Exploring Genetic Interactions in Colombian Women with Polycystic Ovarian Syndrome: A Study on SNP-SNP Associations
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Study Sample
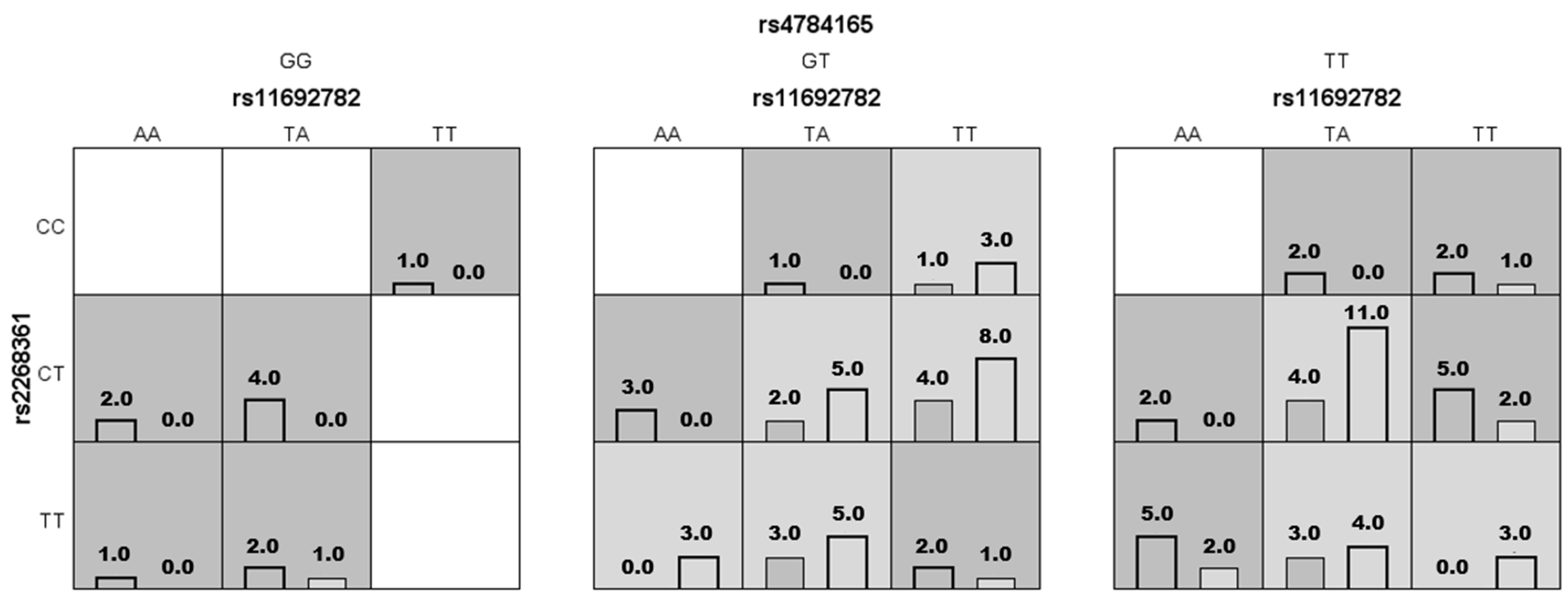
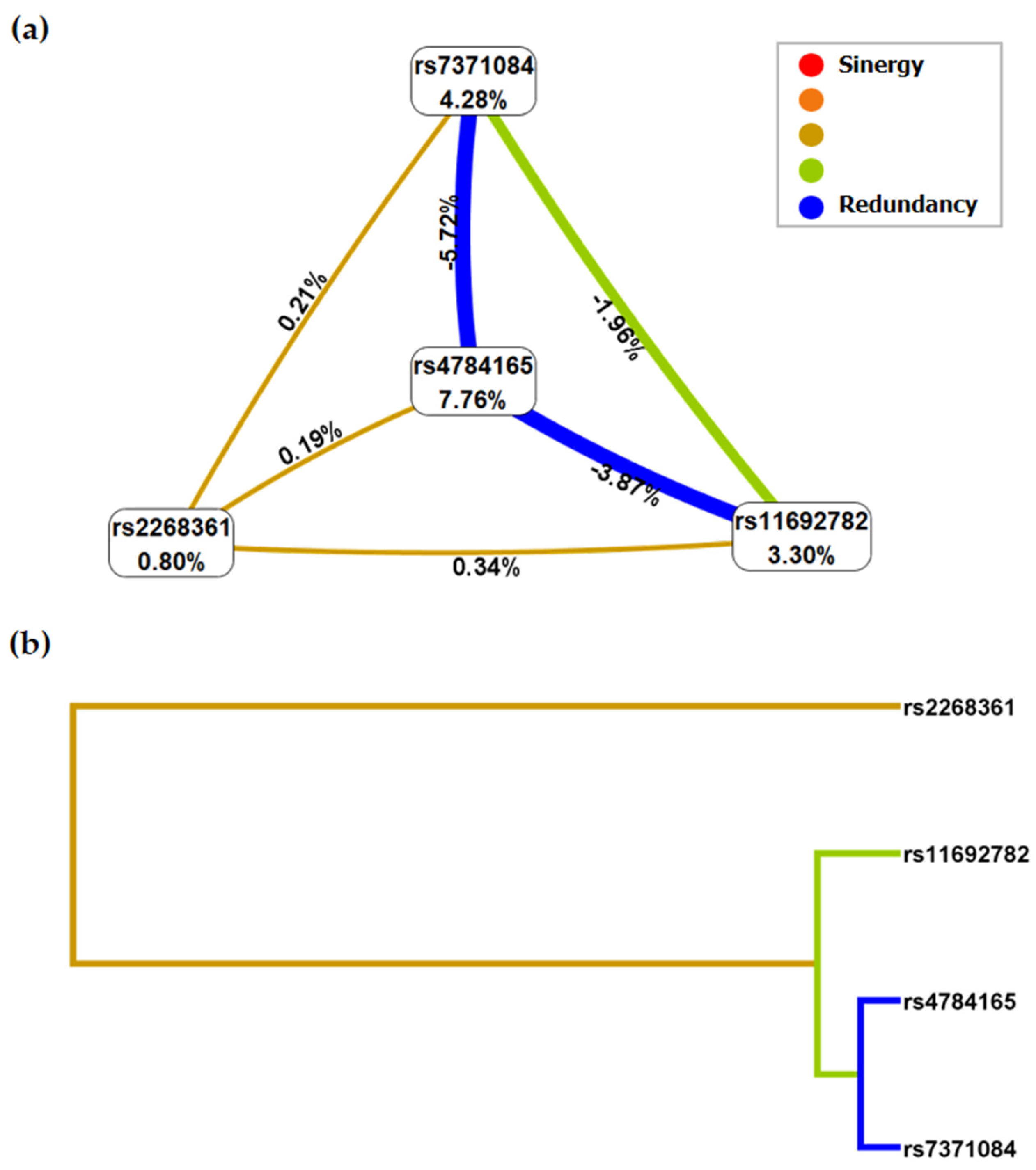
2.2. Epistasis Analysis
3. Discussion
4. Materials and Methods
4.1. Study Participants
4.2. SNP Selection and Genotyping
4.3. Statistic and SNP-SNP Interaction Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rasquin, L.I.; Anastasopoulou, C.; Mayrin, J.V. Polycystic Ovarian Disease. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Siddiqui, S.; Mateen, S.; Ahmad, R.; Moin, S. A Brief Insight into the Etiology, Genetics, and Immunology of Polycystic Ovarian Syndrome (PCOS). J. Assist. Reprod. Genet. 2022, 39, 2439–2473. [Google Scholar] [CrossRef] [PubMed]
- Thien Tay, C.; Garrad, R.; Mousa, A.; Bahri, M.; Joham, A.; Teede, H. Polycystic Ovary Syndrome (PCOS): International Collaboration to Translate Evidence and Guide Future Research. J. Endocrinol. 2023, 257, e220232. [Google Scholar] [CrossRef]
- Zhu, T.; Goodarzi, M.O. Causes and Consequences of Polycystic Ovary Syndrome: Insights From Mendelian Randomization. J. Clin. Endocrinol. Metab. 2022, 107, e899–e911. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Senapati, S.; Goyal, L.D.; Kaur, B.; Kamra, P.; Khetarpal, P. Genome-Wide Association Study (GWAS) Identified PCOS Susceptibility Variants and Replicates Reported Risk Variants. Arch. Gynecol. Obstet. 2024, 309, 2009–2019. [Google Scholar] [CrossRef]
- Lee, J.W.; Lee, S. A Comparative Study on the Unified Model Based Multifactor Dimensionality Reduction Methods for Identifying Gene-Gene Interactions Associated with the Survival Phenotype. BioData Min. 2021, 14, 17. [Google Scholar] [CrossRef] [PubMed]
- Gola, D.; Mahachie John, J.M.; Van Steen, K.; König, I.R. A Roadmap to Multifactor Dimensionality Reduction Methods. Brief. Bioinform. 2016, 17, 293–308. [Google Scholar] [CrossRef]
- Moore, J.H.; Andrews, P.C. Epistasis Analysis Using Multifactor Dimensionality Reduction. Methods Mol. Biol. 2015, 1253, 301–314. [Google Scholar] [CrossRef]
- Ritchie, M.D.; Hahn, L.W.; Roodi, N.; Bailey, L.R.; Dupont, W.D.; Parl, F.F.; Moore, J.H. Multifactor-Dimensionality Reduction Reveals High-Order Interactions among Estrogen-Metabolism Genes in Sporadic Breast Cancer. Am. J. Hum. Genet. 2001, 69, 138–147. [Google Scholar] [CrossRef]
- Pan, Q.; Hu, T.; Moore, J.H. Epistasis, Complexity, and Multifactor Dimensionality Reduction. In Genome-Wide Association Studies and Genomic Prediction; Gondro, C., van der Werf, J., Hayes, B., Eds.; Humana Press: Totowa, NJ, USA, 2013; pp. 465–477. ISBN 978-1-62703-447-0. [Google Scholar]
- He, H.; Oetting, W.S.; Brott, M.J.; Basu, S. Pair-Wise Multifactor Dimensionality Reduction Method to Detect Gene-Gene Interactions in a Case-Control Study. Hum. Hered. 2010, 69, 60–70. [Google Scholar] [CrossRef]
- Motsinger, A.A.; Ritchie, M.D. The Effect of Reduction in Cross-Validation Intervals on the Performance of Multifactor Dimensionality Reduction. Genet. Epidemiol. 2006, 30, 546–555. [Google Scholar] [CrossRef]
- Alarcón-Granados, M.C.; Moreno-Ortíz, H.; Esteban-Pérez, C.I.; Ferrebuz-Cardozo, A.; Camargo-Villalba, G.E.; Forero-Castro, M. Assessment of THADA Gene Polymorphisms in a Sample of Colombian Women with Polycystic Ovary Syndrome: A Pilot Study. Heliyon 2022, 8, e09673. [Google Scholar] [CrossRef] [PubMed]
- Alarcón-Granados, M.C.; Moreno-Ortíz, H.; Rondón-Lagos, M.; Camargo-Villalba, G.E.; Forero-Castro, M. Study of LHCGR Gene Variants in a Sample of Colombian Women with Polycystic Ovarian Syndrome: A Pilot Study. J. King Saud Univ. Sci. 2022, 34, 102202. [Google Scholar] [CrossRef]
- Fang, G.; Wang, W.; Paunic, V.; Heydari, H.; Costanzo, M.; Liu, X.; Liu, X.; VanderSluis, B.; Oately, B.; Steinbach, M.; et al. Discovering Genetic Interactions Bridging Pathways in Genome-Wide Association Studies. Nat. Commun. 2019, 10, 4274. [Google Scholar] [CrossRef] [PubMed]
- Watkinson, J.; Anastassiou, D. Synergy Disequilibrium Plots: Graphical Visualization of Pairwise Synergies and Redundancies of SNPs with Respect to a Phenotype. Bioinformatics 2009, 25, 1445–1446. [Google Scholar] [CrossRef] [PubMed]
- Itriyeva, K. The Normal Menstrual Cycle. Curr. Probl. Pediatr. Adolesc. Health Care 2022, 52, 101183. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, R.L. The Endocrinology of the Menstrual Cycle. Methods Mol. Biol. 2014, 1154, 145–169. [Google Scholar] [CrossRef] [PubMed]
- Constantin, S.; Bjelobaba, I.; Stojilkovic, S.S. Pituitary Gonadotroph-Specific Patterns of Gene Expression and Hormone Secretion. Curr. Opin. Pharmacol. 2022, 66, 102274. [Google Scholar] [CrossRef] [PubMed]
- Wide, L.; Eriksson, K. Low-Glycosylated Forms of Both FSH and LH Play Major Roles in the Natural Ovarian Stimulation. Upsala J. Med. Sci. 2018, 123, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Malini, N.A.; Roy George, K. Evaluation of Different Ranges of LH:FSH Ratios in Polycystic Ovarian Syndrome (PCOS)—Clinical Based Case Control Study. Gen. Comp. Endocrinol. 2018, 260, 51–57. [Google Scholar] [CrossRef]
- Pratama, G.; Wiweko, B.; Asmarinah; Widyahening, I.S.; Andraini, T.; Bayuaji, H.; Hestiantoro, A. Mechanism of Elevated LH/FSH Ratio in Lean PCOS Revisited: A Path Analysis. Sci. Rep. 2024, 14, 8229. [Google Scholar] [CrossRef]
- Ruddenklau, A.; Campbell, R.E. Neuroendocrine Impairments of Polycystic Ovary Syndrome. Endocrinology 2019, 160, 2230–2242. [Google Scholar] [CrossRef]
- Walker, K.; Decherney, A.H.; Saunders, R. Menstrual Dysfunction in PCOS. Clin. Obstet. Gynecol. 2021, 64, 119–125. [Google Scholar] [CrossRef]
- Zehravi, M.; Maqbool, M.; Ara, I. Polycystic Ovary Syndrome and Infertility: An Update. Int. J. Adolesc. Med. Health 2021, 34, 1–9. [Google Scholar] [CrossRef]
- Han, C.-C.; Yue, L.-L.; Yang, Y.; Jian, B.-Y.; Ma, L.-W.; Liu, J.-C. TOX3 Protein Expression Is Correlated with Pathological Characteristics in Breast Cancer. Oncol. Lett. 2016, 11, 1762–1768. [Google Scholar] [CrossRef][Green Version]
- Tian, Y.; Li, J.; Su, S.; Cao, Y.; Wang, Z.; Zhao, S.; Zhao, H. PCOS-GWAS Susceptibility Variants in THADA, INSR, TOX3, and DENND1A Are Associated With Metabolic Syndrome or Insulin Resistance in Women With PCOS. Front. Endocrinol. 2020, 11, 274. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Chan, K.C.C.; Huang, Z.; Wang, J. Determining Dependency and Redundancy for Identifying Gene-Gene Interaction Associated with Complex Disease. J. Bioinform. Comput. Biol. 2020, 18, 2050035. [Google Scholar] [CrossRef]
- Pérez-Pérez, J.M.; Candela, H.; Micol, J.L. Understanding Synergy in Genetic Interactions. Trends Genet. 2009, 25, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.P.; Chen, J. Nonsense mutations and genetic compensation response. Yi Chuan 2019, 41, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Thathapudi, S.; Jayashankar, E.; Putcha, U.K.; Mullapudi, S.; Challa, S. Multifactor Dimensionality Reduction Analysis for Detecting SNP-SNP, SNP-Environment Interactions Associated with Polycystic Ovarian Syndrome among South Indian Women. Int. J. Mol. Biol. Open Access 2019, 4, 59–65. [Google Scholar] [CrossRef]
- Castillo-Higuera, T.; Alarcón-Granados, M.C.; Marin-Suarez, J.; Moreno-Ortiz, H.; Esteban-Pérez, C.I.; Ferrebuz-Cardozo, A.J.; Forero-Castro, M.; Camargo-Vill alba, G. A Comprehensive Overview of Common Polymorphic Variants in Genes Related to Polycystic Ovary Syndrome. Reprod. Sci. 2020, 28, 2399–2412. [Google Scholar] [CrossRef]
- Crespo, R.P.; Bachega, T.A.S.S.; Mendonça, B.B.; Gomes, L.G. An Update of Genetic Basis of PCOS Pathogenesis. Arch. Endocrinol. Metab. 2018, 62, 352–361. [Google Scholar] [CrossRef]
- Oeth, P.; Mistro, G.D.; Marnellos, G.; Shi, T.; Boom, D. Qualitative and Quantitative Genotyping Using Single Base Primer Extension Coupled with Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MassARRAY®). In Single Nucleotide Polymorphisms; Komar, A.A., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 578, pp. 307–343. ISBN 978-1-60327-410-4. [Google Scholar]
- Park, M.; Kim, S.A.; Shin, J.; Joo, E.-J. Investigation of Gene-Gene Interactions of Clock Genes for Chronotype in a Healthy Korean Population. Genom. Inform. 2020, 18, e38. [Google Scholar] [CrossRef]
| PCOS Group | Control Group | p-Value | |
|---|---|---|---|
| Case number | 49 | 49 | |
| Age (years) | 28 (24–33) | 27 (24–30) | 0.448 † |
| Weight (kg) | 60.8 (55–74) | 60 (52–64) | 0.037 † |
| Height (m) | 1.62 (1.59–1.66) | 1.6 (1.56–1.64) | 0.064 † |
| BMI (kg/m2) | 23.16 (21.48–25.6) | 22.6 (20–24.98) | 0.22 † |
| Menarche (years) | 13 (12–14) | 12 (11.5–14) | 0.185 † |
| Menstrual cycle length (days) | 31 (29.5–45) | 28 (28–30) | <0.0001 † |
| Period length (days) | 5 (4–8) | 5 (4–5) | 0.129 † |
| FSH (mUl/mL) | 5.95 ± 3.47 | 9.5 ± 5 | <0.0001 †† |
| AMH (ng/mL) | 8.02 (5.07–12.55) | 4.87 (3.05–6.77) | <0.0001 † |
| LH (mUl/mL) | 6.8 (4.55–10.3) | 3.2 (2.12–5.17) | <0.0001 † |
| LH/FSH ratio | 1.27 (0.83–1.74) | 0.38 (0.18–0.64) | <0.0001 † |
| TSH (mUl/mL) | 1.67 (1.29–2.69) | 1.65 (1.05–2.47) | 0.284 † |
| E2 (pg/mL) | 53.3 (32.72–72.87) | 29.7 (15–40.6) | <0.0001 † |
| Total ovarian volume (cm3) | 12.25 (9.62–18.75) | 7.61 (6.63–9.47) | <0.0001 † |
| Total AFC (number of follicles) | 27 (23–34,75) | 16 (13–20) | <0.0001 † |
| Family History | |||
| Family history of polycystic ovaries | 22 (44.8%) | 6 (12.24%) | <0.0001 ††† |
| Family history of endometriosis | 10 (20.4%) | 4 (8.16%) | 0.013 ††† |
| Family history of breast and ovarian cancer | 10 (20.4%) | 6 (12.24%) | 0.196 ††† |
| Reproductive traits | |||
| Pregnancies | 12 (24.48%) | 33 (67.34%) | <0.0001 ††† |
| Early pregnancy loss | 8 (16.32%) | 2 (4.08%) | 0.045 ††† |
| Spontaneous abortion | 7 (14.28%) | 2 (4.08%) | 0.091 ††† |
| Endocrine–Metabolic Parameters | Value † |
| Androstenedione (ng/mL) | 1.49 ± 0.59 |
| DHEAS (μg/dL) | 152.8 ± 64.51 |
| Free testosterone (pg/mL) | 1.34 (0.91–2.40) |
| Fasting insulin (μUl/mL) | 4.68 (2.62–9.16) |
| Post-meal insulin (μUl/mL) | 28.3 (13.1–43.6) |
| Fasting blood glucose (mg/dL) | 83.91 ± 8.51 |
| Post-meal glucose (mg/dL) | 80.5 (72.5–95) |
| HOMA-IR | 0.84 (0.48–1.95) |
| HOMA-IS | 0.49 (0.02–0.08) |
| Glycosylated hemoglobin (%) | 5.24 (5.01–5.74) |
| Clinical Parameters | n (%) †† |
| Acne | 30 (60%) |
| Hair loss | 43 (86%) |
| Facial hair | 34 (68%) |
| Abdominal hair | 30 (60%) |
| Fatty discharge from scalp and face | 33 (66%) |
| Acanthosis nigricans | 10 (20%) |
| Cystic lesion resection | 2 (4%) |
| Menstrual bleeding stopped for more than 3 months | 30 (60%) |
| Multiple menstrual bleeds in one month | 25 (50%) |
| Postcoital bleeding | 5 (10%) |
| Dysmenorrhea | 29(58%) |
| Gene | SNP ID | Chr | Position | Consequence | Alleles | MAF | HWE-p | OR (95% CI) | p-Value | |
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | |||||||||
| THADA | rs13429458 | 2 | 43411699 | Intron Variant | A > C | 0.12 | 0.1 | 0.34 | 1.23 (0.50–2.99) | 0.65 |
| THADA | rs12478601 | 2 | 43494369 | Intron Variant | C > T | 0.43 | 0.39 | 0.53 | 1.18 (0.67–2.09) | 0.56 |
| THADA | rs12468394 | 2 | 43334022 | Intron Variant | C > A | 0.32 | 0.32 | 0.49 | 1.01 (0.55–1.86) | 0.97 |
| THADA | rs6544661 | 2 | 43484786 | Intron Variant | A > G | 0.44 | 0.4 | 0.84 | 1.18 (0.67–2.09) | 0.56 |
| THADA | rs11891936 | 2 | 43305163 | Intron Variant | C > T | 0.12 | 0.13 | 0.65 | 0.91 (0.39–2.11) | 0.83 |
| LHCGR | rs13405728 | 2 | 48751020 | Intron Variant | A > G | 0.1 | 0.14 | 0.63 | 0.68 (0.2–1.6) | 0.38 |
| LHCGR | rs7371084 | 2 | 48712814 | Intron Variant | T > C | 0.13 | 0.21 | 0.29 | 0.56 (0.26–1.19) | 0.13 |
| LHCGR | rs4953616 | 2 | 48714289 | Intron Variant | T > C | 0.32 | 0.28 | 0.63 | 1.21 (0.6–2.25) | 0.53 |
| LHCGR | rs2293275 | 2 | 48694236 | Missense Variant | C > T | 0.36 | 0.29 | 0.49 | 1.52 (0.30–7.53) | 0.28 |
| LHCGR | rs6732721 | 2 | 48738464 | Intron Variant | T > C | 0.12 | 0.16 | 0.4 | 0.71 (0.32–1.60) | 0.41 |
| FSHR | rs2268361 | 2 | 48974473 | Intron Variant | T > C | 0.35 | 0.41 | 0.28 | 0.77 (0.43–1.37) | 0.38 |
| FSHR | rs2349415 | 2 | 49020693 | Intron Variant | C > T | 0.4 | 0.31 | 1 | 1.50 (0.83–2.70) | 0.18 |
| FSHR | rs11692782 | 2 | 49064754 | Intron Variant | T > A | 0.37 | 0.48 | 0.84 | 0.63 (0.36–1.12) | 0.11 |
| DENND1A | rs2479106 | 9 | 123762933 | Intron Variant | A > G | 0.22 | 0.23 | 0.77 | 0.94 (0.49–1.84) | 0.87 |
| DENND1A | rs10818854 | 9 | 123684499 | Intron Variant | G > A | 0.09 | 0.04 | 0.35 | 2.38 (0.71–7.99) | 0.15 |
| DENND1A | rs10986105 | 9 | 123787676 | Intron Variant | T > G | 0.09 | 0.03 | 0.3 | 3.20 (0.84–12.21) | 0.07 |
| DENND1A | rs12337273 | 9 | 123804666 | Intron Variant | A > G | 0.08 | 0.03 | 0.26 | 2.81 (0.72–10.94) | 0.12 |
| DENND1A | rs1778890 | 9 | 123769476 | Intron Variant | T > C | 0.15 | 0.14 | 1 | 1.08 (0.49–2.39) | 0.84 |
| DENND1A | rs1627536 | 9 | 123780425 | Intron Variant | A > T | 0.23 | 0.24 | 1 | 0.95 (0.49–1.82) | 0.87 |
| DENND1A | rs7857605 | 9 | 123745334 | Intron Variant | T > C | 0.09 | 0.04 | 0.35 | 2.38 (0.71–7.99) | 0.15 |
| YAP1 | rs1894116 | 11 | 102199908 | Intron Variant | A > G | 0.03 | 0.05 | 1 | 0.59 (0.14–2.53) | 0.47 |
| HMGA2 | rs2272046 | 12 | 65830681 | Intron Variant | A > C | 0.03 | 0.01 | 1 | 3.06 (0.31–29.97) | 0.31 |
| ERBB3 | rs2292239 | 12 | 56088396 | Intron Variant | G > T | 0.21 | 0.22 | 1 | 0.94 (0.48–1.85) | 0.86 |
| AMHR2 | rs2272002 | 12 | 53424132 | Intron Variant | T > A | 0.07 | 0.1 | 1 | 0.68 (0.25–1.86) | 0.45 |
| TOX3 | rs4784165 | 16 | 52313907 | Intron Variant | T > G | 0.28 | 0.37 | 0.65 | 0.66 (0.36–1.20) | 0.17 |
| INSR | rs2059807 | 19 | 7166098 | Intron Variant | G > A | 0.45 | 0.5 | 1 | 0.82 (0.47–1.43) | 0.47 |
| AMH | rs10407022 | 19 | 2249478 | Missense Variant | T > G | 0.16 | 0.19 | 0.5 | 0.81 (0.39–1.69) | 0.58 |
| Model | Bal. Acc. CV Training | Bal. Acc. CV Testing | CV Consistency | OR (95% CI) | p-Value |
|---|---|---|---|---|---|
| rs7371084 | 0.61 | 0.4184 | 5/10 | 2.59 (1.077–6.232) | 0.0312 |
| rs11692782, rs4784165 | 0.6667 | 0.4082 | 3/10 | 3.78 (1.6–8.949) | 0.002 |
| rs11692782, rs2268361, rs4784165 | 0.7574 | 0.6327 | 7/10 | 11.29 (4.183–30.49) | p < 0.0001 |
| SNP_ID | 2nd-PCRP | 1st-PCRP | AMP_LEN | UP_CONF | MP_CONF | Tm (NN) | PcGC | UEP_DIR | UEP_MASS | UEP_SEQ | EXT1_CALL | EXT1_MASS | EXT1_SEQ | EXT2_CALL | EXT2_MASS | EXT2_SEQ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10407022 | ACGTTGGATGTCTTCCGAGAAGACTTGGAC | ACGTTGGATGAGCTGCTGCCATTGCTGTC | 110 | 95.6 | 72.3 | 53.7 | 66.7 | F | 4538.0 | ACTGGCCTCCAGGCA | G | 4825.2 | ACTGGCCTCCAGGCAG | T | 4865.1 | ACTGGCCTCCAGGCAT |
| rs10818854 | ACGTTGGATGGTGCTTAAAGGTGGGAATGC | ACGTTGGATGCACTGCCTTCTGTAAGACAC | 90 | 99.6 | 72.3 | 47.5 | 60.0 | R | 4664.0 | GGGAATGCTTGCTGG | G | 4911.2 | GGGAATGCTTGCTGGC | A | 4991.1 | GGGAATGCTTGCTGGT |
| rs2349415 | ACGTTGGATGAAAAACAGGTGTCAGGCTGG | ACGTTGGATGACAACTCCACGATCTAGGAC | 92 | 99.7 | 72.3 | 46.0 | 50.0 | F | 4952.2 | GTCAGGCTGGATTTGA | C | 5199.4 | GTCAGGCTGGATTTGAC | T | 5279.3 | GTCAGGCTGGATTTGAT |
| rs2272002 | ACGTTGGATGGTAAGGGTGAAGGATAGAGC | ACGTTGGATGTATGGTAAAGCCACAGGAGG | 111 | 99.5 | 72.3 | 55.1 | 73.3 | F | 5162.4 | ttTCCCCATGGCAGGGC | A | 5433.6 | ttTCCCCATGGCAGGGCA | T | 5489.5 | ttTCCCCATGGCAGGGCT |
| rs12468394 | ACGTTGGATGTCTGTGGCTAACTGCAGAAG | ACGTTGGATGAATGCTGTTTTCAGCTGTTG | 88 | 93.8 | 72.3 | 45.9 | 50.0 | R | 5241.4 | gCTGCAGAAGTTCTGGT | C | 5528.6 | gCTGCAGAAGTTCTGGTG | A | 5568.5 | gCTGCAGAAGTTCTGGTT |
| rs1627536 | ACGTTGGATGCATGGCAATAGTAAGTGCTC | ACGTTGGATGCATCCAGTGAATGATGGTGC | 116 | 97.6 | 72.3 | 45.9 | 44.4 | R | 5360.5 | CTTCCCTTCTTAATCCGA | T | 5631.7 | CTTCCCTTCTTAATCCGAA | A | 5687.6 | CTTCCCTTCTTAATCCGAT |
| rs13405728 | ACGTTGGATGCTTCAATATCCTGGGCTTAC | ACGTTGGATGGATTTAGAAACCTGCTCTGG | 120 | 95.6 | 72.3 | 49.2 | 42.1 | R | 5762.8 | CCATAATGCAGCCATTTGT | G | 6010.0 | CCATAATGCAGCCATTTGTC | A | 6089.9 | CCATAATGCAGCCATTTGTT |
| rs6544661 | ACGTTGGATGAACACATATAGGTGCTCCTC | ACGTTGGATGTCCTCTCATTAGAACATCTC | 93 | 92.8 | 72.3 | 45.6 | 52.9 | F | 5770.7 | gcGGTGCTCCTCTTAGTAC | A | 6042.0 | gcGGTGCTCCTCTTAGTACA | G | 6058.0 | gcGGTGCTCCTCTTAGTACG |
| rs2293275 | ACGTTGGATGCAATGTGAAAGCACAGTAAG | ACGTTGGATGCACACAGAACAAGATACGAC | 111 | 92.6 | 72.3 | 47.1 | 44.4 | R | 5934.9 | gGCACAGTAAGGAAAGTGA | T | 6206.1 | gGCACAGTAAGGAAAGTGAA | C | 6222.1 | gGCACAGTAAGGAAAGTGAG |
| rs12337273 | ACGTTGGATGAGTGGCTGATACATTGGCTC | ACGTTGGATGACATCTCCACTTGACGTCTC | 109 | 99.7 | 72.3 | 47.3 | 35.0 | R | 6140.0 | AAAGATCAGGAGTTCCATTT | G | 6387.2 | AAAGATCAGGAGTTCCATTTC | A | 6467.1 | AAAGATCAGGAGTTCCATTTT |
| rs2268361 | ACGTTGGATGTTGATGCTGTGAGACGAAGG | ACGTTGGATGTTCTTACCAAGAGCTCCCTC | 110 | 99.6 | 72.3 | 46.4 | 50.0 | F | 6173.0 | gtgcGACGAAGGCATCTTGT | C | 6420.2 | gtgcGACGAAGGCATCTTGTC | T | 6500.1 | gtgcGACGAAGGCATCTTGTT |
| rs2059807 | ACGTTGGATGATGTGAATCAGACCTCTTGC | ACGTTGGATGAGCCAATAACCATATCAAGG | 98 | 93.0 | 72.3 | 48.0 | 33.3 | R | 6355.2 | AATCAGACCTCTTGCTTTTAA | G | 6602.3 | AATCAGACCTCTTGCTTTTAAC | A | 6682.3 | AATCAGACCTCTTGCTTTTAAT |
| rs2272046 | ACGTTGGATGGGATTCAGTAATTGGCCTTG | ACGTTGGATGACATTCTGCATGCATTGTCC | 109 | 96.8 | 72.3 | 50.4 | 52.9 | F | 6533.2 | ggagTGGCCTTGGGACATTTG | C | 6780.4 | ggagTGGCCTTGGGACATTTGC | A | 6804.4 | ggagTGGCCTTGGGACATTTGA |
| rs11692782 | ACGTTGGATGACAGTTTCTCAGATCCCTTG | ACGTTGGATGTGGTGTTGTACTTCAGTACG | 97 | 97.1 | 72.3 | 50.1 | 40.9 | R | 6642.3 | TTCTCAGATCCCTTGGTTATTC | T | 6913.5 | TTCTCAGATCCCTTGGTTATTCA | A | 6969.4 | TTCTCAGATCCCTTGGTTATTCT |
| rs12478601 | ACGTTGGATGAGAGCTGGAAGTAAAGCCCG | ACGTTGGATGTTCTTTCATTCCTGCTGGTC | 93 | 97.0 | 72.3 | 48.4 | 38.1 | R | 6740.4 | gCGGGTCCTAACATTTTATTGA | T | 7011.6 | gCGGGTCCTAACATTTTATTGAA | C | 7027.6 | gCGGGTCCTAACATTTTATTGAG |
| rs4953616 | ACGTTGGATGACTTCATCAGCCACTCTATG | ACGTTGGATGCTACATAACCACACTGAGGG | 116 | 97.6 | 72.3 | 47.1 | 34.8 | F | 6868.5 | CCTCATCATCATTTCCATTATAC | C | 7115.7 | CCTCATCATCATTTCCATTATACC | T | 7195.6 | CCTCATCATCATTTCCATTATACT |
| rs1778890 | ACGTTGGATGGAATGTTAAGAATGGTATGG | ACGTTGGATGATGTGGACAGGTAGTGTCAG | 116 | 86.9 | 72.3 | 46.1 | 26.1 | F | 7058.6 | ATTTTCTATAGCAGGTTTATTGA | C | 7305.8 | ATTTTCTATAGCAGGTTTATTGAC | T | 7385.7 | ATTTTCTATAGCAGGTTTATTGAT |
| rs6732721 | ACGTTGGATGGACATAGCAGGAGTTGTCAG | ACGTTGGATGTTCCTGTCACTCCATCGTTG | 90 | 99.6 | 72.3 | 45.7 | 40.0 | R | 7152.7 | cggTGTCAGGAAGAGTAATCTAG | T | 7423.9 | cggTGTCAGGAAGAGTAATCTAGA | C | 7439.9 | cggTGTCAGGAAGAGTAATCTAGG |
| rs11891936 | ACGTTGGATGCACTCTTAACGTCAATGTCC | ACGTTGGATGGTTCCTATGGTTTCCTTTTC | 100 | 93.0 | 72.3 | 45.4 | 36.8 | F | 7234.7 | tcattTCCTGTTATGCAATTTCTC | C | 7481.9 | tcattTCCTGTTATGCAATTTCTCC | T | 7561.8 | tcattTCCTGTTATGCAATTTCTCT |
| rs2479106 | ACGTTGGATGGACTCCTGTCCTTTTGGTTC | ACGTTGGATGACAGGGCACTGGGTTGTTTC | 120 | 97.0 | 72.3 | 47.9 | 36.4 | R | 7348.8 | tgTTGGTTCCTTGATCATAACTAG | G | 7596.0 | tgTTGGTTCCTTGATCATAACTAGC | A | 7675.9 | tgTTGGTTCCTTGATCATAACTAGT |
| rs7857605 | ACGTTGGATGAAAGCCCATGAGATCTAGGT | ACGTTGGATGTAGCAACACCTCTGCAAACG | 104 | 97.3 | 72.3 | 47.1 | 30.4 | R | 7525.9 | gaCCTTATTTACTTCTCCAAACATT | T | 7797.1 | gaCCTTATTTACTTCTCCAAACATTA | C | 7813.1 | gaCCTTATTTACTTCTCCAAACATTG |
| rs7371084 | ACGTTGGATGCAGTCCCACTATTTAACAGC | ACGTTGGATGCAAGCCTATTATTGGATCCAT | 120 | 85.2 | 72.3 | 47.7 | 38.1 | R | 7634.0 | agacGCAAGTTACTTAACCGATCTA | T | 7905.2 | agacGCAAGTTACTTAACCGATCTAA | C | 7921.2 | agacGCAAGTTACTTAACCGATCTAG |
| rs13429458 | ACGTTGGATGATGCACAATGGAGACTGCTG | ACGTTGGATGTAATTAGTGGCAGGGTATAG | 99 | 94.4 | 72.3 | 46.9 | 33.3 | F | 7738.1 | gcttTGCAAAGTTAGAAGATGAAAC | C | 7985.3 | gcttTGCAAAGTTAGAAGATGAAACC | A | 8009.3 | gcttTGCAAAGTTAGAAGATGAAACA |
| rs2292239 | ACGTTGGATGGCTATCACCCTTACTTCTGC | ACGTTGGATGACCCTAGATCCCTTAAGTGC | 106 | 99.9 | 72.3 | 45.5 | 33.3 | F | 7761.1 | gggcGTGAAGAGACTTTTGAATCTA | G | 8048.3 | gggcGTGAAGAGACTTTTGAATCTAG | T | 8088.2 | gggcGTGAAGAGACTTTTGAATCTAT |
| rs1894116 | ACGTTGGATGAAATTTAGTTGCATTGAGG | ACGTTGGATGAAGGATTGACCACTGTCAAG | 113 | 77.8 | 72.3 | 46.7 | 22.2 | R | 8231.4 | TCTACATAATATTGATTCTAGACAATT | G | 8478.6 | TCTACATAATATTGATTCTAGACAATTC | A | 8558.5 | TCTACATAATATTGATTCTAGACAATTT |
| rs10986105 | ACGTTGGATGTCCATCACAATTAGCCTGAG | ACGTTGGATGCACTATAGGCAGTTAAACAA | 116 | 84.5 | 72.3 | 50.0 | 36.4 | F | 8363.4 | gggagTTAGCCTGAGTTATGCAACATA | G | 8650.7 | gggagTTAGCCTGAGTTATGCAACATAG | T | 8690.5 | gggagTTAGCCTGAGTTATGCAACATAT |
| rs4784165 | ACGTTGGATGGAGCCAGCCGTACATTAATC | ACGTTGGATGGGAATTTAAGTTATTTTCCC | 115 | 78.6 | 72.3 | 49.3 | 28.6 | R | 8612.7 | GTCACATAATAACTTGAAAAACTATGAG | G | 8859.8 | GTCACATAATAACTTGAAAAACTATGAGC | T | 8883.9 | GTCACATAATAACTTGAAAAACTATGAGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alarcón-Granados, M.C.; Camargo-Villalba, G.E.; Forero-Castro, M. Exploring Genetic Interactions in Colombian Women with Polycystic Ovarian Syndrome: A Study on SNP-SNP Associations. Int. J. Mol. Sci. 2024, 25, 9212. https://doi.org/10.3390/ijms25179212
Alarcón-Granados MC, Camargo-Villalba GE, Forero-Castro M. Exploring Genetic Interactions in Colombian Women with Polycystic Ovarian Syndrome: A Study on SNP-SNP Associations. International Journal of Molecular Sciences. 2024; 25(17):9212. https://doi.org/10.3390/ijms25179212
Chicago/Turabian StyleAlarcón-Granados, Maria Camila, Gloria Eugenia Camargo-Villalba, and Maribel Forero-Castro. 2024. "Exploring Genetic Interactions in Colombian Women with Polycystic Ovarian Syndrome: A Study on SNP-SNP Associations" International Journal of Molecular Sciences 25, no. 17: 9212. https://doi.org/10.3390/ijms25179212
APA StyleAlarcón-Granados, M. C., Camargo-Villalba, G. E., & Forero-Castro, M. (2024). Exploring Genetic Interactions in Colombian Women with Polycystic Ovarian Syndrome: A Study on SNP-SNP Associations. International Journal of Molecular Sciences, 25(17), 9212. https://doi.org/10.3390/ijms25179212






