative Splicing. FEBS Lett. 1991, 290, 157–158. [Google Scholar] [CrossRef]
- Harteneck, C.; Wedel, B.; Koesling, D.; Malkewitz, J.; Böhme, E.; Schultz, G. Molecular Cloning and Expression of a New Alpha-Subunit of Soluble Guanylyl Cyclase. Interchangeability of the Alpha-Subunits of the Enzyme. FEBS Lett. 1991, 292, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Giuili, G.; Scholl, U.; Bulle, F.; Guellaën, G. Molecular Cloning of the cDNAs Coding for the Two Subunits of Soluble Guanylyl Cyclase from Human Brain. FEBS Lett. 1992, 304, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, D.A.; O’Halloran, D.M.; Burnell, A.M. Multiple Lineage Specific Expansions within the Guanylyl Cyclase Gene Family. BMC Evol. Biol. 2006, 6, 26. [Google Scholar] [CrossRef]
- Sharina, I.G.; Jelen, F.; Bogatenkova, E.P.; Thomas, A.; Martin, E.; Murad, F. Alpha1 Soluble Guanylyl Cyclase (sGC) Splice Forms as Potential Regulators of Human sGC Activity. J. Biol. Chem. 2008, 283, 15104–15113. [Google Scholar] [CrossRef] [PubMed]
- Cote, G.J.; Zhu, W.; Thomas, A.; Martin, E.; Murad, F.; Sharina, I.G. Hydrogen Peroxide Alters Splicing of Soluble Guanylyl Cyclase and Selectively Modulates Expression of Splicing Regulators in Human Cancer Cells. PLoS ONE 2012, 7, e41099. [Google Scholar] [CrossRef]
- Martin, E.; Golunski, E.; Laing, S.T.; Estrera, A.L.; Sharina, I.G. Alternative Splicing Impairs Soluble Guanylyl Cyclase Function in Aortic Aneurysm. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H1565–H1575. [Google Scholar] [CrossRef] [PubMed]
- Wobst, J.; Rumpf, P.M.; Dang, T.A.; Segura-Puimedon, M.; Erdmann, J.; Schunkert, H. Molecular Variants of Soluble Guanylyl Cyclase Affecting Cardiovascular Risk. Circ. J. Off. J. Jpn. Circ. Soc. 2015, 79, 463–469. [Google Scholar] [CrossRef]
- Mittal, C.K.; Murad, F. Properties and Oxidative Regulation of Guanylate Cyclase. J. Cyclic Nucleotide Res. 1977, 3, 381–391. [Google Scholar] [PubMed]
- Behrends, S.; Harteneck, C.; Schultz, G.; Koesling, D. A Variant of the Alpha 2 Subunit of Soluble Guanylyl Cyclase Contains an Insert Homologous to a Region within Adenylyl Cyclases and Functions as a Dominant Negative Protein. J. Biol. Chem. 1995, 270, 21109–21113. [Google Scholar] [CrossRef]
- Wilson, E.M.; Chinkers, M. Identification of Sequences Mediating Guanylyl Cyclase Dimerization. Biochemistry 1995, 34, 4696–4701. [Google Scholar] [CrossRef]
- Zabel, U.; Weeger, M.; La, M.; Schmidt, H.H. Human Soluble Guanylate Cyclase: Functional Expression and Revised Isoenzyme Family. Biochem. J. 1998, 335 Pt 1, 51–57. [Google Scholar] [CrossRef]
- Smigrodzki, R.; Levitt, P. The Alpha 1 Subunit of Soluble Guanylyl Cyclase Is Expressed Prenatally in the Rat Brain. Brain Res. Dev. Brain Res. 1996, 97, 226–234. [Google Scholar] [CrossRef]
- Bidmon, H.-J.; Starbatty, J.; Görg, B.; Zilles, K.; Behrends, S. Cerebral Expression of the Alpha2-Subunit of Soluble Guanylyl Cyclase Is Linked to Cerebral Maturation and Sensory Pathway Refinement during Postnatal Development. Neurochem. Int. 2004, 45, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Sharin, V.G.; Mujoo, K.; Kots, A.Y.; Martin, E.; Murad, F.; Sharina, I.G. Nitric Oxide Receptor Soluble Guanylyl Cyclase Undergoes Splicing Regulation in Differentiating Human Embryonic Cells. Stem Cells Dev. 2011, 20, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Sharina, I.G.; Cote, G.J.; Martin, E.; Doursout, M.-F.; Murad, F. RNA Splicing in Regulation of Nitric Oxide Receptor Soluble Guanylyl Cyclase. Nitric Oxide Biol. Chem. 2011, 25, 265–274. [Google Scholar] [CrossRef]
- Hildebrand, S.; Ibrahim, M.; Schlitzer, A.; Maegdefessel, L.; Röll, W.; Pfeifer, A. PDGF Regulates Guanylate Cyclase Expression and cGMP Signaling in Vascular Smooth Muscle. Commun. Biol. 2022, 5, 197. [Google Scholar] [CrossRef]
- Marro, M.L.; Peiró, C.; Panayiotou, C.M.; Baliga, R.S.; Meurer, S.; Schmidt, H.H.H.W.; Hobbs, A.J. Characterization of the Human Alpha1 Beta1 Soluble Guanylyl Cyclase Promoter: Key Role for NF-kappaB(P50) and CCAAT-Binding Factors in Regulating Expression of the Nitric Oxide Receptor. J. Biol. Chem. 2008, 283, 20027–20036. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.C.Y.; Fu, Y.; Garside, V.C.; Niessen, K.; Chang, L.; Fuller, M.; Setiadi, A.; Smrz, J.; Kyle, A.; Minchinton, A.; et al. Notch Initiates the Endothelial-to-Mesenchymal Transition in the Atrioventricular Canal through Autocrine Activation of Soluble Guanylyl Cyclase. Dev. Cell 2011, 21, 288–300. [Google Scholar] [CrossRef]
- Jiang, Y.; Stojilkovic, S.S. Molecular Cloning and Characterization of Alpha1-Soluble Guanylyl Cyclase Gene Promoter in Rat Pituitary Cells. J. Mol. Endocrinol. 2006, 37, 503–515. [Google Scholar] [CrossRef]
- Vazquez-Padron, R.I.; Pham, S.M.; Mateu, D.; Khan, S.; Aitouche, A. An Internal Ribosome Entry Site Mediates the Initiation of Soluble Guanylyl Cyclase Beta2 mRNA Translation. FEBS J. 2008, 275, 3598–3607. [Google Scholar] [CrossRef]
- Cai, C.; Chen, S.-Y.; Zheng, Z.; Omwancha, J.; Lin, M.-F.; Balk, S.P.; Shemshedini, L. Androgen Regulation of Soluble Guanylyl Cyclasealpha1 Mediates Prostate Cancer Cell Proliferation. Oncogene 2007, 26, 1606–1615. [Google Scholar] [CrossRef] [PubMed]
- Galley, J.C.; Durgin, B.G.; Miller, M.P.; Hahn, S.A.; Yuan, S.; Wood, K.C.; Straub, A.C. Antagonism of Forkhead Box Subclass O Transcription Factors Elicits Loss of Soluble Guanylyl Cyclase Expression. Mol. Pharmacol. 2019, 95, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Galley, J.C.; Hahn, S.A.; Miller, M.P.; Durgin, B.G.; Jackson, E.K.; Stocker, S.D.; Straub, A.C. Angiotensin II Augments Renal Vascular Smooth Muscle Soluble GC Expression via an AT1 Receptor-Forkhead Box Subclass O Transcription Factor Signalling Axis. Br. J. Pharmacol. 2022, 179, 2490–2504. [Google Scholar] [CrossRef]
- Galley, J.C.; Miller, M.P.; Sanker, S.; Liu, M.; Sharina, I.; Martin, E.; Gomez, D.; Straub, A.C. FoxO4 Controls sGCβ Transcription in Vascular Smooth Muscle. Am. J. Physiol. Heart Circ. Physiol. 2022, 322, H417–H426. [Google Scholar] [CrossRef]
- Zhou, F.; Gao, S.; Han, D.; Han, W.; Chen, S.; Patalano, S.; Macoska, J.A.; He, H.H.; Cai, C. TMPRSS2-ERG Activates NO-cGMP Signaling in Prostate Cancer Cells. Oncogene 2019, 38, 4397–4411. [Google Scholar] [CrossRef]
- Velázquez, E.; Blázquez, E.; Ruiz-Albusac, J.M. Glucagon-like Peptide-2 (GLP-2) Modulates the cGMP Signalling Pathway by Regulating the Expression of the Soluble Guanylyl Cyclase Receptor Subunits in Cultured Rat Astrocytes. Mol. Neurobiol. 2012, 46, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Cabilla, J.P.; Díaz, M.d.C.; Machiavelli, L.I.; Poliandri, A.H.; Quinteros, F.A.; Lasaga, M.; Duvilanski, B.H. 17 Beta-Estradiol Modifies Nitric Oxide-Sensitive Guanylyl Cyclase Expression and down-Regulates Its Activity in Rat Anterior Pituitary Gland. Endocrinology 2006, 147, 4311–4318. [Google Scholar] [CrossRef] [PubMed]
- Cabilla, J.P.; Ronchetti, S.A.; Nudler, S.I.; Miler, E.A.; Quinteros, F.A.; Duvilanski, B.H. Nitric Oxide Sensitive-Guanylyl Cyclase Subunit Expression Changes during Estrous Cycle in Anterior Pituitary Glands. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E731–E737. [Google Scholar] [CrossRef]
- Ronchetti, S.A.; Novack, G.V.; Bianchi, M.S.; Crocco, M.C.; Duvilanski, B.H.; Cabilla, J.P. In Vivo Xenoestrogenic Actions of Cadmium and Arsenic in Anterior Pituitary and Uterus. Reprod. Camb. Engl. 2016, 152, 1–10. [Google Scholar] [CrossRef]
- Pino, M.T.L.; Ronchetti, S.A.; Cordeiro, G.; Bollani, S.; Duvilanski, B.H.; Cabilla, J.P. Soluble Guanylyl Cyclase Alpha1 Subunit: A New Marker for Estrogenicity of Endocrine Disruptor Compounds. Environ. Toxicol. Chem. 2019, 38, 2719–2728. [Google Scholar] [CrossRef]
- Marino, M.; Acconcia, F.; Bresciani, F.; Weisz, A.; Trentalance, A. Distinct Nongenomic Signal Transduction Pathways Controlled by 17beta-Estradiol Regulate DNA Synthesis and Cyclin D(1) Gene Transcription in HepG2 Cells. Mol. Biol. Cell 2002, 13, 3720–3729. [Google Scholar] [CrossRef] [PubMed]
- O’Lone, R.; Frith, M.C.; Karlsson, E.K.; Hansen, U. Genomic Targets of Nuclear Estrogen Receptors. Mol. Endocrinol. Baltim. Md 2004, 18, 1859–1875. [Google Scholar] [CrossRef]
- Vrtačnik, P.; Ostanek, B.; Mencej-Bedrač, S.; Marc, J. The Many Faces of Estrogen Signaling. Biochem. Medica 2014, 24, 329–342. [Google Scholar] [CrossRef]
- Fuentes, N.; Silveyra, P. Estrogen Receptor Signaling Mechanisms. Adv. Protein Chem. Struct. Biol. 2019, 116, 135–170. [Google Scholar] [CrossRef]
- Papapetropoulos, A.; Cziraki, A.; Rubin, J.W.; Stone, C.D.; Catravas, J.D. cGMP Accumulation and Gene Expression of Soluble Guanylate Cyclase in Human Vascular Tissue. J. Cell. Physiol. 1996, 167, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Filippov, G.; Bloch, D.B.; Bloch, K.D. Nitric Oxide Decreases Stability of mRNAs Encoding Soluble Guanylate Cyclase Subunits in Rat Pulmonary Artery Smooth Muscle Cells. J. Clin. Investig. 1997, 100, 942–948. [Google Scholar] [CrossRef]
- Kloss, S.; Furneaux, H.; Mülsch, A. Post-Transcriptional Regulation of Soluble Guanylyl Cyclase Expression in Rat Aorta. J. Biol. Chem. 2003, 278, 2377–2383. [Google Scholar] [CrossRef]
- Liu, H.; Force, T.; Bloch, K.D. Nerve Growth Factor Decreases Soluble Guanylate Cyclase in Rat Pheochromocytoma PC12 Cells. J. Biol. Chem. 1997, 272, 6038–6043. [Google Scholar] [CrossRef]
- Klöss, S.; Srivastava, R.; Mülsch, A. Down-Regulation of Soluble Guanylyl Cyclase Expression by Cyclic AMP Is Mediated by mRNA-Stabilizing Protein HuR. Mol. Pharmacol. 2004, 65, 1440–1451. [Google Scholar] [CrossRef]
- Bachiller, P.R.; Nakanishi, H.; Roberts, J.D. Transforming Growth Factor-Beta Modulates the Expression of Nitric Oxide Signaling Enzymes in the Injured Developing Lung and in Vascular Smooth Muscle Cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2010, 298, L324–L334. [Google Scholar] [CrossRef]
- Sidali, A.; Teotia, V.; Solaiman, N.S.; Bashir, N.; Kanagaraj, R.; Murphy, J.J.; Surendranath, K. AU-Rich Element RNA Binding Proteins: At the Crossroads of Post-Transcriptional Regulation and Genome Integrity. Int. J. Mol. Sci. 2021, 23, 96. [Google Scholar] [CrossRef]
- Gallouzi, I.E.; Brennan, C.M.; Stenberg, M.G.; Swanson, M.S.; Eversole, A.; Maizels, N.; Steitz, J.A. HuR Binding to Cytoplasmic mRNA Is Perturbed by Heat Shock. Proc. Natl. Acad. Sci. USA 2000, 97, 3073–3078. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.M.; Steitz, J.A. HuR and mRNA Stability. Cell. Mol. Life Sci. CMLS 2001, 58, 266–277. [Google Scholar] [CrossRef]
- Kullmann, M.; Göpfert, U.; Siewe, B.; Hengst, L. ELAV/Hu Proteins Inhibit P27 Translation via an IRES Element in the P27 5’UTR. Genes Dev. 2002, 16, 3087–3099. [Google Scholar] [CrossRef]
- Mazan-Mamczarz, K.; Galbán, S.; López de Silanes, I.; Martindale, J.L.; Atasoy, U.; Keene, J.D.; Gorospe, M. RNA-Binding Protein HuR Enhances P53 Translation in Response to Ultraviolet Light Irradiation. Proc. Natl. Acad. Sci. USA 2003, 100, 8354–8359. [Google Scholar] [CrossRef]
- Klöss, S.; Rodenbach, D.; Bordel, R.; Mülsch, A. Human-Antigen R (HuR) Expression in Hypertension: Downregulation of the mRNA Stabilizing Protein HuR in Genetic Hypertension. Hypertension 2005, 45, 1200–1206. [Google Scholar] [CrossRef]
- Loflin, P.T.; Chen, C.Y.; Xu, N.; Shyu, A.B. Transcriptional Pulsing Approaches for Analysis of mRNA Turnover in Mammalian Cells. Methods San Diego Calif 1999, 17, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Kiledjian, M.; DeMaria, C.T.; Brewer, G.; Novick, K. Identification of AUF1 (Heterogeneous Nuclear Ribonucleoprotein D) as a Component of the Alpha-Globin mRNA Stability Complex. Mol. Cell. Biol. 1997, 17, 4870–4876. [Google Scholar] [CrossRef] [PubMed]
- Citron, F.; Armenia, J.; Franchin, G.; Polesel, J.; Talamini, R.; D’Andrea, S.; Sulfaro, S.; Croce, C.M.; Klement, W.; Otasek, D.; et al. An Integrated Approach Identifies Mediators of Local Recurrence in Head and Neck Squamous Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 3769–3780. [Google Scholar] [CrossRef]
- Yoon, J.S.J.; Wu, M.K.; Zhu, T.H.; Zhao, H.; Cheung, S.T.; Chamberlain, T.C.; Mui, A.L.-F. Interleukin-10 Control of Pre-miR155 Maturation Involves CELF2. PLoS ONE 2020, 15, e0231639. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Xu, X.; Wang, S.; Liu, J.; Dou, D.; Liu, L.; Chen, Z.; Ye, L.; Liu, H.; He, Q.; Raj, J.U.; et al. Hypoxia Induces Downregulation of Soluble Guanylyl Cyclase Β1 by miR-34c-5p. J. Cell Sci. 2012, 125 Pt 24, 6117–6126. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Choi, S.; Kim, S.; Kim, J.; Lee, D.-K.; Park, W.; Kim, T.; Jung, J.; Hwang, J.Y.; Won, M.-H.; et al. NF-κB-Responsive miR-155 Induces Functional Impairment of Vascular Smooth Muscle Cells by Downregulating Soluble Guanylyl Cyclase. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yue, C.; Chen, L. Post-Transcriptional Regulation of Soluble Guanylate Cyclase That Governs Neuropathic Pain in Alzheimer’s Disease. J. Alzheimers Dis. JAD 2019, 71, 1331–1338. [Google Scholar] [CrossRef]
- Satoh, T.; Wang, L.; Espinosa-Diez, C.; Wang, B.; Hahn, S.A.; Noda, K.; Rochon, E.R.; Dent, M.R.; Levine, A.R.; Baust, J.J.; et al. Metabolic Syndrome Mediates ROS-miR-193b-NFYA-Dependent Downregulation of Soluble Guanylate Cyclase and Contributes to Exercise-Induced Pulmonary Hypertension in Heart Failure With Preserved Ejection Fraction. Circulation 2021, 144, 615–637. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and Recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Pyriochou, A.; Papapetropoulos, A. Soluble Guanylyl Cyclase: More Secrets Revealed. Cell. Signal. 2005, 17, 407–413. [Google Scholar] [CrossRef]
- Meurer, S.; Pioch, S.; Gross, S.; Müller-Esterl, W. Reactive Oxygen Species Induce Tyrosine Phosphorylation of and Src Kinase Recruitment to NO-Sensitive Guanylyl Cyclase. J. Biol. Chem. 2005, 280, 33149–33156. [Google Scholar] [CrossRef]
- Murthy, K.S. Inhibitory Phosphorylation of Soluble Guanylyl Cyclase by Muscarinic M2 Receptors via Gbetagamma-Dependent Activation of c-Src Kinase. J. Pharmacol. Exp. Ther. 2008, 325, 183–189. [Google Scholar] [CrossRef]
- Zhou, Z.; Sayed, N.; Pyriochou, A.; Roussos, C.; Fulton, D.; Beuve, A.; Papapetropoulos, A. Protein Kinase G Phosphorylates Soluble Guanylyl Cyclase on Serine 64 and Inhibits Its Activity. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1803–1810. [Google Scholar] [CrossRef] [PubMed]
- Kostic, T.S.; Andric, S.A.; Stojilkovic, S.S. Receptor-Controlled Phosphorylation of Alpha 1 Soluble Guanylyl Cyclase Enhances Nitric Oxide-Dependent Cyclic Guanosine 5’-Monophosphate Production in Pituitary Cells. Mol. Endocrinol. Baltim. Md 2004, 18, 458–470. [Google Scholar] [CrossRef]
- Marino, S.M.; Gladyshev, V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010, 404, 902–916. [Google Scholar] [CrossRef]
- Pace, N.J.; Weerapana, E. Diverse functional roles of reactive cysteines. ACS Chem. Biol. 2013, 8, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Beuve, A. Thiol-Based Redox Modulation of Soluble Guanylyl Cyclase, the Nitric Oxide Receptor. Antioxid. Redox Signal. 2017, 26, 137–149. [Google Scholar] [CrossRef]
- Furuta, S. Basal S-Nitrosylation Is the Guardian of Tissue Homeostasis. Trends Cancer 2017, 3, 744–748. [Google Scholar] [CrossRef]
- Beuve, A.; Wu, C.; Cui, C.; Liu, T.; Jain, M.R.; Huang, C.; Yan, L.; Kholodovych, V.; Li, H. Identification of Novel S-Nitrosation Sites in Soluble Guanylyl Cyclase, the Nitric Oxide Receptor. J. Proteomics 2016, 138, 40–47. [Google Scholar] [CrossRef]
- Oppermann, M.; Suvorava, T.; Freudenberger, T.; Dao, V.T.-V.; Fischer, J.W.; Weber, M.; Kojda, G. Regulation of Vascular Guanylyl Cyclase by Endothelial Nitric Oxide-Dependent Posttranslational Modification. Basic Res. Cardiol. 2011, 106, 539–549. [Google Scholar] [CrossRef]
- Rajagopal, S.; Nalli, A.D.; Kumar, D.P.; Bhattacharya, S.; Hu, W.; Mahavadi, S.; Grider, J.R.; Murthy, K.S. Cytokine-Induced S-Nitrosylation of Soluble Guanylyl Cyclase and Expression of Phosphodiesterase 1A Contribute to Dysfunction of Longitudinal Smooth Muscle Relaxation. J. Pharmacol. Exp. Ther. 2015, 352, 509–518. [Google Scholar] [CrossRef]
- Sayed, N.; Kim, D.D.; Fioramonti, X.; Iwahashi, T.; Durán, W.N.; Beuve, A. Nitroglycerin-Induced S-Nitrosylation and Desensitization of Soluble Guanylyl Cyclase Contribute to Nitrate Tolerance. Circ. Res. 2008, 103, 606–614. [Google Scholar] [CrossRef]
- Huang, C.; Alapa, M.; Shu, P.; Nagarajan, N.; Wu, C.; Sadoshima, J.; Kholodovych, V.; Li, H.; Beuve, A. Guanylyl Cyclase Sensitivity to Nitric Oxide Is Protected by a Thiol Oxidation-Driven Interaction with Thioredoxin-1. J. Biol. Chem. 2017, 292, 14362–14370. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Wu, C.; Shu, P.; Liu, T.; Li, H.; Beuve, A. Soluble Guanylyl Cyclase Mediates Noncanonical Nitric Oxide Signaling by Nitrosothiol Transfer under Oxidative Stress. Redox Biol. 2022, 55, 102425. [Google Scholar] [CrossRef]
- Heckler, E.J.; Crassous, P.-A.; Baskaran, P.; Beuve, A. Protein Disulfide-Isomerase Interacts with Soluble Guanylyl Cyclase via a Redox-Based Mechanism and Modulates Its Activity. Biochem. J. 2013, 452, 161–169. [Google Scholar] [CrossRef]
- Balashova, N.; Chang, F.-J.; Lamothe, M.; Sun, Q.; Beuve, A. Characterization of a Novel Type of Endogenous Activator of Soluble Guanylyl Cyclase. J. Biol. Chem. 2005, 280, 2186–2196. [Google Scholar] [CrossRef] [PubMed]
- Stuehr, D.J.; Misra, S.; Dai, Y.; Ghosh, A. Maturation, Inactivation, and Recovery Mechanisms of Soluble Guanylyl Cyclase. J. Biol. Chem. 2021, 296, 100336. [Google Scholar] [CrossRef]
- Ghosh, A.; Stuehr, D.J. Soluble Guanylyl Cyclase Requires Heat Shock Protein 90 for Heme Insertion during Maturation of the NO-Active Enzyme. Proc. Natl. Acad. Sci. USA 2012, 109, 12998–13003. [Google Scholar] [CrossRef] [PubMed]
- Pifarré, P.; Baltrons, M.A.; Földi, I.; García, A. NO-Sensitive Guanylyl Cyclase Beta1 Subunit Is Peripherally Associated to Chromosomes during Mitosis. Novel Role in Chromatin Condensation and Cell Cycle Progression. Int. J. Biochem. Cell Biol. 2009, 41, 1719–1730. [Google Scholar] [CrossRef]
- Schaap, P. Guanylyl Cyclases across the Tree of Life. Front. Biosci. J. Virtual Libr. 2005, 10, 1485–1498. [Google Scholar] [CrossRef]
- Wen, H.-C.; Chuu, C.-P.; Chen, C.-Y.; Shiah, S.-G.; Kung, H.-J.; King, K.-L.; Su, L.-C.; Chang, S.-C.; Chang, C.-H. Elevation of Soluble Guanylate Cyclase Suppresses Proliferation and Survival of Human Breast Cancer Cells. PLoS ONE 2015, 10, e0125518. [Google Scholar] [CrossRef]
- Sotolongo, A.; Mónica, F.Z.; Kots, A.; Xiao, H.; Liu, J.; Seto, E.; Bian, K.; Murad, F. Epigenetic Regulation of Soluble Guanylate Cyclase (sGC) Β1 in Breast Cancer Cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016, 30, 3171–3180. [Google Scholar] [CrossRef]
- Ivanova, K.; Das, P.K.; van den Wijngaard, R.M.; Lenz, W.; Klockenbring, T.; Malcharzyk, V.; Drummer, C.; Gerzer, R. Differential Expression of Functional Guanylyl Cyclases in Melanocytes: Absence of Nitric-Oxide-Sensitive Isoform in Metastatic Cells. J. Investig. Dermatol. 2001, 116, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Friebe, A.; Mergia, E.; Dangel, O.; Lange, A.; Koesling, D. Fatal Gastrointestinal Obstruction and Hypertension in Mice Lacking Nitric Oxide-Sensitive Guanylyl Cyclase. Proc. Natl. Acad. Sci. USA 2007, 104, 7699–7704. [Google Scholar] [CrossRef] [PubMed]
- Mauersberger, C.; Sager, H.B.; Wobst, J.; Dang, T.A.; Lambrecht, L.; Koplev, S.; Stroth, M.; Bettaga, N.; Schlossmann, J.; Wunder, F.; et al. Loss of Soluble Guanylyl Cyclase in Platelets Contributes to Atherosclerotic Plaque Formation and Vascular Inflammation. Nat. Cardiovasc. Res. 2022, 1, 1174–1186. [Google Scholar] [CrossRef] [PubMed]
- Bachiller, P.R.; Cornog, K.H.; Kato, R.; Buys, E.S.; Roberts, J.D. Soluble Guanylate Cyclase Modulates Alveolarization in the Newborn Lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 305, L569–L581. [Google Scholar] [CrossRef] [PubMed]
- Mergia, E.; Friebe, A.; Dangel, O.; Russwurm, M.; Koesling, D. Spare Guanylyl Cyclase NO Receptors Ensure High NO Sensitivity in the Vascular System. J. Clin. Investig. 2006, 116, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Li, J.T.; Zheng, F.; Martin, E.; Kots, A.Y.; Krumenacker, J.S.; Choi, B.-K.; McCutcheon, I.E.; Weisbrodt, N.; Bögler, O.; et al. Restoring Soluble Guanylyl Cyclase Expression and Function Blocks the Aggressive Course of Glioma. Mol. Pharmacol. 2011, 80, 1076–1084. [Google Scholar] [CrossRef] [PubMed]
- Postovit, L.-M.; Adams, M.A.; Lash, G.E.; Heaton, J.P.; Graham, C.H. Oxygen-Mediated Regulation of Tumor Cell Invasiveness. Involvement of a Nitric Oxide Signaling Pathway. J. Biol. Chem. 2002, 277, 35730–35737. [Google Scholar] [CrossRef] [PubMed]
- Cabilla, J.P.; Nudler, S.I.; Ronchetti, S.A.; Quinteros, F.A.; Lasaga, M.; Duvilanski, B.H. Nitric Oxide-Sensitive Guanylyl Cyclase Is Differentially Regulated by Nuclear and Non-Nuclear Estrogen Pathways in Anterior Pituitary Gland. PLoS ONE 2011, 6, e29402. [Google Scholar] [CrossRef]
- Xiao, H.; Zhu, H.; Bögler, O.; Mónica, F.Z.; Kots, A.Y.; Murad, F.; Bian, K. Soluble Guanylate Cyclase Β1 Subunit Represses Human Glioblastoma Growth. Cancers 2023, 15, 1567. [Google Scholar] [CrossRef]
- Acosta, L.H.; Pino, M.T.L.; Rocca, M.V.; Cabilla, J.P. Soluble Guanylyl Cyclase Beta1 Subunit Targets Epithelial-to-Mesenchymal Transition and Downregulates Akt Pathway in Human Endometrial and Cervical Cancer Cells. Heliyon 2024, 10, e23927. [Google Scholar] [CrossRef]
- Mintz, J.; Vedenko, A.; Rosete, O.; Shah, K.; Goldstein, G.; Hare, J.M.; Ramasamy, R.; Arora, H. Current Advances of Nitric Oxide in Cancer and Anticancer Therapeutics. Vaccines 2021, 9, 94. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, Z.; Zhang, M.; Hu, H.; Li, J.; Zhang, D.; Guo, K.; Sha, H. Targeting the L-arginine-nitric oxide pathway for cancer treatment. Curr. Pharm. Des. 2010, 16, 392–410. [Google Scholar] [CrossRef]
- Burke, A.J.; Sullivan, F.J.; Giles, F.J.; Glynn, S.A. The yin and yang of nitric oxide in cancer progression. Carcinogenesis 2013, 34, 503–512. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Wang, Y.; Yang, M.; Hong, F.; Yang, S. Protein Phosphorylation in Cancer: Role of Nitric Oxide Signaling Pathway. Biomolecules 2021, 11, 1009. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.M.; Ridnour, L.A.; McGinity, C.L.; Bhattacharyya, D.; Wink, D.A. Nitric Oxide and Cancer: When to Give and When to Take Away? Inorg. Chem. 2021, 60, 15941–15947. [Google Scholar] [CrossRef] [PubMed]
- Reddy, T.P.; Glynn, S.A.; Billiar, T.R.; Wink, D.A.; Chang, J.C. Targeting Nitric Oxide: Say NO to Metastasis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2023, 29, 1855–1868. [Google Scholar] [CrossRef]
- Gao, D.; Asghar, S.; Hu, R.; Chen, S.; Niu, R.; Liu, J.; Chen, Z.; Xiao, Y. Recent Advances in Diverse Nanosystems for Nitric Oxide Delivery in Cancer Therapy. Acta Pharm. Sin. B 2023, 13, 1498–1521. [Google Scholar] [CrossRef]
- Hu, Y.; Xiang, J.; Su, L.; Tang, X. The Regulation of Nitric Oxide in Tumor Progression and Therapy. J. Int. Med. Res. 2020, 48, 300060520905985. [Google Scholar] [CrossRef] [PubMed]
- Fallahian, F.; Karami-Tehrani, F.; Salami, S.; Aghaei, M. Cyclic GMP Induced Apoptosis via Protein Kinase G in Oestrogen Receptor-Positive and -Negative Breast Cancer Cell Lines. FEBS J. 2011, 278, 3360–3369. [Google Scholar] [CrossRef]
- Mujoo, K.; Sharin, V.G.; Martin, E.; Choi, B.-K.; Sloan, C.; Nikonoff, L.E.; Kots, A.Y.; Murad, F. Role of Soluble Guanylyl Cyclase-Cyclic GMP Signaling in Tumor Cell Proliferation. Nitric Oxide Biol. Chem. 2010, 22, 43–50. [Google Scholar] [CrossRef]
- Tuttle, T.R.; Mierzwa, M.L.; Wells, S.I.; Fox, S.R.; Ben-Jonathan, N. The Cyclic GMP/Protein Kinase G Pathway as a Therapeutic Target in Head and Neck Squamous Cell Carcinoma. Cancer Lett. 2016, 370, 279–285. [Google Scholar] [CrossRef] [PubMed]
- McDonald, P.C.; Fielding, A.B.; Dedhar, S. Integrin-Linked Kinase—Essential Roles in Physiology and Cancer Biology. J. Cell Sci. 2008, 121 Pt 19, 3121–3132. [Google Scholar] [CrossRef]
- Serrano, I.; De Frutos, S.; Griera, M.; Medrano, D.; Rodríguez-Puyol, M.; Dedhar, S.; Ruiz-Torres, M.P.; Rodríguez-Puyol, D. Ilk Conditional Deletion in Adult Animals Increases Cyclic GMP-Dependent Vasorelaxation. Cardiovasc. Res. 2013, 99, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Vetter, M.; Che, D.; Liu, S.; Tsai, M.L.; Chang, C.H. The Bradykinin/Soluble Guanylate Cyclase Signaling Pathway Is Impaired in Androgen-Independent Prostate Cancer Cells. Cancer Lett. 2002, 177, 181–187. [Google Scholar] [CrossRef]
- Zhang, L.; Troccoli, C.I.; Mateo-Victoriano, B.; Lincheta, L.M.; Jackson, E.; Shu, P.; Plastini, T.; Tao, W.; Kwon, D.; Chen, X.; et al. The Soluble Guanylyl Cyclase Pathway Is Inhibited to Evade Androgen Deprivation-Induced Senescence and Enable Progression to Castration Resistance. BioRxiv 2023. [Google Scholar] [CrossRef]
- Korkmaz, Y.; Roggendorf, H.C.; Siefer, O.G.; Seehawer, J.; Imhof, T.; Plomann, M.; Bloch, W.; Friebe, A.; Huebbers, C.U. Downregulation of the A1- and Β1-Subunit of sGC in Arterial Smooth Muscle Cells of OPSCC Is HPV-Independent. J. Dent. Res. 2018, 97, 1214–1221. [Google Scholar] [CrossRef]
- Eggen, T.; Sager, G.; Berg, T.; Nergaard, B.; Moe, B.T.G.; Ørbo, A. Increased Gene Expression of the ABCC5 Transporter without Distinct Changes in the Expression of PDE5 in Human Cervical Cancer Cells during Growth. Anticancer Res. 2012, 32, 3055–3061. [Google Scholar]
- König, J.; Hartel, M.; Nies, A.T.; Martignoni, M.E.; Guo, J.; Büchler, M.W.; Friess, H.; Keppler, D. Expression and Localization of Human Multidrug Resistance Protein (ABCC) Family Members in Pancreatic Carcinoma. Int. J. Cancer 2005, 115, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Hagmann, W.; Jesnowski, R.; Faissner, R.; Guo, C.; Löhr, J.M. ATP-Binding Cassette C Transporters in Human Pancreatic Carcinoma Cell Lines. Upregulation in 5-Fluorouracil-Resistant Cells. Pancreatology 2009, 9, 136–144. [Google Scholar] [CrossRef]
- Chen, J.; Guo, L.; Peiffer, D.A.; Zhou, L.; Chan, O.T.M.; Bibikova, M.; Wickham-Garcia, E.; Lu, S.-H.; Zhan, Q.; Wang-Rodriguez, J.; et al. Genomic Profiling of 766 Cancer-Related Genes in Archived Esophageal Normal and Carcinoma Tissues. Int. J. Cancer 2008, 122, 2249–2254. [Google Scholar] [CrossRef]
- Gazzaniga, P.; Gradilone, A.; Petracca, A.; Nicolazzo, C.; Raimondi, C.; Iacovelli, R.; Naso, G.; Cortesi, E. Molecular Markers in Circulating Tumour Cells from Metastatic Colorectal Cancer Patients. J. Cell. Mol. Med. 2010, 14, 2073–2077. [Google Scholar] [CrossRef] [PubMed]
- Samidurai, A.; Xi, L.; Das, A.; Kukreja, R.C. Beyond Erectile Dysfunction: cGMP-Specific Phosphodiesterase 5 Inhibitors for Other Clinical Disorders. Annu. Rev. Pharmacol. Toxicol. 2023, 63, 585–615. [Google Scholar] [CrossRef] [PubMed]
- Schenk, M.W.; Humphrey, S.; Hossain, A.S.M.M.; Revill, M.; Pearsall, S.; Lallo, A.; Brown, S.; Bratt, S.; Galvin, M.; Descamps, T.; et al. Soluble Guanylate Cyclase Signalling Mediates Etoposide Resistance in Progressing Small Cell Lung Cancer. Nat. Commun. 2021, 12, 6652. [Google Scholar] [CrossRef]
- El-Sehemy, A.; Chang, A.C.; Azad, A.K.; Gupta, N.; Xu, Z.; Steed, H.; Karsan, A.; Fu, Y. Notch Activation Augments Nitric Oxide/Soluble Guanylyl Cyclase Signaling in Immortalized Ovarian Surface Epithelial Cells and Ovarian Cancer Cells. Cell. Signal. 2013, 25, 2780–2787. [Google Scholar] [CrossRef]
- Bian, K.; Murad, F. sGC-cGMP Signaling: Target for Anticancer Therapy. Adv. Exp. Med. Biol. 2014, 814, 5–13. [Google Scholar] [CrossRef]
- Bian, K.; Murad, F. What Is next in Nitric Oxide Research? From Cardiovascular System to Cancer Biology. Nitric Oxide Biol. Chem. 2014, 43, 3–7. [Google Scholar] [CrossRef]
- El-Sehemy, A.; Postovit, L.-M.; Fu, Y. Nitric Oxide Signaling in Human Ovarian Cancer: A Potential Therapeutic Target. Nitric Oxide Biol. Chem. 2016, 54, 30–37. [Google Scholar] [CrossRef]
- Zhu, Z.; Tang, W.; Qiu, X.; Xin, X.; Zhang, J. Advances in Targeting Phosphodiesterase 1: From Mechanisms to Potential Therapeutics. Eur. J. Med. Chem. 2024, 263, 115967. [Google Scholar] [CrossRef]
- ElHady, A.K.; El-Gamil, D.S.; Abdel-Halim, M.; Abadi, A.H. Advancements in Phosphodiesterase 5 Inhibitors: Unveiling Present and Future Perspectives. Pharm. Basel Switz. 2023, 16, 1266. [Google Scholar] [CrossRef]
- Stehle, D.; Barresi, M.; Schulz, J.; Feil, R. Heterogeneity of cGMP Signalling in Tumour Cells and the Tumour Microenvironment: Challenges and Chances for Cancer Pharmacology and Therapeutics. Pharmacol. Ther. 2023, 242, 108337. [Google Scholar] [CrossRef]
- Hsieh, C.-L.; Cai, C.; Giwa, A.; Bivins, A.; Chen, S.-Y.; Sabry, D.; Govardhan, K.; Shemshedini, L. Expression of a Hyperactive Androgen Receptor Leads to Androgen-Independent Growth of Prostate Cancer Cells. J. Mol. Endocrinol. 2008, 41, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Bhansali, M.; Shemshedini, L. COP9 Subunits 4 and 5 Target Soluble Guanylyl Cyclase A1 and P53 in Prostate Cancer Cells. Mol. Endocrinol. Baltim. Md 2014, 28, 834–845. [Google Scholar] [CrossRef]
- Cai, C.; Hsieh, C.-L.; Gao, S.; Kannan, A.; Bhansali, M.; Govardhan, K.; Dutta, R.; Shemshedini, L. Soluble Guanylyl Cyclase A1 and P53 Cytoplasmic Sequestration and Down-Regulation in Prostate Cancer. Mol. Endocrinol. Baltim. Md 2012, 26, 292–307. [Google Scholar] [CrossRef]
- Gao, S.; Hsieh, C.-L.; Bhansali, M.; Kannan, A.; Shemshedini, L. A Peptide against Soluble Guanylyl Cyclase A1: A New Approach to Treating Prostate Cancer. PLoS ONE 2013, 8, e64189. [Google Scholar] [CrossRef]
- Mohammadoo Khorasani, M.; Karami Tehrani, F.; Parizadeh, S.M.R.; Atri, M. Differential Expression of Alternative Transcripts of Soluble Guanylyl Cyclase, GYCY1a3 and GUCY1b3 Genes, in the Malignant and Benign Breast Tumors. Nitric Oxide Biol. Chem. 2019, 83, 65–71. [Google Scholar] [CrossRef]
- Mohammadoo-Khorasani, M.; Karami Tehrani, F.; Atri, M. Soluble Guanylate Cyclase Isoenzymes: The Expression of A1, A2, Β1, and Β2 Subunits in the Benign and Malignant Breast Tumors. J. Cell. Physiol. 2020, 235, 1358–1365. [Google Scholar] [CrossRef]
- Mitra, S.; Lami, M.S.; Ghosh, A.; Das, R.; Tallei, T.E.; Fatimawali; Islam, F.; Dhama, K.; Begum, M.Y.; Aldahish, A.; et al. Hormonal Therapy for Gynecological Cancers: How Far Has Science Progressed toward Clinical Applications? Cancers 2022, 14, 759. [Google Scholar] [CrossRef]
- Bardhi, O.; Palmer, B.F.; Clegg, D.J. The Evolutionary Impact and Influence of Oestrogens on Adipose Tissue Structure and Function. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2023, 378, 20220207. [Google Scholar] [CrossRef]
- Ramírez-López, I.G.; Ramírez de Arellano, A.; Jave-Suárez, L.F.; Hernández-Silva, C.D.; García-Chagollan, M.; Hernández-Bello, J.; Lopez-Pulido, E.I.; Macias-Barragan, J.; Montoya-Buelna, M.; Muñoz-Valle, J.F.; et al. Interaction between 17β-Estradiol, Prolactin and Human Papillomavirus Induce E6/E7 Transcript and Modulate the Expression and Localization of Hormonal Receptors. Cancer Cell Int. 2019, 19, 227. [Google Scholar] [CrossRef]
- Rojo-León, V.; García, C.; Valencia, C.; Méndez, M.-A.; Wood, C.; Covarrubias, L. The E6/E7 Oncogenes of Human Papilloma Virus and Estradiol Regulate Hedgehog Signaling Activity in a Murine Model of Cervical Cancer. Exp. Cell Res. 2019, 381, 311–322. [Google Scholar] [CrossRef]
- Ronchetti, S.A.; Pino, M.T.L.; Cordeiro, G.; Bollani, S.N.; Ricci, A.G.; Duvilanski, B.H.; Cabilla, J.P. Soluble Guanylyl Cyclase A1 Subunit Is a Key Mediator of Proliferation, Survival, and Migration in ECC-1 and HeLa Cell Lines. Sci. Rep. 2019, 9, 14797. [Google Scholar] [CrossRef]
- Eggen, T.; Sager, G.; Arnes, M.; Pettersen, I.; Ørbo, A. Expression of iNOS—A Favourable Prognostic Marker for Early-Stage Carcinoma of the Uterine Cervix. Anticancer Res. 2011, 31, 2319–2325. [Google Scholar] [PubMed]
- Itkonen, H.M.; Minner, S.; Guldvik, I.J.; Sandmann, M.J.; Tsourlakis, M.C.; Berge, V.; Svindland, A.; Schlomm, T.; Mills, I.G. O-GlcNAc Transferase Integrates Metabolic Pathways to Regulate the Stability of c-MYC in Human Prostate Cancer Cells. Cancer Res. 2013, 73, 5277–5287. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Xu, Y.; Wang, J.; Qiao, Y.; Zhang, Y.; Zhang, X.; Chen, Y.; Wu, Q.; Zhao, Y.; Zhu, G.; et al. Reciprocal Regulation between O-GlcNAcylation and Tribbles Pseudokinase 2 (TRIB2) Maintains Transformative Phenotypes in Liver Cancer Cells. Cell. Signal. 2016, 28, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- De Martino, L.; Errico, M.E.; Ruotolo, S.; Cascone, D.; Chiaravalli, S.; Collini, P.; Ferrari, A.; Muto, P.; Cinalli, G.; Quaglietta, L. Pediatric Lung Adenocarcinoma Presenting with Brain Metastasis: A Case Report. J. Med. Case Rep. 2018, 12, 243. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, X.; Hu, X.; Shen, Y.; Xu, R.; Wu, L.; Shen, X. Overexpression of GUCY1A2 Correlates With Poor Prognosis in Gastric Cancer Patients. Front. Oncol. 2021, 11, 632172. [Google Scholar] [CrossRef]
- Behrends, S.; Vehse, K. The Beta(2) Subunit of Soluble Guanylyl Cyclase Contains a Human-Specific Frameshift and Is Expressed in Gastric Carcinoma. Biochem. Biophys. Res. Commun. 2000, 271, 64–69. [Google Scholar] [CrossRef]
- Li, H.; Yu, B.; Li, J.; Su, L.; Yan, M.; Zhang, J.; Li, C.; Zhu, Z.; Liu, B. Characterization of Differentially Expressed Genes Involved in Pathways Associated with Gastric Cancer. PLoS ONE 2015, 10, e0125013. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pino, M.T.L.; Rocca, M.V.; Acosta, L.H.; Cabilla, J.P. Challenging the Norm: The Unrecognized Impact of Soluble Guanylyl Cyclase Subunits in Cancer. Int. J. Mol. Sci. 2024, 25, 10053. https://doi.org/10.3390/ijms251810053
Pino MTL, Rocca MV, Acosta LH, Cabilla JP. Challenging the Norm: The Unrecognized Impact of Soluble Guanylyl Cyclase Subunits in Cancer. International Journal of Molecular Sciences. 2024; 25(18):10053. https://doi.org/10.3390/ijms251810053
Chicago/Turabian StylePino, María Teresa L., María Victoria Rocca, Lucas H. Acosta, and Jimena P. Cabilla. 2024. "Challenging the Norm: The Unrecognized Impact of Soluble Guanylyl Cyclase Subunits in Cancer" International Journal of Molecular Sciences 25, no. 18: 10053. https://doi.org/10.3390/ijms251810053
APA StylePino, M. T. L., Rocca, M. V., Acosta, L. H., & Cabilla, J. P. (2024). Challenging the Norm: The Unrecognized Impact of Soluble Guanylyl Cyclase Subunits in Cancer. International Journal of Molecular Sciences, 25(18), 10053. https://doi.org/10.3390/ijms251810053





