Transcriptome Profiling of Phenylalanine-Treated Human Neuronal Model: Spotlight on Neurite Impairment and Synaptic Connectivity
Abstract
1. Introduction
2. Results
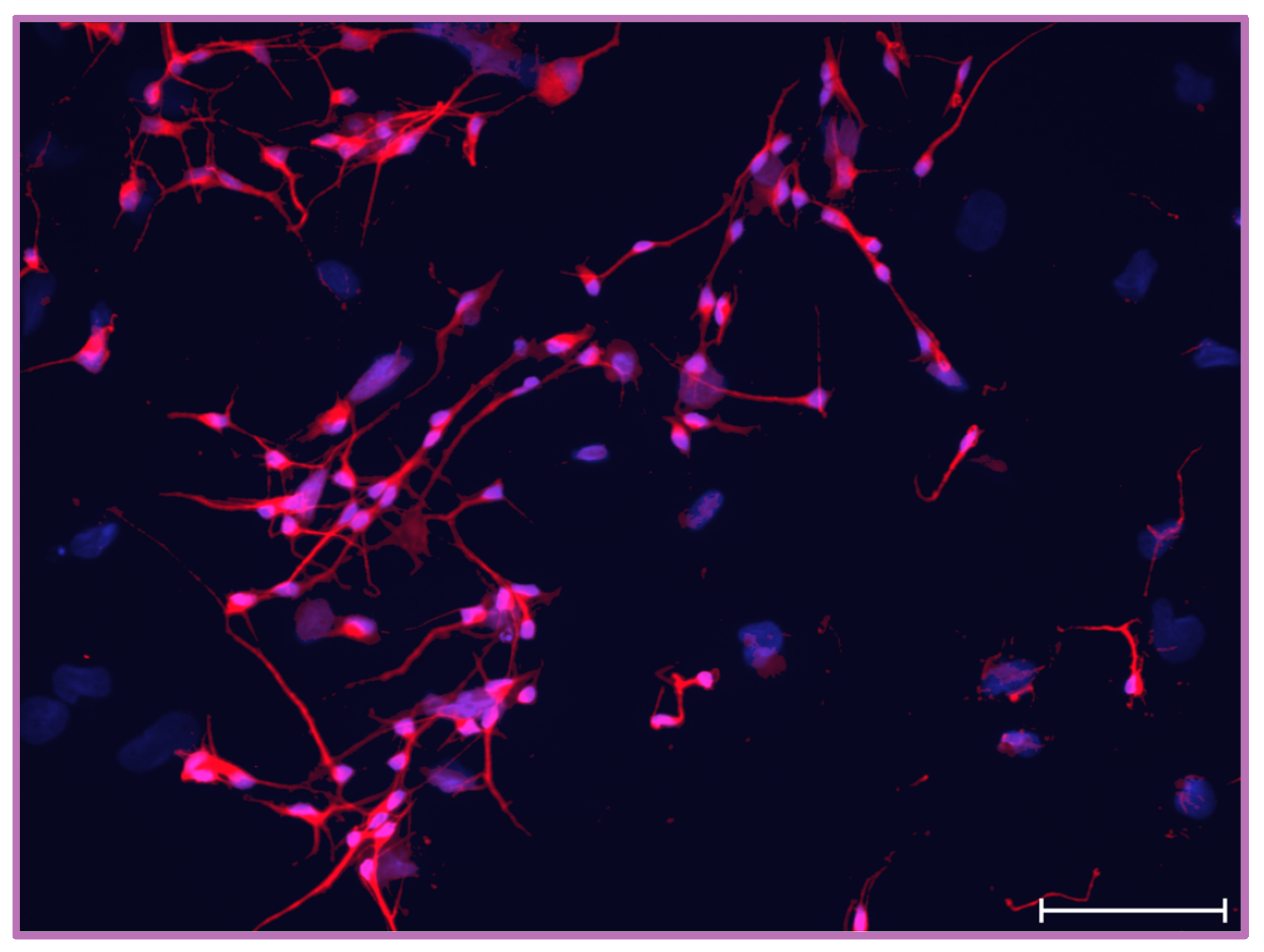
2.1. Characterization of NT2/D1 Cell Differentiation into NT2/N In Vitro
2.2. Phenylalanine Treatment of NT2/N and Cell Viability Assay
2.3. RNA Sequencing Quality Control and Mapping to the Reference Genome
2.4. Co-Expressed and Uniquely Expressed Genes
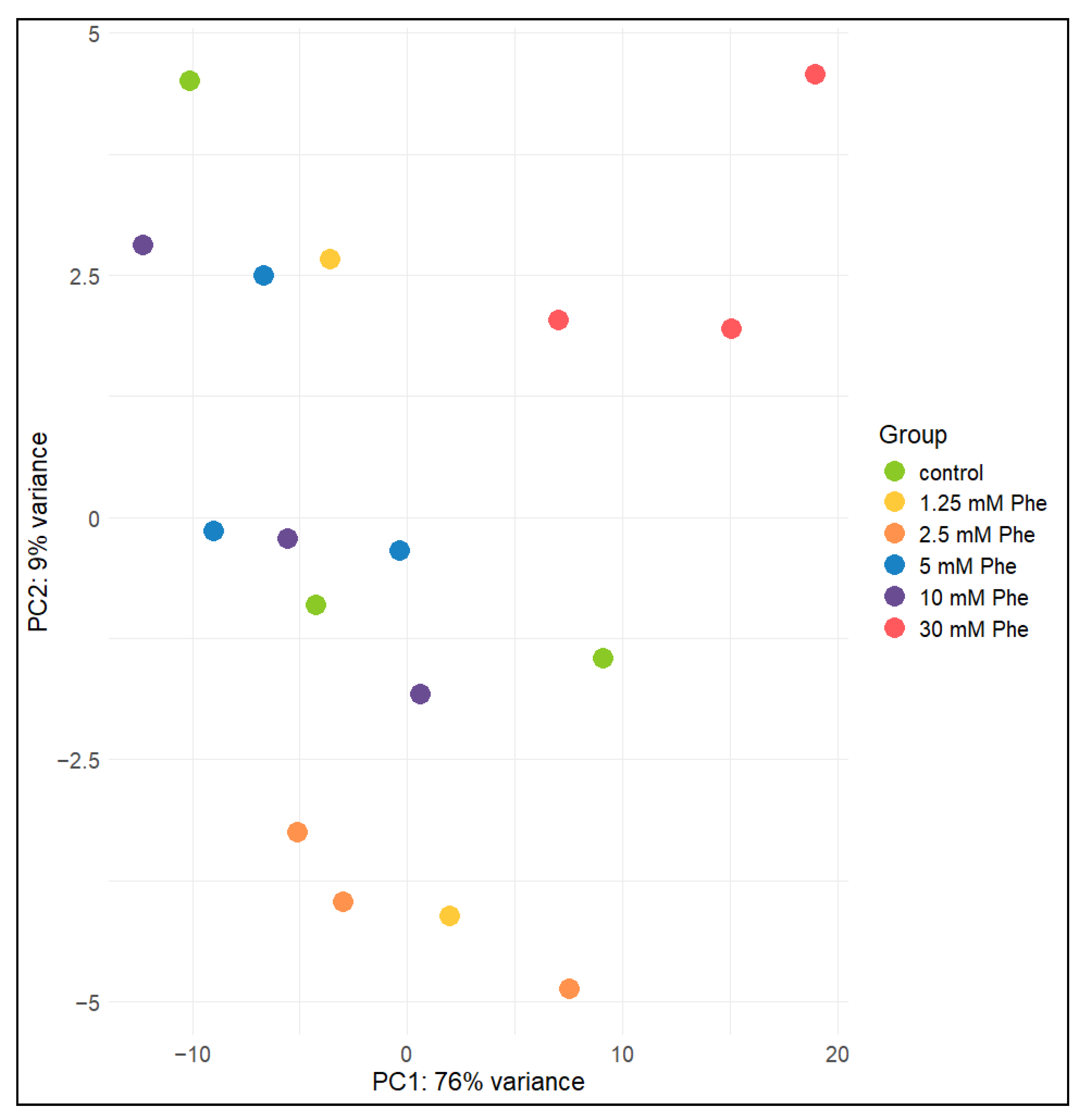
2.5. Principal Component Analysis
2.6. Analysis of Differentially Expressed Genes
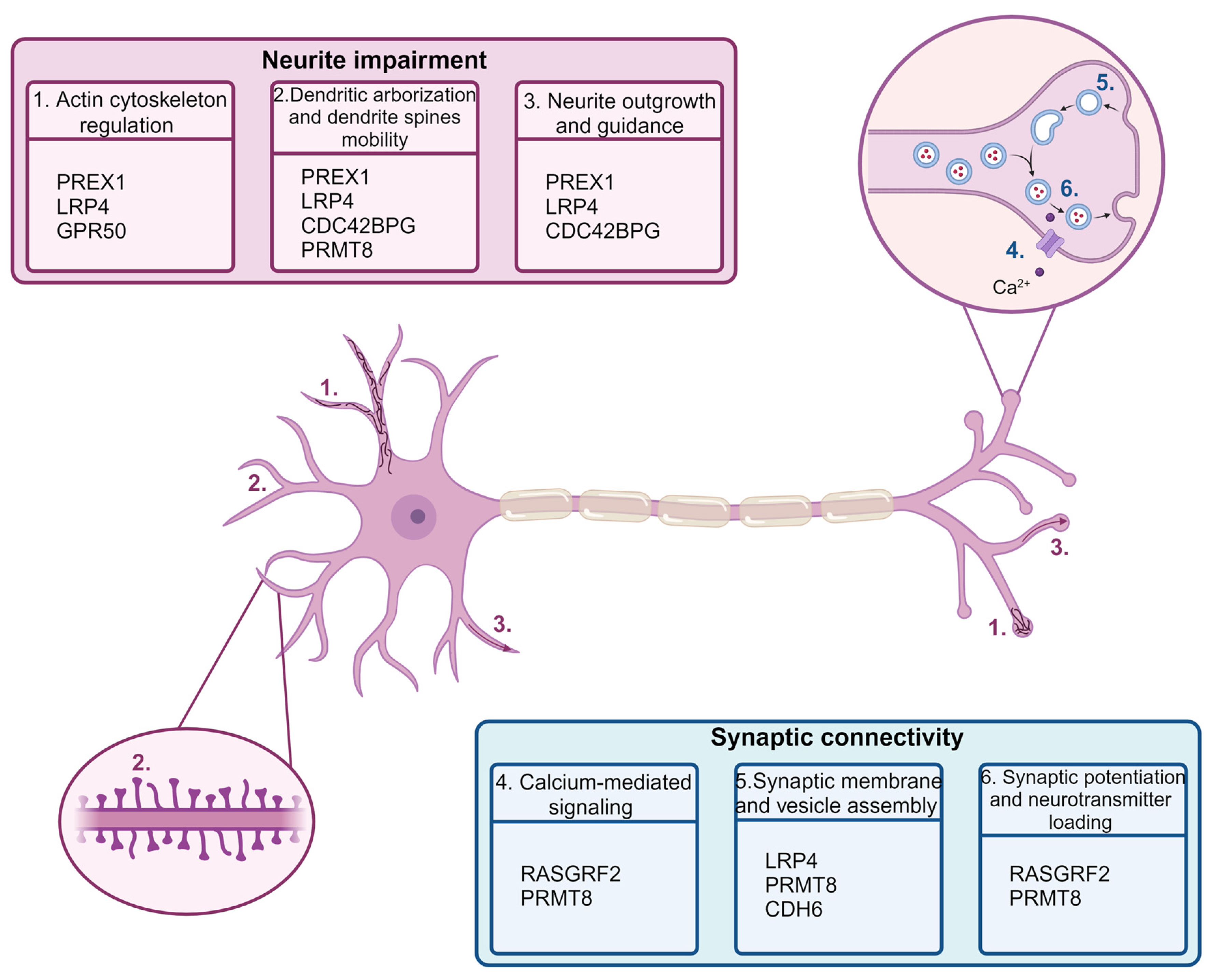
2.7. Gene Set Enrichment Analysis and Overrepresentation Analysis of DEGs
3. Discussion
4. Materials and Methods
4.1. NT2 Differentiation to NT2-Derived Neurons
4.2. Immunocytochemistry
4.3. Cell Viability Assay
4.4. Phenylalanine Treatment
4.5. RNA Extraction and mRNA Sequencing
4.6. Quality Control, Mapping to Reference Genome, and Differential Expression Analysis
4.7. Gene Set Enrichment Analysis and Overrepresentation Analysis
4.8. QRT-PCR
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van Spronsen, F.J.; Blau, N.; Harding, C.; Burlina, A.; Longo, N.; Bosch, A.M. Phenylketonuria. Nat. Rev. Dis. Primers 2021, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- van Wegberg, A.M.J.; MacDonald, A.; Ahring, K.; Bélanger-Quintana, A.; Blau, N.; Bosch, A.M.; Burlina, A.; Campistol, J.; Feillet, F.; Giżewska, M.; et al. The Complete European Guidelines on Phenylketonuria: Diagnosis and Treatment. Orphanet J. Rare Dis. 2017, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Elhawary, N.A.; AlJahdali, I.A.; Abumansour, I.S.; Elhawary, E.N.; Gaboon, N.; Dandini, M.; Madkhali, A.; Alosaimi, W.; Alzahrani, A.; Aljohani, F.; et al. Genetic Etiology and Clinical Challenges of Phenylketonuria. Hum. Genom. 2022, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Klaassen, K.; Djordjevic, M.; Skakic, A.; Desviat, L.R.; Pavlovic, S.; Perez, B.; Stojiljkovic, M. Functional Characterization of Novel Phenylalanine Hydroxylase p.Gln226Lys Mutation Revealed Its Non-Responsiveness to Tetrahydrobiopterin Treatment in Hepatoma Cellular Model. Biochem. Genet. 2018, 56, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, M.; Klaassen, K.; Sarajlija, A.; Tosic, N.; Zukic, B.; Kecman, B.; Ugrin, M.; Spasovski, V.; Pavlovic, S.; Stojiljkovic, M. Molecular Genetics and Genotype-Based Estimation of BH4-Responsiveness in Serbian PKU Patients: Spotlight on Phenotypic Implications of p.L48S. In JIMD Reports—Case and Research Reports, 2012/6; Zschocke, J., Gibson, K.M., Brown, G., Morava, E., Peters, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 49–58. ISBN 978-3-642-35518-9. [Google Scholar]
- Pilotto, A.; Zipser, C.M.; Leks, E.; Haas, D.; Gramer, G.; Freisinger, P.; Schaeffer, E.; Liepelt-Scarfone, I.; Brockmann, K.; Maetzler, W.; et al. Phenylalanine Effects on Brain Function in Adult Phenylketonuria. Neurology 2021, 96, e399–e411. [Google Scholar] [CrossRef]
- Hillert, A.; Anikster, Y.; Belanger-Quintana, A.; Burlina, A.; Burton, B.K.; Carducci, C.; Chiesa, A.E.; Christodoulou, J.; Đorđević, M.; Desviat, L.R.; et al. The Genetic Landscape and Epidemiology of Phenylketonuria. Am. J. Hum. Genet. 2020, 107, 234–250. [Google Scholar] [CrossRef]
- Klaassen, K.; Djordjevic, M.; Skakic, A.; Kecman, B.; Drmanac, R.; Pavlovic, S.; Stojiljkovic, M. Untreated PKU Patients without Intellectual Disability: SHANK Gene Family as a Candidate Modifier. Mol. Genet. Metab. Rep. 2021, 29, 100822. [Google Scholar] [CrossRef]
- van Vliet, D.; van Wegberg, A.M.J.; Ahring, K.; Bik-Multanowski, M.; Blau, N.; Bulut, F.D.; Casas, K.; Didycz, B.; Djordjevic, M.; Federico, A.; et al. Can Untreated PKU Patients Escape from Intellectual Disability? A Systematic Review. Orphanet J. Rare Dis. 2018, 13, 149. [Google Scholar] [CrossRef]
- van Vliet, D.; van Wegberg, A.M.J.; Ahring, K.; Bik-Multanowski, M.; Casas, K.; Didycz, B.; Djordjevic, M.; Hertecant, J.L.; Leuzzi, V.; Mathisen, P.; et al. Untreated PKU Patients without Intellectual Disability: What Do They Teach Us? Nutrients 2019, 11, 2572. [Google Scholar] [CrossRef]
- Hartwig, C.; Gal, A.; Santer, R.; Ullrich, K.; Finckh, U.; Kreienkamp, H.-J. Elevated Phenylalanine Levels Interfere with Neurite Outgrowth Stimulated by the Neuronal Cell Adhesion Molecule L1 in Vitro. FEBS Lett. 2006, 580, 3489–3492. [Google Scholar] [CrossRef]
- Hörster, F.; Schwab, M.A.; Sauer, S.W.; Pietz, J.; Hoffmann, G.F.; Okun, J.G.; Kölker, S.; Kins, S. Phenylalanine Reduces Synaptic Density in Mixed Cortical Cultures from Mice. Pediatr. Res. 2006, 59, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Wasserstein, M.P.; Snyderman, S.E.; Sansaricq, C.; Buchsbaum, M.S. Cerebral Glucose Metabolism in Adults with Early Treated Classic Phenylketonuria. Mol. Genet. Metab. 2006, 87, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Martynyuk, A.E.; van Spronsen, F.J.; Van der Zee, E.A. Animal Models of Brain Dysfunction in Phenylketonuria. Mol. Genet. Metab. 2010, 99, S100–S105. [Google Scholar] [CrossRef]
- Harding, C.O.; Blau, N. Advances and Challenges in Phenylketonuria. J. Inherit. Metab. Dis. 2010, 33, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Zhu, T.; Zheng, S.; Zhan, X.; Xu, F.; Gu, X.; Liang, L. Gene Expression Profiles in the Brain of Phenylketonuria Mouse Model Reversed by the Low Phenylalanine Diet Therapy. Metab. Brain Dis. 2021, 36, 2405–2414. [Google Scholar] [CrossRef]
- Wyse, A.T.S.; dos Santos, T.M.; Seminotti, B.; Leipnitz, G. Insights from Animal Models on the Pathophysiology of Hyperphenylalaninemia: Role of Mitochondrial Dysfunction, Oxidative Stress and Inflammation. Mol. Neurobiol. 2021, 58, 2897–2909. [Google Scholar] [CrossRef]
- Zhang, H.; Gu, X.F. A Study of Gene Expression Profiles of Cultured Embryonic Rat Neurons Induced by Phenylalanine. Metab. Brain Dis. 2005, 20, 61–72. [Google Scholar] [CrossRef]
- Lu, L.; Ben, X.; Xiao, L.; Peng, M.; Zhang, Y. AMP-Activated Protein Kinase Activation in Mediating Phenylalanine-Induced Neurotoxicity in Experimental Models of Phenylketonuria. J. Inherit. Metab. Dis. 2018, 41, 679–687. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq Methods for Transcriptome Analysis. WIREs RNA 2017, 8, e1364. [Google Scholar] [CrossRef]
- Watson, J.N.; Seagraves, N.J. RNA-Seq Analysis in an Avian Model of Maternal Phenylketonuria. Mol. Genet. Metab. 2019, 126, 23–29. [Google Scholar] [CrossRef]
- Manek, R.; Zhang, Y.V.; Berthelette, P.; Hossain, M.; Cornell, C.S.; Gans, J.; Anarat-Cappillino, G.; Geller, S.; Jackson, R.; Yu, D.; et al. Blood Phenylalanine Reduction Reverses Gene Expression Changes Observed in a Mouse Model of Phenylketonuria. Sci. Rep. 2021, 11, 22886. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, S.; Lee, J.; Park, J.-C.; Kim, K.H.; Ko, J.M.; Park, S.-H.; Kim, S.-K.; Mook-Jung, I.; Lee, J.Y. Neurotoxicity of Phenylalanine on Human iPSC-Derived Cerebral Organoids. Mol. Genet. Metab. 2022, 136, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.C.; Broersen, K.; Leandro, P.; Fernandes, T.G. Engineering Organoids for in Vitro Modeling of Phenylketonuria. Front. Mol. Neurosci. 2022, 14, 787242. [Google Scholar] [CrossRef]
- Kim, J.; Sullivan, G.J.; Park, I.-H. How Well Do Brain Organoids Capture Your Brain? IScience 2021, 24, 102063. [Google Scholar] [CrossRef]
- Li, D.; Gu, X.; Lu, L.; Liang, L. Effects of Phenylalanine on the Survival and Neurite Outgrowth of Rat Cortical Neurons in Primary Cultures: Possible Involvement of Brain-Derived Neurotrophic Factor. Mol. Cell. Biochem. 2010, 339, 1–7. [Google Scholar] [CrossRef]
- Szigetvari, P.D.; Patil, S.; Birkeland, E.; Kleppe, R.; Haavik, J. The Effects of Phenylalanine and Tyrosine Levels on Dopamine Production in Rat PC12 Cells. Implications for Treatment of Phenylketonuria, Tyrosinemia Type 1 and Comorbid Neurodevelopmental Disorders. Neurochem. Int. 2023, 171, 105629. [Google Scholar] [CrossRef]
- Schumacher, U.; Lukacs, Z.; Kaltschmidt, C.; Freudlsperger, C.; Schulz, D.; Kompisch, K.; Müller, R.; Rudolph, T.; Santer, R.; Lorke, D.E.; et al. High Concentrations of Phenylalanine Stimulate Peroxisome Proliferator-Activated Receptor γ: Implications for the Pathophysiology of Phenylketonuria. Neurobiol. Dis. 2008, 32, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Haile, Y.; Fu, W.; Shi, B.; Westaway, D.; Baker, G.; Jhamandas, J.; Giuliani, F. Characterization of the NT2-Derived Neuronal and Astrocytic Cell Lines as Alternative in Vitro Models for Primary Human Neurons and Astrocytes. J. Neurosci. Res. 2014, 92, 1187–1198. [Google Scholar] [CrossRef]
- Tegenge, M.A.; Roloff, F.; Bicker, G. Rapid Differentiation of Human Embryonal Carcinoma Stem Cells (NT2) into Neurons for Neurite Outgrowth Analysis. Cell. Mol. Neurobiol. 2011, 31, 635–643. [Google Scholar] [CrossRef]
- Hill, E.J.; Jiménez-González, C.; Tarczyluk, M.; Nagel, D.A.; Coleman, M.D.; Parri, H.R. NT2 Derived Neuronal and Astrocytic Network Signalling. PLoS ONE 2012, 7, e36098. [Google Scholar] [CrossRef]
- Popović, J.; Klajn, A.; Paunesku, T.; Ma, Q.; Chen, S.; Lai, B.; Stevanović, M.; Woloschak, G.E. Neuroprotective Role of Selected Antioxidant Agents in Preventing Cisplatin-Induced Damage of Human Neurons In Vitro. Cell. Mol. Neurobiol. 2019, 39, 619–636. [Google Scholar] [CrossRef] [PubMed]
- González-Burguera, I.; Ricobaraza, A.; Aretxabala, X.; Barrondo, S.; del Caño, G.G.; de Jesús, M.L.; Sallés, J. Highly Efficient Generation of Glutamatergic/Cholinergic NT2-Derived Postmitotic Human Neurons by Short-Term Treatment with the Nucleoside Analogue Cytosine β-d-Arabinofuranoside. Stem Cell Res. 2016, 16, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Tamagno, E.; Aragno, M.; Parola, M.; Parola, S.; Brignardello, E.; Boccuzzi, G.; Danni, O. NT2 Neurons, a Classical Model for Alzheimer’s Disease, Are Highly Susceptible to Oxidative Stress. Neuroreport 2000, 11, 1865. [Google Scholar] [CrossRef] [PubMed]
- Esteves, A.R.F.; Domingues, A.F.; Ferreira, I.L.; Januário, C.; Swerdlow, R.H.; Oliveira, C.R.; Cardoso, S.M. Mitochondrial Function in Parkinson’s Disease Cybrids Containing an Nt2 Neuron-like Nuclear Background. Mitochondrion 2008, 8, 219–228. [Google Scholar] [CrossRef]
- Hill, E.J.; Woehrling, E.K.; Prince, M.; Coleman, M.D. Differentiating Human NT2/D1 Neurospheres as a Versatile in Vitro 3D Model System for Developmental Neurotoxicity Testing. Toxicology 2008, 249, 243–250. [Google Scholar] [CrossRef]
- Pleasure, S.J.; Page, C.; Lee, V.M. Pure, Postmitotic, Polarized Human Neurons Derived from NTera 2 Cells Provide a System for Expressing Exogenous Proteins in Terminally Differentiated Neurons. J. Neurosci. 1992, 12, 1802–1815. [Google Scholar] [CrossRef]
- Pleasure, S.J.; Lee, V.M.-Y. NTera 2 Cells: A Human Cell Line Which Displays Characteristics Expected of a Human Committed Neuronal Progenitor Cell. J. Neurosci. Res. 1993, 35, 585–602. [Google Scholar] [CrossRef]
- Drakulic, D.; Krstic, A.; Stevanovic, M. Establishment and Initial Characterization of SOX2-Overexpressing NT2/D1 Cell Clones. Genet. Mol. Res. 2012, 11, 1385–1400. [Google Scholar] [CrossRef]
- Newman, M.B.; Misiuta, I.; Willing, A.E.; Zigova, T.; Karl, R.C.; Borlongan, C.V.; Sanberg, P.R. Tumorigenicity Issues of Embryonic Carcinoma-Derived Stem Cells: Relevance to Surgical Trials Using NT2 and hNT Neural Cells. Stem Cells Dev. 2005, 14, 29–43. [Google Scholar] [CrossRef]
- Kylies, J.; Brunne, B.; Rune, G.M. A Culture Model for the Assessment of Phenylalanine Neurotoxicity in Phenylketonuria. In Vitro Models 2022, 1, 103–114. [Google Scholar] [CrossRef]
- Schlegel, G.; Scholz, R.; Ullrich, K.; Santer, R.; Rune, G.M. Phenylketonuria: Direct and Indirect Effects of Phenylalanine. Exp. Neurol. 2016, 281, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.E.; Astle, M.V.; Ooms, L.M.; Balamatsias, D.; Gurung, R.; Mitchell, C.A. P-Rex1—A Multidomain Protein That Regulates Neurite Differentiation. J. Cell Sci. 2008, 121, 2892–2903. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Myers, K.R.; Casanova, J.E. Regulation of Actin Cytoskeleton Dynamics by Arf-Family GTPases. Trends Cell Biol. 2008, 18, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Hampson, E.; Tsonou, E.; Baker, M.J.; Hornigold, D.C.; Hubbard, R.E.; Massey, A.; Welch, H.C.E. P-Rex1 Controls Sphingosine 1-Phosphate Receptor Signalling, Morphology, and Cell-Cycle Progression in Neuronal Cells. Cells 2021, 10, 2474. [Google Scholar] [CrossRef] [PubMed]
- DePew, A.T.; Mosca, T.J. Conservation and Innovation: Versatile Roles for LRP4 in Nervous System Development. J. Dev. Biol. 2021, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Karakatsani, A.; Marichal, N.; Urban, S.; Kalamakis, G.; Ghanem, A.; Schick, A.; Zhang, Y.; Conzelmann, K.-K.; Rüegg, M.A.; Berninger, B.; et al. Neuronal LRP4 Regulates Synapse Formation in the Developing CNS. Development 2017, 144, 4604–4615. [Google Scholar] [CrossRef]
- Selimi, F.; Cristea, I.M.; Heller, E.; Chait, B.T.; Heintz, N. Proteomic Studies of a Single CNS Synapse Type: The Parallel Fiber/Purkinje Cell Synapse. PLoS Biol. 2009, 7, e1000083. [Google Scholar] [CrossRef]
- Grünewald, E.; Kinnell, H.L.; Porteous, D.J.; Thomson, P.A. GPR50 Interacts with Neuronal NOGO-A and Affects Neurite Outgrowth. Mol. Cell. Neurosci. 2009, 42, 363–371. [Google Scholar] [CrossRef]
- Dong, R.; Li, X.; Lai, K.-O. Activity and Function of the PRMT8 Protein Arginine Methyltransferase in Neurons. Life 2021, 11, 1132. [Google Scholar] [CrossRef]
- Lee, J.; Sayegh, J.; Daniel, J.; Clarke, S.; Bedford, M.T. PRMT8, a New Membrane-Bound Tissue-Specific Member of the Protein Arginine Methyltransferase Family. J. Biol. Chem. 2005, 280, 32890–32896. [Google Scholar] [CrossRef]
- Andolina, D.; Conversi, D.; Cabib, S.; Trabalza, A.; Ventura, R.; Puglisi-Allegra, S.; Pascucci, T. 5-Hydroxytryptophan during Critical Postnatal Period Improves Cognitive Performances and Promotes Dendritic Spine Maturation in Genetic Mouse Model of Phenylketonuria. Int. J. Neuropsychopharmacol. 2011, 14, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Horling, K.; Schlegel, G.; Schulz, S.; Vierk, R.; Ullrich, K.; Santer, R.; Rune, G.M. Hippocampal Synaptic Connectivity in Phenylketonuria. Hum. Mol. Genet. 2015, 24, 1007–1018. [Google Scholar] [CrossRef]
- De Jaco, A.; Mango, D.; De Angelis, F.; Favaloro, F.L.; Andolina, D.; Nisticò, R.; Fiori, E.; Colamartino, M.; Pascucci, T. Unbalance between Excitation and Inhibition in Phenylketonuria, a Genetic Metabolic Disease Associated with Autism. Int. J. Mol. Sci. 2017, 18, 941. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.M.; Froemke, R.C.; Burden, S.J. Synaptic Plasticity and Cognitive Function Are Disrupted in the Absence of Lrp4. ELife 2014, 3, e04287. [Google Scholar] [CrossRef] [PubMed]
- Schwechter, B.; Rosenmund, C.; Tolias, K.F. RasGRF2 Rac-GEF Activity Couples NMDA Receptor Calcium Flux to Enhanced Synaptic Transmission. Proc. Natl. Acad. Sci. USA 2013, 110, 14462–14467. [Google Scholar] [CrossRef]
- Miller, M.B.; Yan, Y.; Eipper, B.A.; Mains, R.E. Neuronal Rho GEFs in Synaptic Physiology and Behavior. Neuroscientist 2013, 19, 255–273. [Google Scholar] [CrossRef]
- Penney, J.; Seo, J.; Kritskiy, O.; Elmsaouri, S.; Gao, F.; Pao, P.-C.; Su, S.C.; Tsai, L.-H. Loss of Protein Arginine Methyltransferase 8 Alters Synapse Composition and Function, Resulting in Behavioral Defects. J. Neurosci. 2017, 37, 8655–8666. [Google Scholar] [CrossRef]
- Park, S.-W.; Jun, Y.-W.; Choi, H.-E.; Lee, J.-A.; Jang, D.-J. Deciphering the Molecular Mechanisms Underlying the Plasma Membrane Targeting of PRMT8. BMB Rep. 2019, 52, 601–606. [Google Scholar] [CrossRef]
- Yamagata, M.; Duan, X.; Sanes, J.R. Cadherins Interact With Synaptic Organizers to Promote Synaptic Differentiation. Front. Mol. Neurosci. 2018, 11, 142. [Google Scholar] [CrossRef]
- Bortolasci, C.C.; Jaehne, E.J.; Hernández, D.; Spolding, B.; Connor, T.; Panizzutti, B.; Dean, O.M.; Crowley, T.M.; Yung, A.R.; Gray, L.; et al. Metergoline Shares Properties with Atypical Antipsychotic Drugs Identified by Gene Expression Signature Screen. Neurotox. Res. 2023, 41, 502–513. [Google Scholar] [CrossRef]
- Dobrowolski, S.F.; Lyons-Weiler, J.; Spridik, K.; Vockley, J.; Skvorak, K.; Biery, A. DNA Methylation in the Pathophysiology of Hyperphenylalaninemia in the PAHenu2 Mouse Model of Phenylketonuria. Mol. Genet. Metab. 2016, 119, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tzartos, J.S.; Zisimopoulou, P.; Rentzos, M.; Karandreas, N.; Zouvelou, V.; Evangelakou, P.; Tsonis, A.; Thomaidis, T.; Lauria, G.; Andreetta, F.; et al. LRP4 Antibodies in Serum and CSF from Amyotrophic Lateral Sclerosis Patients. Ann. Clin. Transl. Neurol. 2014, 1, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-H.; Lin, Z.-Y.; Zeng, X.-X.; Jiang, Y.-H.; Geng, F. LRP4-Related Signalling Pathways and Their Regulatory Role in Neurological Diseases. Brain Res. 2024, 1825, 148705. [Google Scholar] [CrossRef]
- O’Connell, E.M.; Lohoff, F.W. Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) in the Brain and Relevance for Neuropsychiatric Disorders. Front. Neurosci. 2020, 14, 609. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.S.; Wagner, J.; Rosoff, D.B.; Lohoff, F.W. Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) in the Central Nervous System. Neurosci. Biobehav. Rev. 2023, 149, 105155. [Google Scholar] [CrossRef]
- Zhu, X.-B.; Xu, Y.-Y.; Li, L.-C.; Sun, J.-B.; Wang, Y.-Z.; Chen, J.; Wang, C.; Zhang, S.; Jin, L.-Y. Function of Proprotein Convertase Subtilisin/Kexin Type 9 and Its Role in Central Nervous System Diseases: An Update on Clinical Evidence. Drug Dev. Res. 2024, 85, e22131. [Google Scholar] [CrossRef]
- Andrews, P.W. Retinoic Acid Induces Neuronal Differentiation of a Cloned Human Embryonal Carcinoma Cell Line in Vitro. Dev. Biol. 1984, 103, 285–293. [Google Scholar] [CrossRef]
- Stanisavljevic, D.; Popovic, J.; Petrovic, I.; Davidovic, S.; Atkinson, M.J.; Anastasov, N.; Stevanovic, M. Radiation Effects on Early Phase of NT2/D1 Neural Differentiation in Vitro. Int. J. Radiat. Biol. 2019, 95, 1627–1639. [Google Scholar] [CrossRef]
- Wickham, H. Data Analysis. In ggplot2: Elegant Graphics for Data Analysis; Wickham, H., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 189–201. ISBN 978-3-319-24277-4. [Google Scholar]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Sva. Available online: http://bioconductor.org/packages/sva/ (accessed on 30 August 2024).
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Nat. Preced. 2010, 11, R106. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Rothfels, K.; Milacic, M.; Matthews, L.; Haw, R.; Sevilla, C.; Gillespie, M.; Stephan, R.; Gong, C.; Ragueneau, E.; May, B.; et al. Using the Reactome Database. Curr. Protoc. 2023, 3, e722. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Yan, G.-R.; He, Q.-Y. DOSE: An R/Bioconductor Package for Disease Ontology Semantic and Enrichment Analysis. Bioinformatics 2015, 31, 608–609. [Google Scholar] [CrossRef]
- Enrichplot. Available online: http://bioconductor.org/packages/enrichplot/ (accessed on 22 July 2024).
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Kassambara, A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests; R package version 0.7.2; 2023. Available online: https://CRAN.R-project.org/package=rstatix (accessed on 14 August 2024).
- Signorell, A.; Aho, K.; Alfons, A.; Anderegg, N.; Aragon, T.; Arachchige, C.; Arppe, A.; Baddeley, A.; Barton, K.; Bolker, B.; et al. DescTools: Tools for Descriptive Statistics, R Package Version 0.99; 2024. Available online: https://CRAN.R-project.org/package=DescTools (accessed on 14 August 2024).

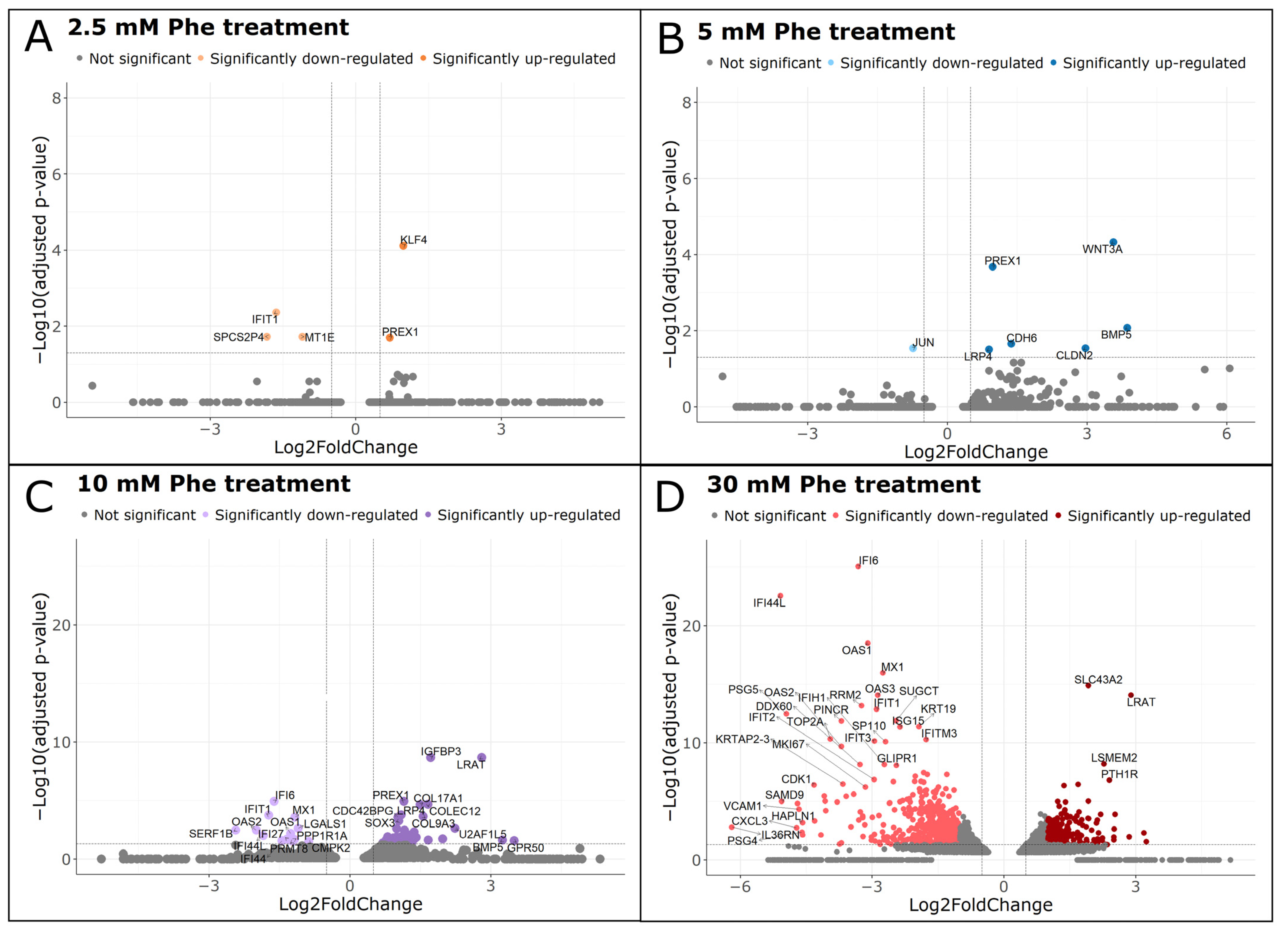

| Treatment Group | Gene Name | Function | Differential Expression | Fold Change |
|---|---|---|---|---|
| 2.5 mM Phe | KLF4 | Transcription factor | Up-regulated | 1.98 **** |
| MT1E | Metal-binding protein | Down-regulated | 0.47 ** | |
| PREX1 | Signaling protein (guanine nucleotide exchange factor) | Up-regulated | 1.63 * | |
| 5 mM Phe | WNT3A | Signaling protein | Up-regulated | 11.88 **** |
| PREX1 | Signaling protein (guanine nucleotide exchange factor) | Up-regulated | 1.96 *** | |
| CDH6 | Cell adhesion protein | Up-regulated | 2.60 * | |
| LRP4 | Low-density lipoprotein receptor-related protein | Up-regulated | 1.86 * | |
| 10 mM Phe | IGFBP3 | Binding protein (transcription regulator) | Up-regulated | 3.29 **** |
| PREX1 | Signaling protein (guanine nucleotide exchange factor) | Up-regulated | 2.22 **** | |
| LRP4 | Low-density lipoprotein receptor-related protein | Up-regulated | 2.10 *** | |
| CDC42BPG | Enzyme (protein kinase) | Up-regulated | 2.03 *** | |
| SOX3 | Transcription factor | Up-regulated | 2.06 *** | |
| CDH6 | Cell adhesion protein | Up-regulated | 2.24 ** | |
| AEBP1 | Enzyme (carboxypeptidase) | Up-regulated | 2.24 ** | |
| PPP1R13L | Enzyme (protein phosphatase) | Up-regulated | 2.10 * | |
| PCSK9 | Enzyme (subtilisin-like proprotein convertase) | Up-regulated | 2.66 * | |
| NTNG2 | Cell adhesion protein | Up-regulated | 1.83 * | |
| GPR50 | G-protein coupled receptor | Up-regulated | 11.31 * | |
| RASGRF2 | Nucleotide exchange factor | Up-regulated | 1.89 * | |
| PRMT8 | Enzyme (arginine methyltransferase) | Down-regulated | 0.43 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stankovic, S.; Lazic, A.; Parezanovic, M.; Stevanovic, M.; Pavlovic, S.; Stojiljkovic, M.; Klaassen, K. Transcriptome Profiling of Phenylalanine-Treated Human Neuronal Model: Spotlight on Neurite Impairment and Synaptic Connectivity. Int. J. Mol. Sci. 2024, 25, 10019. https://doi.org/10.3390/ijms251810019
Stankovic S, Lazic A, Parezanovic M, Stevanovic M, Pavlovic S, Stojiljkovic M, Klaassen K. Transcriptome Profiling of Phenylalanine-Treated Human Neuronal Model: Spotlight on Neurite Impairment and Synaptic Connectivity. International Journal of Molecular Sciences. 2024; 25(18):10019. https://doi.org/10.3390/ijms251810019
Chicago/Turabian StyleStankovic, Sara, Andrijana Lazic, Marina Parezanovic, Milena Stevanovic, Sonja Pavlovic, Maja Stojiljkovic, and Kristel Klaassen. 2024. "Transcriptome Profiling of Phenylalanine-Treated Human Neuronal Model: Spotlight on Neurite Impairment and Synaptic Connectivity" International Journal of Molecular Sciences 25, no. 18: 10019. https://doi.org/10.3390/ijms251810019
APA StyleStankovic, S., Lazic, A., Parezanovic, M., Stevanovic, M., Pavlovic, S., Stojiljkovic, M., & Klaassen, K. (2024). Transcriptome Profiling of Phenylalanine-Treated Human Neuronal Model: Spotlight on Neurite Impairment and Synaptic Connectivity. International Journal of Molecular Sciences, 25(18), 10019. https://doi.org/10.3390/ijms251810019






