Abstract
One of the many unresolved obstacles in the field of cardiovascular research is an uncompromising in vitro cardiac model. While primary cell sources from animal models offer both advantages and disadvantages, efforts over the past half-century have aimed to reduce their use. Additionally, obtaining a sufficient quantity of human primary cardiomyocytes faces ethical and legal challenges. As the practically unlimited source of human cardiomyocytes from induced pluripotent stem cells (hiPSC-CM) is now mostly resolved, there are great efforts to improve their quality and applicability by overcoming their intrinsic limitations. The greatest bottleneck in the field is the in vitro ageing of hiPSC-CMs to reach a maturity status that closely resembles that of the adult heart, thereby allowing for more appropriate drug developmental procedures as there is a clear correlation between ageing and developing cardiovascular diseases. Here, we review the current state-of-the-art techniques in the most realistic heart models used in disease modelling and toxicity evaluations from hiPSC-CM maturation through heart-on-a-chip platforms and in silico models to the in vitro models of certain cardiovascular diseases.
1. Introduction
Cardiovascular diseases (CVDs) are the most prevalent diseases and the leading cause of death worldwide. In recent decades, most of the phase 3 cardiac clinical trials failed due to safety concerns, lack of efficiency, or economic considerations as we described previously [1]. This was best illustrated by the disappointing outcomes of the Cardiac Arrhythmia Suppression Trial (CAST), CAST II, and Survival with oral d-sotalol (SWORD) trials. In these studies, antiarrhythmic drugs (encainide, flecainide, moricizine, or d-sotalol) were administered; however, in the CAST trial, the number of deaths due to arrhythmia or shock after recurrent myocardial infarction was significantly higher in the treated patient groups [2], while the CAST II and SWORD trials resulted in higher risk of drug-associated mortality (cardiac arrests due to arrhythmias) in patients assigned to the drug treatment [3,4,5]. Unexpected adverse effects (e.g., hidden cardiotoxicity), therefore, will lead to the discontinuation of clinical studies and withdrawal of the drug from the market, leading to the failure of new therapies and an immense financial burden [6].
As a consequence of the unsuccessful clinical attempts, only nine drugs targeting CVDs were approved by the U.S. Food and Drug Administration (FDA) in the last decade [1]. To date, ticagrelor was approved in 2011 to reduce cardiovascular death and heart attack in patients with acute coronary syndromes [7]; vorapaxar in 2014 to reduce the risk of heart attacks and stroke in high-risk patients [8]; ivabradine and sacubitril/valsartan in 2015 to reduce hospitalisation from worsening heart failure [9] and to treat heart failure [10], respectively; tafamidis meglumine in 2019 to treat cardiomyopathy caused by transthyretin-mediated amyloidosis in adults [11]; vericiguat in 2021 to mitigate the risk of cardiovascular death and hospitalisation for chronic heart failure [12]; mavacamten in 2022 to cure patients with obstructive hypertrophic cardiomyopathy [13]; sotagliflozin in 2023 to treat heart failure [14]; and aprocitentan in 2024 to treat resistant hypertension [15].
The development of realistic cardiac models for preclinical drug testing and cardiotoxicity assessment could significantly reduce the above-mentioned failure rates in clinical trials by effectively addressing safety concerns. Furthermore, these advanced models have the potential to enhance drug development, leading to the production of more effective treatments for cardiovascular diseases. There is an urgent need for novel and more realistic heart models that could both replace animal research in preclinical studies and serve as tools of human origin to better identify drug candidates that can reach production and large-scale treatment of patients. This toolset will not only allow the selection of potential drug molecules but can save a tremendous amount of time and expense by identifying adverse drug effects as early risk prediction before the clinical studies. Recently, human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) have been considered as potential options for in vitro cytotoxicity evaluations, high-throughput drug screenings, and complex systems, such as organ-on-a-chip models. Besides assessing the safety profiles of a compound in the preclinical phase, hiPSC-CMs are anticipated to serve as an ideal cellular source for regenerative and precision or personalised medicine. Ongoing clinical trials attempt, for example, implanting clinical-grade hiPSC-CMs as surgical grafts to treat patients [16] or directly injecting allogeneic hiPSC-CM spheroids to remuscularise the injured myocardium [17]. The hiPSC-CMs have also been considered as valuable resources for modelling certain genetic heart diseases such as long QT syndrome (LQTS) and cardiomyopathies (hypertrophic or dilated cardiomyopathy and arrhythmogenic cardiomyopathy) or atrial fibrillation (AF), enabling the study of their molecular pathologies [18]. On the other hand, the maturity status of hiPSC-CMs is still a main bottleneck in the field. To better understand the real pathophysiology, especially considering the often late in-life onset of cardiac diseases, and to translate these findings into improved drug development and more evolved disease modelling, it is key to pay special attention to pioneering maturation studies. In this review, we summarise the current state-of-the-art in the field of hiPSC-CMs, focusing on the maturation level of these cells also from a bioengineering point of view, while highlighting the in silico and disease modelling applications.
2. The Potential of hiPSC-CMs and the Substantial Issues with Maturation
Reprogramming of somatic cells to a pluripotent state was a groundbreaking discovery that led to the generation of human induced pluripotent stem cells (hiPSCs) [19]. The pluripotency of hiPSCs offers the possibility of creating virtually every cell type of the human body through directed differentiation protocols. Soon after the creation of hiPSCs, the first publications were released on the generation of hiPSC-CMs. The iPSC technology and, moreover, the possible applications of human-based cardiac models are expected to revolutionise and transform the field and regenerative medicine. Heart models in research and in drug screening take advantage of the unlimited human cardiomyocytes (CMs) without the ethical debates over the application of embryonic stem cells (ESCs) or without the translational challenges of animal models.
The de novo CMs are generated by cardiac differentiation. The cardiac induction is initiated on the hiPSC cultures and can be performed in both 2D and 3D arrangements. Early attempts to produce CMs followed the hanging drop culture technique, while later studies applied 2D plate formats or low attachment plates for 3D aggregates. There is also evidence that co-culture with other cell types can help the survival of hiPSC-CMs. The different approaches of CM differentiation were summarised elsewhere [1,20]. In general, several protocols follow a 14-day cultivation with the application of small molecules or growth factors or alternatively, a combination of the two (Figure 1) [1]. These techniques allow researchers to collect viable beating cells at Day 14 (D14) with around 70–90% of the cell population positive to cardiac troponin T (cTnT), a clear marker of CM fate (Figure 2).
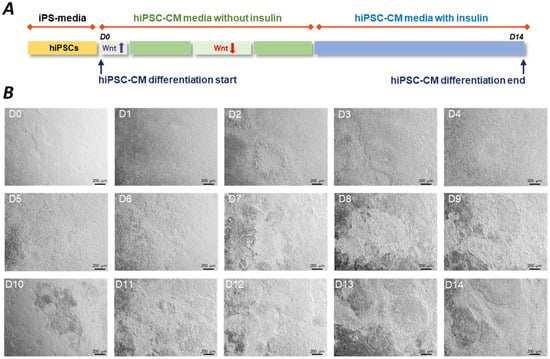
Figure 1.
The process of hiPSC-CM differentiation. (A) Conventional cardiac differentiation protocols use CHIR99021 and IWP-2 or IWR-1 small molecules to sequentially activate and inhibit the Wnt/β-catenin pathway, respectively. This panel shows a representative example of a typically applied hiPSC-CM differentiation protocol. (B) Representative images show the process of cardiac induction from Day 1 hiPSCs (D1) to Day 14 (D14) beating hiPSC-CMs. Small molecules were applied to a confluent hiPSC culture on D0. Cardiomyocytes showed spontaneous beating from D7. At D14, harvesting of the hiPSC-CMs is followed by functional readouts and cryopreservation. Images were taken on hiPSC-CMs generated by BioTalentum Ltd., at 4× magnification; scale bar, 200 μm.
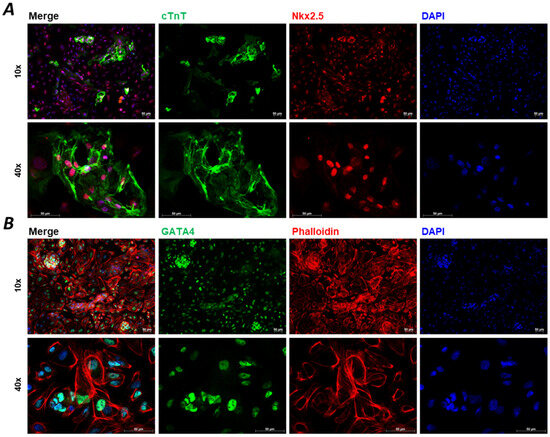
Figure 2.
Immunocytochemistry staining of D15 2D hiPSC-CMs. Immunocytochemistry images of specific biomarkers for hiPSC-CM characterisation at Day 15 (D15). Cardiac troponin T (cTnT) is the most generally used marker for CMs, which shows apparent striations ((A), green) in the case of the development of CM sarcomere structures. Other biomarkers, such as mesodermal marker homeobox protein Nkx2.5 ((A), red), transcription factor GATA4 ((B), green), and phalloidin ((B), red) are shown, while 4′,6-diamidino-2-phenylindole (DAPI) shows nuclear staining ((A,B), blue). Images were taken on hiPSC-CMs generated by BioTalentum Ltd., at 10× and 40× magnification; scale bar, 50 μm.
Allogeneic or autologous cell transplantation paved the way for cardiac personalised and regenerative medicine already with several clinical trials in diseases such as ischaemic cardiomyopathy and heart failure [16,21,22,23,24,25,26]. These clinical-grade hiPSC-CMs are transplanted into patients following cardiac differentiation, where the purity of the culture is key. Undoubtedly, superior CM maturation can be observed in vivo and besides the numerous factors that drive this process, time is also a crucial aspect of CM maturation [27]. While a study in guinea pigs showed reduced arrhythmia incidence after transplanting human ESC-derived CMs (hESC-CMs) [28], transplanted de novo hESC-CMs in non-human primate hearts or allogeneic iPSC-CMs in cynomolgus hearts might carry the risk of post-transplant arrhythmogenicity with idioventricular rhythms or even with sustained ventricular tachycardia, respectively [29,30].
On the other hand, those hiPSC-CMs that are used for in vitro drug testing and heart-on-a-chip (HOC) platforms in healthy conditions and disease modelling lack a pivotal element: the maturation of the cells. Using existing protocols, hiPSC-CMs are developmentally immature, with foetal-like CM properties, such as glycolytic metabolism, circular cellular shape, lack of sarcomere alignment, and expression of foetal genes in their sarcomere components (e.g., TNNI1 rather than TNNI3). On the other hand, mature CMs express a different set of maturation markers (e.g., MYH7, TNNI3, connexin-43, and TTNN2B), while characterised by a distinct morphology, including hypertrophic, rod-shaped cellular body, higher myofibril density and alignment, and polyploidy (Figure 3). There are also characteristic differences in their electrophysiology; during maturation, the unstable membrane potential—constant diastolic depolarisation leading to spontaneous activity—changes to a stable resting membrane potential at around −80–90 mV.
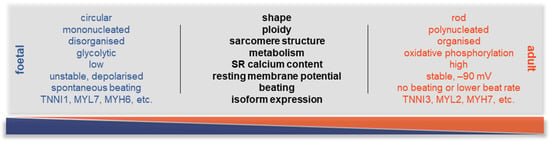
Figure 3.
Key differences between immature and mature CMs. Summary of the main physiological parameters that are different in a foetal CM compared to an adult one. During maturation, many processes evolve, from the shape of the cells to the metabolic switch. One of the most important molecular changes is the development of the adult ion channel pool and the adult-like electrophysiologic properties, allowing a stable resting membrane potential and, therefore, a physiological AP and non-spontaneous contractions.
Although the currently available protocols give a high yield of viable cells, the usually applied 2–3 weeks of cardiac differentiation is not sufficient to reach an adult phenotype in these cells (Figure 2). Additionally, it has been shown that several factors are needed to age hiPSC-CMs (Figure 4) [1]. In the drug development pipeline and disease modelling, maturity of the cells is crucial as in most of the cases the older the patient, the higher the likelihood of developing CVDs, the more essential it is to reach an adult phenotype in drug testing platforms [31]. Several approaches have been tested in recent years to overcome this developmental stalemate, but there is no fully adequate answer yet to this unresolved scientific question [32]. On the other hand, it must be noted that the full potential of early hiPSC-CMs can be exploited if the overall goal is to study neonatal diseases or to develop neonatal disease models and drug screening platforms and establish methods for developmental toxicity [33,34,35].
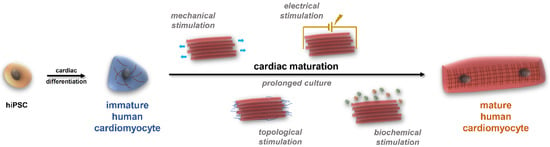
Figure 4.
The multifactorial process of cardiac maturation. HiPSCs can differentiate into virtually all cell types. To produce beating muscle cells during cardiac differentiation, small molecules are applied; however, these cells usually do not reach the desired maturity status. To achieve maturation, several approaches are available—besides the time factor—in biomaterial sciences, including various stimuli (mechanical, electrical, topological, and biochemical stimulation, detailed in the text).
2.1. Standardisation Procedures
Despite their immense potential, harnessing hiPSC-CMs for research and therapeutic purposes needs to overcome several challenges. A primary challenge lies in the necessity to establish standardised procedures for the differentiation process, aiming to assure reproducibility and consistency across various experiments and laboratory settings. A significant achievement in standardisation efforts has been defined through the use of chemically defined protocols for generating functional hiPSC-CMs [36,37]. Nonetheless, in hiPSC-CM cultures, the presence of endothelial cells and fibroblasts alongside CMs underscores the inherent cellular heterogeneity, while further complexity is introduced by the differential expression of CM subtypes, with distinct characteristics observed between atrial and ventricular populations [28,38,39]. This cellular heterogeneity is a prominent feature in these cultures and it is known that factors such as substrate rigidity, the duration and dosage of growth factors, or oxygen levels in the culture milieu contribute substantially to cell subtype heterogeneity [40,41,42] and manifest both advantages and disadvantages. On the one hand, this diversity mirrors the cellular environment observed in the native myocardium [43], thereby rendering 2D plate-format hiPSC-CM cultures invaluable tools for biomimetic inquiries. The broad spectrum of cardiac phenotypes and responses under diverse physiological and pathological conditions can be explored to identify disease mechanisms and drug reactions [44,45]. Studies utilising hiPSC-derived cultures containing diverse CM subtypes also provide insights into cardiac development, maturation kinetics, and subtype-specific disease manifestations [38,39]. Additionally, heterogeneous cultures may synergistically contribute to tissue regeneration and functional recovery following cardiac injury, which is beneficial in the context of regenerative medicine [28]. On the other hand, this cellular heterogeneity of hiPSC-CM cultures presents challenges, particularly when purity is paramount for specific applications like cell-specific drug interactions. Efforts have continued for the optimisation of CM differentiation protocols including the reprogramming and expanding of hiPSC colonies under feeder-free conditions, facilitated by specific culture media and substrates, and have contributed to isolating fully reprogrammed hiPSC colonies, thus enhancing the purity of the resulting CM populations [46]. Other efforts have considered factors such as cell seeding density and low concentrations of Wnt-agonists that have been shown to significantly impact the purity of hiPSC-CMs [47]. The pursuit of homogeneous hiPSC-CM cultures highlights the importance of refining co-culture conditions to replicate the intricate microenvironment of tissues or organs more effectively.
2.2. The Need for Co-Cultures and Subtype-Specific Cultures
While the necessity for purification and homogeneity is evident, recent insights suggest that the success of such endeavours may hinge upon co-culture conditions that are only beginning to emerge, thus introducing a layer of complexity similar to the heart [48]. Co-culture systems have demonstrated effectiveness in enhancing the differentiation of hiPSC-CMs. One approach involves co-culturing hiPSCs with cardiac fibroblasts (CF) and cardiac endothelial cells, which facilitates their differentiation into mature CMs [49]. Integrating these various cell types into co-culture systems holds promise for enhancing the tissue-like features and functionality of differentiated cardiac cells. However, the precise role of these additional cell types in co-culture systems remains poorly defined, and their integration presents practical challenges and complexities [45].
Much interest is placed in subtype-directed differentiation of hiPSCs into atrial and ventricular CMs, highlighting the importance of tailored differentiation strategies [50,51,52]. Coupling with advanced culture techniques such as microfluidic platforms or 3D tissue engineering systems offers enhanced control over CM subtype specification and maturation [45,50,53]. Therefore, the need to obtain CM cultures that are homogeneous and are subtype-specific is particularly important as there are electrophysiological variances among cardiac subtypes, such as atrial, ventricular, and nodal-like cells [54]. Each subtype displays distinctive action potential (AP) waveforms, indicative of their specialised functions within the heart’s electrical conduction system [54]. Much interest is placed in subtype-directed differentiation of hiPSCs into atrial and ventricular CMs, highlighting the importance of tailored differentiation strategies [50,51,52]. For example, CRISPR-Cas9 technology coupled with fluorescent cell sorting is an effective method for purifying atrial and ventricular populations from the same hiPSC line [55]. Several studies show the faithful recapitulation of disease-specific electrophysiological phenotypes in patient-derived cells [48,56]. As such, aberrant AP morphologies in specific CM subtypes may indicate underlying pathological conditions, such as arrhythmias, hypertrophic cardiomyopathy, or conduction defects [57,58]. Therefore, elucidating the molecular underpinnings governing CM subtype specification is essential for directing differentiation towards desired subtypes, thereby enhancing the fidelity of disease modelling and drug screening applications [59].
2.3. The Benefits of Gene-Editing Technology
Ultimately, the continued improvements in hiPSC-CM cultures, coupled with gene editing approaches like CRISPR-Cas9, present a powerful advancement for understanding specific gene mutations underlying cardiomyopathies. For example, such studies may help further our understanding of conditions such as hypertrophic cardiomyopathy, dilated cardiomyopathy, and arrhythmogenic cardiomyopathy, which are thought to be inherited and result from mutations in genes that encode proteins crucial for CM structure and function [60]. By introducing specific disease-associated mutations into healthy hiPSC-CMs, researchers can recapitulate and even discern spatiotemporal gene functioning of the disease phenotype in vitro. Studies have already shown the utility of introducing disease-associated mutations into healthy cells using CRISPR-Cas9 to investigate inherited cardiac channelopathies such as LQTS, Brugada syndrome, and short QT syndrome (SQTS) [61,62,63]. Much research is aimed at exploring the effects of these mutations on ion-channel function and on the electrophysiology profiles of CMs [63,64,65,66]. More recent advances show that hiPSC-derived cardiac disease models using precision gene editing and bioengineered 3D tissue models accurately recapitulated clinical phenotypes of inherited cardiac arrhythmias, providing robust validation of gene mutations and offering a powerful tool for dissecting pathophysiology in vitro [67]. Continued research efforts toward refining hiPSC-derived cultures, whether mixed, cell-specific, or even subtype-specific, will undoubtedly remain an ongoing endeavour in cardiovascular research.
3. In Vitro Cardiotoxicity
Cardiac safety is the main concern related to drug development during clinical trials and after release to the market. Many drugs have been withdrawn from the market due to cardiotoxicity, although a wider list of drugs is known to cause cardiac side effects. Antineoplastic drugs, central nervous system agents, genitourinary system agents, and anti-inflammatory, anti-infective, and cardiovascular agents are the most common to induce cardiotoxicity [68]. There are different mechanisms of drug-induced cardiotoxicity, including arrhythmias, disruption of mitochondrial function, apoptosis, altered growth factor signalling, or oxidative stress [68]. Correlation between animal models and human cardiac physiology is often complex due to the significant differences in heart rate (rats and mice have 6–10 times higher heart rate than humans) and the ability to increase heart rate and due to the dissimilarities in the main ion currents [69,70]. However, non-rodent models (rabbits, guinea pigs, dogs, and non-human primates) have a 90% chance of predicting QT interval changes in humans [71]. Given the current tendencies to reduce the use of animal models in science, in vitro cellular models are increasingly becoming essential tools for drug toxicity testing, including arrhythmias. The Comprehensive in vitro Proarrhythmia Assay (CiPA) collaboration, including regulatory agencies, industry, and academia from U.S. FDA, Europe, Canada, and Japan, created guidelines for cardiotoxic risk evaluation in drug candidates [72]. These include the in vitro assessment of drug effects on ion channels, computer modelling to predict risk based on drug effects on ion channels, effects in hiPSC-CMs, and phase 1 clinical testing in vivo [73,74,75,76].
The HOC platform is an automatic tool for large-scale drug testing, measuring electrophysiological and metabolic responses of CMs to different compounds [77]. HiPSC-CMs are the most promising model for cardiotoxicity tests as the primary cells are of limited availability and dedifferentiate rapidly while animal cell lines might provide false results because of differently functioning ion channels [78,79]. HiPSC-CMs can be used as an autologous or personalised system with specific genetic background. Additionally, patient-specific hiPSC-CMs can reflect the response from an individual patient in a personalised setting or represent the whole patient population, and these can be used for modelling various diseases of interest, including genetic mutations [80,81]. A typical HOC platform includes external electrical or mechanical stimuli, a calcium sensor, or a contraction detector. Validation of the HOC system can include testing drugs with well-known cardiac effects (e.g., isoproterenol) to serve as baseline controls [79]. On the other hand, the main concern about HOC platforms is the insufficient maturity level of CMs; therefore, the functional response might differ from the fully matured adult CMs [82].
Cardiotoxic Effects and Their Detection
The main cardiotoxic effects include drug-induced arrhythmias or changes in cell metabolism or viability, which can lead to heart failure. There are many physiological processes that can be monitored for detecting cardiotoxicity in CMs, including electrophysiological assays, cell metabolism, viability, or oxidative stress (Figure 5). Drug-induced arrhythmias can be caused by many broadly used medications. One of the most serious conditions is QT interval prolongation, leading to torsade de pointes type ventricular tachyarrhythmias, although bradyarrhythmia, AF, tachycardia, or Brugada syndrome can also be caused [83]. It is known that blocking the potassium HERG channel leads to drug-induced prolongation of QT interval and is associated with sudden cardiac death [84].
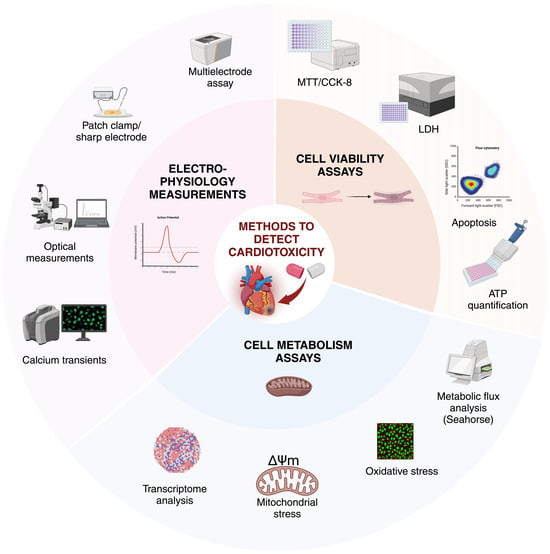
Figure 5.
Methods to detect cardiotoxicity in CMs. In vitro, cardiotoxicity can be evaluated by a range of methods, including cell viability assays, cell metabolism assays, and electrophysiological readouts. Ion channel profiles, AP characteristics, and electrophysiological properties can be analysed by patch clamp, sharp electrode, or multielectrode measurements, in parallel with calcium transient kinetics and parameters. Cell viability can be monitored by ATP-release-based techniques, intracellular lactate dehydrogenase (LDH) activity, colorimetric assays such as MTT and CCK-8, and detected by a plate reader or by measuring apoptotic events. To quantify cell metabolism, the assessment of glycolytic and oxidative metabolism can be measured by Seahorse assays along with oxidative stress by measuring the toxic levels of ROS. Mitochondrial stress tests provide information on the resting respiration of the cells, by determining ATP production and oxygen consumption. Transcriptomic analysis and metabolomics focus on the underlying genes and their expression, the end products of these genes, and on the possible interactions in the metabolic pathways. Created with BioRender.com.
Mitochondria account for almost half of human CM volume and are vitally necessary for energy production. During hiPSC-CM maturation, changes in the cell metabolism include the switch from glycolysis to fatty acid oxidation by oxidative phosphorylation, a decrease in glucose uptake, an increase in mitochondrial DNA content, an enhanced mitochondrial membrane potential, and a higher mitochondrial calcium level [53]. Cardiotoxic drugs might interfere with the mitochondrial respiratory chain, inhibit mitochondrial enzymes, induce loss of membrane potential, and increase oxidative stress [85]. If damaged mitochondria are not degraded, they start to accumulate, and it is associated with the pathogenesis of myocardial dysfunction [86]. For example, doxorubicin causes not only mitochondrial damage but the impairment of iron metabolism as well, resulting in the accumulation of reactive oxygen species (ROS) and leading to oxidative stress [87]. Mitochondrial damage can not only disrupt physiological processes in the myocardium but might also lead to cell death. CM death can be caused by several mechanisms, including autophagy, ferroptosis, apoptosis, pyroptosis, and necroptosis or DNA damage [88,89]. It has been shown that cardiac troponin I release in the medium correlates with cell viability data [90]. Consequently, the reduced viability of CMs can lead to structural damage to the myocardium.
The most important assays for detecting arrhythmias in vitro include multielectrode assays, patch-clamp and sharp electrode measurements, and optical evaluations of calcium transients or cell contractions providing analysis of multiple parameters such as the beat rate, field potential, or AP profiles (Table 1). Cell viability is generally measured by ATP production, troponin release, the metabolism of formazan-based dye (e.g., CCK-8, MTT, MTS), or by the intracellular lactate dehydrogenase activity [91]. These methods correlate with the number of living cells, while repeated measurements can be taken at different time points, making them suitable for long-term experiments. To evaluate drug effects on cell metabolism, cell respiration, oxidative stress, or mitochondrial membrane potential can be measured. Based on these readouts, a set of in vitro assays is available to estimate the potential damage in the myocardium caused by a substance (Figure 5).

Table 1.
Examples of functional readouts for determining cardiotoxicity in cardiomyocyte cellular models. APA, action potential amplitude; APD, action potential duration; BPM, beats per minute; BR, beat rate; CL, cycle length; ECG, electrocardiogram; FPD, field potential duration; MCS, maximum contraction speed; MRS, maximum relaxation speed; OAP, optical action potential; RMP, resting membrane potential; TdP, torsade de pointes type polymorphic ventricular tachyarrhythmia.
Briefly, arrhythmias, altered metabolism, and reduced cell viability are major conditions representing drug cardiotoxicity that can be tested using in vitro cardiac models, including HOC. Furthermore, in vitro cardiac models may contribute to the elucidation of molecular mechanisms leading to drug-induced cardiotoxicity.
4. Scaffolds in Cardiac Applications
The field of cardiac tissue engineering has seen significant progress in recent years, with a focus on developing scaffolds that promote the maturation of cardiac cells. This section discusses the recent developments related to 2D and 3D scaffolds. The fabrication of 2D scaffolds involves the production of a stable and soft substrate with a smooth surface for cell growth and attachment. Several techniques are commonly used for fabricating 2D scaffolds. Soft lithography utilises elastomeric materials to create 2D patterns on a substrate, while micropatterning involves creating specific patterns or geometries using photolithography, microcontact printing, or microfluidics. Layer-by-layer assembly and various deposition and casting techniques are also employed for fabricating 2D scaffolds [107]. Electrospinning is a versatile technique that involves the creation of nanofibers using an electric field. Obtaining aligned fibers using rotating collectors is often the preferred approach [108]. Indeed, aligned fiber scaffolds offer advantages for tissue engineering applications, including cardiac tissue engineering, as they mimic the aligned structure of native tissues, such as in the myocardium, promoting directed cell growth and assisting the functional maturation of the cells. CM alignment can also be controlled using micropatterning techniques to favour cell adhesion along geometric pathways.
Two-dimensional CM cultures can simulate viable environments for cardiac cell maturation, which is a relevant and easy way of drug screening and toxicological testing with high throughput. These 2D cultures are often combined with microelectrode arrays (MEA) to measure the electrophysiology of the cardiac cells completed with optical imaging. Single-cell patch clamp and sharp electrode measurements in electrophysiology are also widely used on 2D cultures as they enable measuring the membrane potential, the upstroke velocity, and ion currents [107]. The disadvantage of these models is the limited complexity in the tissue construction without a physiological extracellular matrix (ECM), especially when using 2D monotypic models [109].
Alternatively, utilising 3D scaffolds offers several advantages over 2D scaffolds in cardiac tissue engineering. Most importantly, 3D scaffolds allow for the recreation of the complex architecture and microenvironment found in native tissues, facilitating appropriate cell–cell interactions, the formation of gap junctions, and tissue organisation. It is now well-established that other cell types such as CFs, macrophages, and endothelial cells are needed to favour the maturation of CMs and to recapitulate specific functions of the native cardiac tissue [110]. Moreover, 3D-vascularised scaffolds improve nutrient and oxygen diffusion, promote cellular organisation and polarisation, and offer a physiologically relevant platform for disease modelling. In addition, 3D scaffolds allow the use of spatial, electrical, and mechanical cues, which are crucial for cardiac tissue development and maturation. The physical properties of 3D scaffolds such as stiffness, topography, and porosity can be precisely controlled to mimic the native mechanical environment. Different construction methods include 3D aggregation, application of hydrogels, and micromold design to obtain engineered cardiac tissues (ECT) around anchor pillars or 3D bioprinting [107,111]. It is important to underline that 3D cardiac tissues can be fabricated with or without scaffolds [112,113]. Layer-by-layer cardiac tissues, cardiac spheroids, and organoids can be obtained without scaffolds while the use of hydrogels, decellularised ECM, and microfabricated 3D scaffolds are alternative strategies to obtain 3D cardiac tissues using 3D scaffolds with different geometries. For the fabrication of cardiac patches, the latter approach is usually preferred [114].
In these applications, a crucial issue is the sufficient supply of oxygen and nutrients without a proper vascularisation of the native 3D tissue. The use of microfluidic systems, sacrificial materials, or 3D printing of vascular channels has been explored to overcome this issue [115]. While 3D scaffolds provide a more biomimetic and functionally relevant platform, shifting toward HOC systems presents significant challenges. These include the complexity of the cardiac microenvironment, integration of multiple cell types, and achieving functional maturity of CMs.
5. Heart-on-a-Chip Platforms
A HOC is a microfluidic device capable of mimicking the complex physiological conditions of the native heart tissue and delivering electrical, mechanical, and/or biochemical cues under a controlled environment [116]. Mechanical (stretch forces and fluidic shear stress), biochemical (e.g., vascular endothelial growth factor and fibroblast growth factor), and electrical stimuli have an influence on cell differentiation, alignment, and physiological behaviour (Figure 6) [117]. Cell culture medium perfusion, i.e., fluidic shear stress, is widely applied in HOC devices with the aim of evaluating drug responses. For instance, Christoffersson et al. attached cardiac spheroids formed by hiPSC-CMs to a microfluidic device channel coated with laminin to assess drug toxicity [118]. The drug effect was quantified by observing the cardiac cell outgrowth from 3D spheroids under perfusion and it was validated using six compounds with well-established effects on cardiac cells. Despite the sensitivity of the assay, it is worth noting that the CMs used in this study had an immature phenotype and the model is not completely representative of a 3D model. However, the easy handling of the system allied to its cost-effectiveness is advantageous, especially if it is used as a complementary tool to the existing assays. In another study, the tissue was stimulated with fluid flow, and the platform contained a vascular endothelial layer to assess the significance of microvessels in drug toxicity experiments [119]. The dynamic conditions showed reduced dextran permeation compared to the static conditions; the viability of both human umbilical vein endothelial cells (HUVECs) and hiPSC-CMs was slightly improved under fluid flow, and HUVECs aligned according to the flow direction. Hence, fluidic shear stress-induced cell alignment and mimicked in vivo forces.
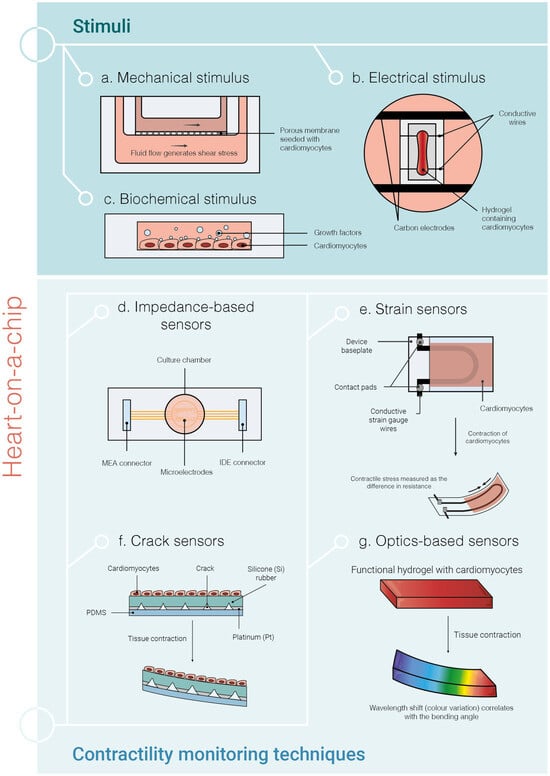
Figure 6.
Current strategies of heart-on-a-chip (HOC) systems using hiPSC-CMs. The microfluidic HOC systems serve as controlled environments to mimic as much as possible the physiological conditions that can be found in the human native heart. The most important stimuli methods (a–c) and contractility monitoring techniques are shown (d–g). Mechanical stimulation (a) provides mechanical cues to the cells, which may be shear stress (by exposing the cells to fluid flow) and/or stretch forces. Biochemical stimulation (b) consists of applying growth factors to the cells and electrical stimulation (c) can be achieved by applying small electrical pulses to the CMs. Impedance-based sensors (d) measure cardiac contraction by reading the impedance changes upon contraction of the CMs. Strain sensors (e) rely on the contraction of CMs to change the curvature of the device baseplate and measure the difference in resistance, which is then converted by a mathematical formula to the stress generated by the tissue. In crack sensors (f), the cracks in the sensor open as the tissue contracts, increasing the resistance. The contraction of the CMs causes the deformation of the hydrogel substrate leading to a wavelength shift (colour variation), which can be measured with optical sensors (g). MEA—multielectrode array; IDE—interdigitated electrodes.
Electrical stimulation in hiPSC-CMs directs the cell differentiation process towards a more mature phenotype [120]. Therefore, this type of stimulation has also been incorporated into HOC platforms. For example, electrical conditioning was used to stimulate the maturation of atrial and ventricular hiPSC-CMs, which were embedded in a hydrogel together with CFs, cultured inside the Biowire II platform [121]. This stimulus improved the sarcomeric organisation in both atrial and ventricular tissues and promoted the expression of maturation genes associated with contraction in the ventricular construct, calcium handling, electrical properties, and lipid metabolism.
In addition to providing different types of stimuli to CMs, HOC systems enable the integration of sensors for continuous monitoring of tissue contractile functions [122]. This function can be measured using impedance-based sensors [123], strain [124,125], and crack sensors [126]. Furthermore, tissue contraction can also be assessed optically by changing the curvature of the cantilever [127] and by capturing images of the tissue [82] or the hydrogel colour change [128,129,130] (Figure 6).
Importantly, the incorporation of iPSCs derived from patients with specific diseases into HOC systems is extremely valuable either to evaluate drug toxicity in a targeted patient or to develop new and tailored treatments [116]. Despite the efforts in developing HOC for different applications (e.g., research on the mechanism of heart diseases, drug development for CVDs, development of medical treatments, and drug toxicity testing) and their promising results, these devices have several limitations. They fail to replicate the complexity of the native heart, only incorporating two or three cell types. Indeed, the incorporation of macrophages, neurons, epicardial cells, and endocardial cells is yet to be successfully accomplished. Moreover, the dilution of secreted biomolecules in the circulating fluid hinders their detection [131]. Currently, hiPSC-CMs remain immature compared to human adult CMs, which hampers their use in pharmacological and toxicological screening. New methodologies for hiPSC-CM maturation must be incorporated in HOCs, e.g., integrating conductive spheroids or scaffolds (hydrogels and membranes) into the platforms. These systems must evolve toward the use of low-cost and disposable materials to be attractive to pharmaceutical companies and research institutions. In the case of more expensive components, such as sensors or motors, these should be reusable and easy to sterilise. The development of multi-organ-on-a-chip and human-on-a-chip seems to be a logical direction to pursue, not only for highly predictable drug toxicology studies but also to study the interactions between different organs. However, the complexity of such platforms together with the need for a universal cell culture medium suitable for all organs present significant challenges. Innovative approaches and solutions are still needed to overcome these obstacles and accelerate the adoption of HOC systems in preclinical testing.
6. In Silico Models
The use of mathematical and computational models plays an important role in comprehending complicated biological processes. These models promote the identification of basic behaviours, laying the groundwork for an extensive characterisation and a more profound understanding of the underlying natural processes. The work from Hodgkin and Huxley is an early example of a mathematical model intended to imitate electrical signal generation [132]. Ever since, the application of in silico modelling has become foundational in cardiac electrophysiology and has shown to be a reliable method that can successfully connect diverse datasets and precisely define cellular characteristics that are responsible for the observed variability in experimental outcomes.
One of the most extensively studied functionalities of CMs is the AP. The collection of AP data is typically carried out using sharp microelectrode or patch clamp techniques. Calcium transient can also be measured through fluorescent imaging and processed to time series [133]. These data are then characterised through computational modelling to depict fundamental behaviours, as it is illustrated by Akwaboah et al. with their implementation of genetic algorithms to accurately reproduce experimental data to validate AP morphology and various ion channel blocking mechanisms [134]. Of great importance, contractile properties of hiPSC-CMs could also be monitored and analysed [135]. Additionally, data on transcription factors have the potential to model the differentiation state of CMs, as their expression changes upon differentiation [136,137].
Usually, primary data from hiPSC-CM cultures undergo processing before model fitting, with derived biomarkers forming the basis for model development. An effective manner to extract biomarkers is through specialised software for automated quantitative analysis of specific CM parameters, which offers a user-friendly approach to analyse data. The ‘Cardio PyMEA’ software (https://github.com/csdunhamUC/cardio_pymea?tab=GPL-3.0-1-ov-file, accessed on 30 July 2024) allows users to analyse MEA data from CMs, from beat detection to biomarker analysis such as beat amplitude and interval [138]. SarcGraph v0.2.1 is an open-source Python-based software that extracts data from CM culture videos of fluorescently labelled contractions [135], which can build upon previous research works [139,140]. These solutions automatically detect and track the sarcomere and z-disc allowing for spatio-temporal post-processing and visualisation. Sarcomere analysis is performed through scanning gradient Fourier transform to the organisation and alignment of sarcomeres [141]. Other contraction tracking options are also available, such as plug-in tools to existing software [142] testing out state-of-the-art methods [143] or focusing on the micro level [144]. Alternatively, images or videos have been directly analysed through deep learning (DL) and machine learning (ML) to generate models for detecting abnormal cultures [145,146]. ML—a subset of artificial intelligence (AI)—enables systems to autonomously learn and enhance from data, utilising various algorithms for tasks such as classification (e.g., decision tree), clustering (e.g., k-nearest neighbours), and prediction (e.g., linear regression). While DL—a subfield of ML—employs layered artificial neural networks to do analogous tasks. The primary limitation of ML methods is their requirement for substantial data, with DL being particularly data intensive. In general, the main drawback of mathematical modelling is that it heavily depends on the quantity and quality of real-world observations.
An important application of hiPSC-CM cultures is the evaluation of candidate drugs in a human model [147]. AP, contraction, and calcium transient are used to monitor drug effects on CM cultures. AP data are particularly valuable for generating computational models of cell cultures, where the effects of drugs can predict the behaviour of the CMs [148,149]. This was illustrated by Jæger et al., who sought to predict the effects of drugs by modelling the AP of cells both with and without the SQT1 mutation associated with SQTS [150,151]. Furthermore, a software tool has been implemented to account for uncertainties in dose–response measurements of drugs in cultures, translating them into prediction models and providing a probability distribution for the model’s outcome [152]. Functional analysis of CMs has also been conducted through video-derived data, such as contraction speed, BPM, and calcium transient. ML models have been developed to assess and classify the effects of drugs in screening assays through the characterisation of AP [143,153,154]. In 2021, Grafton et al. introduced a DL model specifically designed for the rapid identification of cardiotoxicity patterns from images. This model was refined through high-content image analysis of treated hiPSC-CMs across over 1200 compounds [141]. The integration of these models into automatic high-throughput screening processes enables the possibility of conducting large drug screenings efficiently.
HiPSC-CMs exhibit significant versatility in altering their functionality, induced through mutations or specific culture conditions [155,156]. Therefore, developing models capable of rapidly and accurately identifying abnormal changes is essential. Hwang et al. proposed a DL model coupled with an analytical algorithm using images that tracked calcium transient [157]. The model aims to distinguish normal and abnormal behaviour of hiPSC-CMs. These models led to the development of a range of tools commonly applied in hiPSC-CM research, including abnormal AP detectors or tools for identifying LQTS. Moreover, brightfield microscope images and videos, quantifying contraction properties, have been used to train DL models aimed to characterise the hiPSC-CM cultures before commencing an experiment [145,146]. In addition, the iMATURE v1.1.2.3 software is a tool that allows the users to manually select the age of hiPSC-CM post-differentiation and receive predictive readouts of AP morphology and ion channel dynamics over their specified age range [158]. This model is built upon the work of Kernik et al. from 2019, where they collected experimental data from different laboratories to model experimental variability and describe subcellular mechanisms [159].
Characterising cell lines is pivotal in cellular research, and models play a significant role in this regard. Notably, there is a maturity level discrepancy between hiPSC-CMs and primary CMs [137,160], e.g., hiPSC-CMs may lack the completion of critical perinatal processes [161]. Additionally, Paci et al. demonstrated in 2015 that models for hiPSC-CMs and CMs are not interchangeable [162]. Subsequently, it has been recognised that models should be tailored for specific cell lines to enhance predictions of baseline behaviours [149]. For instance, there are differences in gene expression profiles during the differentiation stages [137], in fact, regression models can define CM culture age with gene expression biomarkers data as the input [136]. Therefore, understanding the intricacies of cellular growth, differentiation, and proliferation is also crucial in research, and the models play a supportive role in deciphering the regulatory processes within these crucial aspects of wet lab work. Previous works serve as comprehensive resources for theoretical frameworks to predict differentiation fate or cellular growth [163,164]. Although the adjustment of culture media was a promising method for enhancing hiPSC-CM maturation, no specific modelling approaches have been identified, but there are studies on modelling techniques applied to optimise cell culture formulation [165] and scaffold construction [166].
Typically, biomarkers are derived from raw data to model hiPSC-CM behaviour, and the ability to generalise insights from computational models relies on acquiring substantial amounts of high-quality data. It is important to recognise, though, that gathering this kind of data is hindered significantly by the cost of experimental observations. Today, when modelling approaches like DL are popular and promising, we must keep in mind that their efficacy depends on the availability of significant and reliable data. Although data scarcity in hiPSC-CMs is still an issue, using mathematical and computer models is key to understanding complex biological processes because they make it easier to identify basic behaviours, which lays the groundwork for more detailed characterisation and a better understanding of the underlying events. Thus, the synergy between cellular and computational models offers the potential to streamline processes, reduce the need for animal models, cut down costs, and enhance the reproducibility of scientific experiments.
Besides modelling using hiPSC-CMs, in silico models have also been used to directly simulate primary CMs, thus allowing mathematical modelling of cell function [167] and also dysfunction in heart diseases such as ischaemia or heart failure [168,169]. Regardless of the origin, cellular models carry individual variabilities to some extent, which might be pivotal in predicting diseases or treatments; however, hiPSC-CMs are considered as models with reproducible genetic backgrounds and thus less variability than primary cells. On the other hand for modelling the human population, there is a need for greater genetic diversity among the hiPSC models [170]. This might increase the resources needed and reduce the practical benefits of the in vitro assays. Recently, to overcome the issues of individual differences, ML models were trained on large datasets of CMs to detect individual variability and predict susceptibility to arrhythmias [171]. The integration of in silico models with experimental data leads to the development of digital tools that can be implemented in functional cellular studies and to study the impact of a drug on cardiac tissue. The experimental data serve as an input for a previously trained computational model, which can swiftly identify functional changes without the need for complex experiments and, in parallel, reduce the use of laboratory animals.
7. Disease Modelling
7.1. Atrial Fibrillation
AF is the most prevalent cardiac rhythm disorder, affecting 59 million people globally in 2019 [172]. The incidence and prevalence of AF are rising significantly, considering this disorder as a global epidemic [173]. Despite significant advances in detection and management, AF continues to have a major impact on the morbidity and mortality of millions of patients [174], partly due to unresolved knowledge gaps in AF pathophysiology and therapeutic strategies, including rate/rhythm control and stroke prevention [175]. Additionally, the response in an individual patient to antiarrhythmic drugs is highly variable, in part due to the inability to target the underlying genetic mechanisms of AF. Indeed, current pharmacological options remain inefficient, with substantial adverse side effects, including drug-induced proarrhythmia, and both cardiac and non-cardiac toxicity [175,176]. The limited efficacy of current pharmacological treatment options and the need for a more complete understanding of the pathophysiology of AF require more advanced human models, allowing the development of a powerful personalised treatment for AF. Due to the high interspecies variabilities, in vitro models with human CMs represent a promising human-based tool to study mechanisms and processes involved in AF. While it is possible to isolate adult heart cells from patients after heart surgery, several drawbacks exist, including severe source limitation, the non-proliferative feature of human CMs in culture, and consequently a precipitous functional decline in vitro, severely limiting their application for larger-scale drug screening and disease modelling [38]. In contrast, hiPSCs can be differentiated into atrial CMs (hiPSC-aCMs), providing a nearly unlimited source of cells that can be used to model disease at a single cell level, reproducing the human cardiac electrical phenotype in health and disease [177]. Since gold standard differentiation protocols to obtain hiPSC-CMs yield a mixture of different CM subtypes, such as ventricular-, atrial-, and nodal-like cells [36,43,178], significant efforts have been made to establish differentiation protocols and/or selection strategies to generate and purify a population of chamber-specific CMs [52,179,180]. The potential of generating such chamber-specific cell-type models significantly improved the research for disease modelling and drug testing applications. Atrial-like CMs are particularly suitable for modelling atrial arrhythmias, such as AF. Nevertheless, to generate relevant models for studying the mechanisms involved in AF, the patterns of AP propagation occurring in the human heart during both sinus rhythm and arrhythmia should be properly recapitulated in vitro. Moreover, while it is pivotal to closely mimic the anatomical features of the atria and their function to investigate specific disease pathways or arrhythmia mechanisms, the model should be applicable in a standardised, systemic, and controllable manner [181]. One limitation of using hiPSC-aCMs as a well-established model for AF concerns the method to generate the cells, which, most of the time, is laborious, time-consuming, and costly and, therefore, hard to scale up. To improve the stability and reproducibility of the model, studies focused on the optimisation of the protocol to obtain hiPSC-aCMs in vitro [52,182,183]. Recently, Thorpe et al. investigated the timing of retinoic acid addition during CM differentiation, with the aim of developing a robust and scalable protocol to produce large amounts of atrial-specific CMs [184]. The protocol was successfully applied to six different hiPSC lines, without the need for line-specific optimisation. The authors could elicit arrhythmic activity in response to burst pacing, highlighting the potential of the model as an antiarrhythmic drug screening platform, and investigating human atrial arrhythmias and myopathies. Similarly, Schulz and colleagues highlighted the variability in the expression of atrium-selective currents, suspecting methodological issues in producing hiPSC-aCMs [185]. These results showed that only 1 μM retinoic acid induced enough ultra-rapid delayed rectifier potassium current (IKur) to fully reproduce human atrial AP shape.
Besides the improvement and refinement in protocols to generate hiPSC-aCMs, these cells still phenotypically resemble foetal atrial CMs. This represents another limitation of the model, and numerous maturation strategies have been implemented [1]. Among several methods, co-cultures have been recently implemented in the context of AF models [177]. Because of pivotal cellular interactions, co-culture with CFs has been used to improve hiPSC-CMs maturity [49]. Nevertheless, atrial CFs substantially differ from ventricular CFs, as they produce chamber-specific ECM protein isoforms, respond differently to key growth factors, and have distinct proliferation rates [186]. For these reasons, the effects of chamber-specific fibroblasts on hiPSC-aCM maturation and function remain unclear. Recently, Brown et al. reported that primary adult atrial fibroblasts induced higher functional maturation in hiPSC-aCMs [177]. Together with a soft-lithographic process to generate patterned co-culture of hiPSC-aCMs and ACFs, the work showed a significant improvement in hiPSC-aCM features (i.e., electrophysiology and metabolism) as compared to conventional and randomly distributed co-cultures. Moreover, this method showed a higher sensitivity for drug screening and disease modelling of familial AF [177].
The inclusion of other cell types has also been used in 3D models to generate engineered heart tissues (EHTs). The cellular complexity of these models can produce more accurate results, incorporating cells that can interact and communicate in a 3D environment, which is more representative of the physiological state of the cardiac tissue [187]. Compared to monolayer cultures, the EHT format highlighted atrial versus ventricular differences in vitro, demonstrating the strength of this method to generate atrial-like muscle structures in terms of gene expression, contractile force, contraction kinetics, and AP [188].
Finally, a growing understanding of the genetics underlying AF has enabled new treatment possibilities, allowing a deeper understanding of the disease pathogenesis [189]. Moreover, due to the increased percentage of lone AF cases, efforts have been made to study the underlying genetic contribution to AF, with the aim of facilitating early identification of people at high risk of developing this disease later in their lives [190]. Among several genome editing techniques, the CRISPR-Cas9 system has mainly been used for somatic genome editing of the heart to study disease phenotype and therapeutic interventions [191]. This system allowed the development of isogenic hiPSC disease lines of AF, enabling the study of patient-specific disease mechanisms and setting the stage for a pharmaco-genomic screen. Indeed, the use of unrelated controls and the failure to correct AF-causing genes (to generate an isogenic hiPSCs line) may provide only limited insights into the underlying pathophysiological mechanisms of familial AF [192]. Recent studies investigated underlying genetic mutations linked to AF, including isogenic controls and, hiPSCs from unaffected family members, not harbouring the mutation. For example, in the study of Hong et al. patient-specific AF hiPSC-aCMs exhibited remarkable in vitro phenotypes of AF-linked SCN5A mutations [193]. The use of a hiPSC line not harbouring the SCN5A mutation and the gene corrected-isogenic control line allowed to examine important insight into genotype-phenotype correlation, confirming the E428 variant as the cause of the AF. Similarly, Sumer et al. used hiPSCs to precisely correct heterozygous SHOX2 mutations associated with AF, contributing to elucidating the function of SHOX2 in the genetic network of atrial and nodal CMs and its contribution to the development and progression of AF [194]. A recent review article discussed the use of hiPSC-CMs as a model to study the role of small-conductance calcium-activated potassium channel (SK) variants associated with AF. The authors highlighted the advantages of using the hiPSC-CMs model to investigate single nucleotide polymorphisms (SNPs) associated with AF, addressing limitations and best practices for rigorous hiPSC studies, including the need to minimise off-target mutagenesis and the need to include isogenic controls to better elucidate the impact of a single variant [195]. Consequently, improving gene-editing studies will pave the way to model pre-clinical testing of antiarrhythmic drugs for a more personalised approach to AF therapy.
7.2. Hypertrophy
Cardiac hypertrophy is an adaptive response to pressure or overload, resulting in altered gene expression, metabolism, and cell morphology. Hypertrophy can be physiological or pathological. Physiological hypertrophy is characterised by a 10–20% increase in CM length and width, which is caused by postnatal growth, pregnancy, or high endurance exercise, and it is not related to dysfunction [196]. Whereas, pathological cardiac hypertrophy is caused by myocardial injury, stress induced by hypertension, or neurohumoral regulation leading to fibrotic remodelling, cardiac dysfunction, heart failure, and even death [197]. The main regulators of hypertrophic response are calcineurin/nuclear factor of activated T cells (NFAT), mitogen-activated protein kinase ERK, small guanosine triphosphate (GTP)-binding proteins (Ras, Rho), protein kinase C (PKC), and others [198]. To better understand molecular mechanisms and possible treatment targets of hypertrophy, in vitro models are widely used.
In vitro, hypertrophic phenotypes can be characterised by morphological changes, such as increased cell and nucleus size, upregulated expression of classical hypertrophy markers (foetal genes, ANP, BNP, and ACTA1, β-MHC), cell cycle arrest, and metabolic switch towards the foetal-type under the increased reliance on glucose [199,200,201,202,203,204,205]. Increased glucose consumption is one of the features of hypertrophic response and it can be measured by increased lactate concentration [203]. Multiomic approaches may give a deeper understanding of mechanisms of hypertrophy. For instance, transcriptomic analysis has shown enhanced expression of genes related to muscle contraction, myofibril assembly, and maturity-related structural elements of the cytoskeleton as well as altered expression of genes encoding calcium handling proteins in hypertrophic CMs [203,206]. Elevated expression of COL12A1 and THBS1 genes was also detected in a hiPSC-CM model of endothelin-1-induced hypertrophy, implying a relation to fibrosis and cardiac remodelling [203]. Notably, proteomic analysis revealed that the pathways of cardiac hypertrophy signalling, actin cytoskeleton signalling, and the superpathway of inositol phosphate compounds and PPARα/RXRα activation were changed significantly in hypertrophic CMs [207].
For modelling hypertrophy in vitro, neurohormonal or mechanical approaches are used. Endothelin-1, phenylephrine, isoproterenol, norepinephrine, and angiotensin II are classically used drugs to develop hypertrophy in CM cultures. Increased pressure or overload is introduced using physical stimuli, such as cyclic mechanical stretch [206]. Moderate mechanical stretch is also an option for CM maturation, while high afterload can cause hypertrophic changes in hiPSC-CMs [208]. Activation of glucose metabolism by testosterone or prolonged cultivation under high glucose conditions combined with endothelin-1 and cortisol stimulation can be utilised to induce diabetic hypertrophy in hiPSC-CMs associated with the accumulation and peroxidation of lipids, altered calcium handling, and loss of sarcomere integrity [209,210,211].
Novel approaches including patient-derived materials, such as endothelial cell-derived microvesicles (EMVs) from obese/hypertensive patients can be used to induce hypertrophy with increased levels of hypertrophic markers, such as cTnT, α-actinin and NF-kB, and fibrosis marker TGF-β in hiPSC-CMs [212]. HiPSCs from patients with genetic mutations (i.e., c.478_480del and p.Δ160E) in TNNT2 or hypertrophy-related diastolic dysfunction are also accessible for testing drug responses and for novel drug developments due to their elevated diastolic intracellular calcium levels and altered calcium handling [212,213].
Cardiac organoids serve as promising models to study hypertrophic changes in a mixed 3D culture, even if hiPSC-CM-based models are still lacking. Hypertrophic effects of bisphenol A (BPA) and bisphenol AF (BPAF) were studied in cardiac organoids, composed of hiPSC-CMs, human primary cardiac fibroblasts, and human endothelial cells. Results showed that contraction and calcium transient amplitudes were decreased and the level of proBNP increased [214]. Therefore, data about fibrosis in hypertrophy are still missing in both 2D and 3D models. Taken together, the good response of hiPSC-CMs to hypertrophy-inducing chemical or physical approaches makes them a valuable tool for drug development in hypertrophy-related cardiac diseases. Furthermore, a successful hiPSC-CM maturation strategy for adult phenotypes might provide a more suitable basis for hypertrophy studies.
7.3. Channelopathies
The shape of the APs in CMs is defined by the balance of inward and outward currents flowing through sodium, potassium, and calcium channels [215]. The disruption of functionality in any of these channels, or channelopathies, may lead to arrhythmias or other pathological states [216]. The most common cause of channelopathies is mutations in genes encoding ion channels [216]. The mutation may cause subtle and multiple alterations in ion channel activity, including modulated channel conductance, shifted voltage dependence of activation, or altered channel gating kinetics [215]. This is where the hiPSC technology has provided significant progress by revealing the mechanisms of channelopathies. Electrophysiological investigation in patient-derived hiPSC-CMs enables the determination of patient-specific biophysical consequences of a channel mutation [215,217]. Moreover, it provides the possibility of finding personalised and safe treatment strategies [33].
LQTS is considered one of the most common types of cardiac channelopathies [216]. The delayed repolarisation in LQTS can be caused by different reasons. For instance, LQTS subtype 1 is caused by the mutation in the KCNQ1 channel [216,218], while LQTS subtype 2 is caused by the mutations in the KCNH2 channel, resulting in reduced potassium currents through the respective channel [219]. Brugada syndrome is a rare inherited cardiac arrhythmia, associated with mutations in a number of genes [220,221]. Gene mutations of sodium voltage-gated channel alpha subunit 5 (SCN5A, encoding the ion channel Nav1.5) are responsible for 30% of Brugada syndrome cases. SCN5A mutations result in a constant inward sodium current during the plateau phase of the AP and a prolonged QT interval [222]. Whereas, another mutation associated with Brugada syndrome in plakophilin-2 results in deficits of sodium current [223].
HiPSC-CMs can be implemented for modelling a number of other cardiac channelopathies, including SQTS [224], Timothy syndrome [225], catecholamine-induced polymorphic ventricular tachycardia [226], or channelopathy of small- and intermediate-conductance calcium-activated potassium channels [195,227]. For a more detailed review of the correlation of calcium handling defects and channelopathies, see our previous review by Kistamás et al. [228]. The phenotypic outcomes during the modelling of the cardiac disease may be influenced not just by presumed causative mutations, but the differences in the genetic background as well. The precise introduction of desired mutations in a known hiPSC line can be performed by CRISPR-Cas9 techniques. This approach was used recently to model LQTS and SQTS phenotypes by introducing a specific mutation of KCNH2 in isogenic hiPSC-derived cardiac tissues [67].
One of the main shortcomings in the use of hiPSC-CMs for modelling channelopathies is their immature phenotype. The hiPSC-CMs demonstrate spontaneous activity with a depolarised membrane, low maximal upstroke velocity, and highly variable action potential duration [229]. Also, the expression of the inward rectifier potassium current in hiPSC-CMs is almost negligible in the foetal phenotype [229,230]; however, there are promising studies showing an increased density of this current upon metabolic maturation medium [231]. Therefore, the shape of the AP and the profile of the individual membrane currents contributing to it substantially differ from those observed in adult human CMs. While there were some attempts to facilitate the expression of certain ion channels by adenoviral techniques [232] or enhance them with dynamic clamp [233], the new techniques for enhanced hiPSC-CM maturation will open new possibilities for the investigation of the mechanisms behind channelopathies and develop novel treatment strategies.
7.4. Ischaemia/Hypoxia
A high number of CVD patients experience chronic or acute ischaemia. Chronic ischaemia is caused mostly by coronary artery disease, leading to chronic heart failure and ischaemic cardiomyopathy [234]. Acute myocardial ischaemia is one of the causes of sudden cardiac death, caused by the obstruction of coronary vessels, leading to the lack of oxygen and nutrients and the accumulation of waste products in the myocardium. Ischaemia disrupts cellular metabolism and ion currents leading to lethal arrhythmias [235]. Under ischaemic conditions, metabolism in CMs switches to anaerobic, and the generation of lactic acid is followed by the development of acidosis, which in turn leads to the reduction in ATP availability. If ischaemia persists, bradykinin, histamine, and ROS are released, disrupting CM membranes and leading to changes in electrophysiology [236].
Cardiac ischemia models in hiPSC-CMs can be induced by the application of low oxygen concentration (≤2%) in the absence of glucose and serum. The main responses to ischaemic conditions consist of reduced cell viability, contractility (reduced beating frequency, increased depolarisation time, and field potential propagation), and reduced sarcomere coverage and nuclear size, representing myocardium damage and arrhythmias in vivo [237,238,239]. Exposure to 2% oxygen concentration reduces the viability of hiPSC-CMs, which can be easily indicated by staining of the nuclei with propidium iodide [237]. Modelling acute ischemia (0–1% oxygen and glucose-free) can be combined with reperfusion (recovery in normoxia conditions of 19–20% oxygen, serum-free/glucose-free, or full media), together with hyperkalaemia and acidosis [240,241]. It causes cell apoptosis at the reoxygenation phase corresponding to the injury in the myocardium of patients when circulation is restored following a heart attack or cardiopulmonary bypass surgery [241,242]. The advantage of the application of hiPSC-CMs over the proliferative CM cell lines (e.g., AC16, HL-1, and H9C2) was shown in simulated ischaemia–reperfusion and hypertrophic settings [243]. Ischaemia simulation in hiPSC-CM cultures using 1% oxygen resulted in arrhythmias, which were determined by the analysis of the parameters of calcium transients, such as beat rate, diastolic calcium levels and calcium transient amplitude, irregular phases, double or multiple peaks, prolonged rise, plateau abnormality, and low or high peaks [239]. Whereas, 0% hypoxia-induced electrophysiological changes, such as decreased beating frequency and field potential amplitude, could be reverted during the reoxygenation phase. The reversion of hypoxia-induced morphological changes, such as disruption of the distinct sarcomere structure or a decrease in the nucleus area was not determined [244]. Metabolic purification can also be used for the induction of ischaemia as it increases susceptibility to hypoxia by inhibiting mitochondrial respiration, which can lead to cell death [245,246]. Metabolic purification was shown to reduce sarcoendoplasmic reticulum calcium-ATPase (SERCA) expression, as cells were depolarised and had lower mitochondrial membrane potential [245]. In summary, by reducing the concentration of oxygen in cell cultures and thus recapitulating the microenvironment of ischaemic diseases, it is possible to reproduce acute or chronic changes in cardiac models using hiPSC-CM models, such as alterations in cell viability, electrophysiology, calcium handling, or to study reperfusion-induced injuries after the reversal from hypoxic to physiological oxygen levels in the culture environment.
8. Conclusions and Future Perspectives
We conclude that with the advent of hiPSC-CM technology and tissue engineering models, configurations resembling native tissues may be the optimal direction in drug screening and disease modelling. However, significant limitations persist in the field that first need to be addressed to make it a reliable tool. The most prominent drawback of these cells and models is their insufficient maturity status. Nonetheless, numerous studies demonstrated that these human cells are superior to most animal models, and they can offer several advantages over human primary cells. Importantly, employing these cells is expected to enhance adherence to the principles of 3Rs (Replacement, Reduction, and Refinement) in the use of animals in research. The continuously growing data pool collected from hiPSC-CMs serves as a ground for extensive in silico modelling, which can identify particular patterns and biomarkers associated with CVDs and can also predict drug-induced alterations. On top of that, computational simulations and models might promote diagnostic purposes, either by earlier diagnosis or by easier and more accurate follow-up of patients.
Besides the human origin, the pursuit to establish a pure cell culture has two sides. First, a pure culture having only the desired cell type is a major goal in many studies, but there are studies showing that without co-culturing with other cell types, genuine maturation cannot be achieved. Ongoing clinical trials show encouraging results with pure cultures as a great option for treating heart muscle injuries during myocardial infarction in vivo. In vitro, however, adding supporting cell types to have a mixed population appears to be beneficial for CM maturity. Connecting several organ-on-a-chip platforms to establish a multi-organ-on-a-chip, also known as a human-on-a-chip platform, could allow us to assess not just hidden cardiotoxicity, but to identify effects of complex toxic environments, such as air pollution, on different artificial mini organs. Although hiPSC-CM sources are theoretically unlimited, there are still many ongoing attempts at their large-scale expansion for clinical and in vitro applications. In terms of financial aspects, it is key to establish ready-to-use robust, reproducible, and cost-effective protocols in industrial and medical quality and quantity production.
The demand is high for human disease models as in the case of atrial fibrillation. The key limitations are that there are no such gold standard protocols that would be ideal for all hiPSC lines, the low maturity status, and the imperfections in gene editing. The main limitation of personalised medicine solutions is the limited understanding of the underlying patient-specific mutation linked to the disease. Even if major advancements have been achieved with genome-editing techniques (e.g., CRISPR-Cas9), it is still not straightforward to investigate the phenotype associated with specific mutations. Notably, there is a pivotal need for isogenic controls. However, progress has been made and several works showed that including isogenic and healthy controls (not carrying the mutation) will be extremely relevant for personalised medicine. Next-generation sequencing (NGS) technologies have enabled fast and affordable gene-based diagnostics, but an important challenge persists due to the limited comprehension of genetic variants in detected disease-associated genes. Enhancing our understanding of the molecular pathogenesis of genetic heart diseases can drive efforts to develop novel therapeutic agents. Overall, hiPSC-CMs are considered to revolutionise the field, based on their limitless availability, and on the fact that genomic and proteomic analysis in a human context, high throughput screenings, novel drug discovery, and disease modelling open new horizons. They also provide vital information to improve and validate in silico models. To fulfil this promise, however, we first need to overcome the current roadblocks, including immaturity, homogeneity, and the lack of relevant 3D architectures and complex structures.
In summary, the constant development of hiPSC-CMs and protocols carries immense potential in drug testing and drug discovery, disease modelling, and in regenerative therapies. From bench to bedside, clinical-scale production for regenerative medicine is crucial, while solving risks, such as tumorigenicity of undifferentiated iPSCs or proarrhythmic activities of spontaneously active hiPSC-CMs, are equally important. Current trends show that the purity of hiPSC-CMs is key in vivo, however, drug screening and development requires mature hiPSC-CMs in vitro. Creating more complex systems (e.g., 3D cultures instead of 2D, bioreactors, and EHTs) may help to understand their physiology better, achieve greater maturation, and ultimately, connect basic research with clinical studies to completely exploit the translational potential in the field.
Author Contributions
K.K.: Conceptualisation, Methodology, Formal analysis, Data Curation, Writing—Original Draft, Writing—Review and Editing, Visualisation, and Supervision. F.L. (Federica Lamberto): Investigation and Writing—Original Draft. R.V.: Writing—Original Draft. F.L. (Filipa Leal): Writing—Original Draft. S.M.: Investigation. L.M.: Writing—Original Draft. P.S.-B.: Writing—Original Draft. A.A.: Writing—Original Draft. D.N.A.: Writing—Original Draft. M.Z.: Conceptualisation. P.F.C.: Writing—Original Draft and Supervision. E.B.: Writing—Original Draft and Supervision. C.B.: Writing—Original Draft, Funding acquisition, and Supervision. A.D.: Writing—Review and Editing, Supervision, and Funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the EU Horizon 2020 under Grant agreements No. 953138 (EMAPS-Cardio), No. 812660 (DohART-NET Marie Skłodowska-Curie Programme), the Hungarian Grant No. 2020-1.1.5-GYORSÍTÓSÁV-2021-00016 from the National Research, Development and Innovation Fund, and the Hungarian Ministry for National Economy for the IPCEI Med4Cure “Cardiabetter” project.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding author/s.
Conflicts of Interest
A. Dinnyés is the founder and CEO of BioTalentum Ltd. K. Kistamás, F. Lamberto, S. Muenthaisong, and M. Zana are employed by BioTalentum Ltd. P. F. Costa is the founder and CEO of Biofabics Lda. F. Leal is employed by Biofabics Lda. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Abbreviations
| AF | atrial fibrillation |
| AI | artificial intelligence |
| AP | action potential |
| APA | action potential amplitude |
| APD | action potential duration |
| BPA | bisphenol A |
| BPAF | bisphenol F |
| BPM | beats per minute |
| BR | beat rate |
| CAST | Cardiac Arrhythmia Suppression Trial |
| CF | cardiac fibroblast |
| CiPA | Comprehensive in vitro Proarrhythmia Assay |
| CL | cycle length |
| CM | cardiomyocyte |
| cTnT | cardiac troponin T |
| CVD | cardiovascular disease |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DL | deep learning |
| ECG | electrocardiogram |
| ECM | extracellular matrix |
| ECT | engineered cardiac tissues |
| EHT | engineered heart tissue |
| EMV | endothelial cell-derived microvesicle |
| ESC | embryonic stem cell |
| FDA | US Food and Drug Administration |
| FPD | field potential duration |
| GATA4 | GATA binding protein 4 |
| hESC-CM | human embryonic stem cell-derived cardiomyocyte |
| hiPSC | human induced pluripotent stem cell |
| hiPSC-CM | human induced pluripotent stem cell-derived cardiomyocyte |
| hiPSC-aCMs | human induced pluripotent stem cell-derived atrial cardiomyocyte |
| HOC | heart-on-a-chip |
| HUVEC | human umbilical vein endothelial cells |
| IDE | interdigitated electrodes |
| LDH | lactate dehydrogenase |
| LQTS | long QT syndrome |
| MCS | maximum contraction speed |
| MEA | multielectrode array |
| ML | machine learning |
| MRS | maximum relaxation speed |
| NFAT | nuclear factor of activated T-cells |
| NGS | next-generation sequencing |
| Nkx2.5 | homeobox protein Nkx2.5 |
| OAP | optical action potential |
| PKC | protein kinase C |
| RMP | resting membrane potential |
| ROS | reactive oxygen species |
| SERCA | sarcopendoplasmic reticulum calcium-ATPase |
| SK | small conductance calcium-activated potassium channel |
| SNP | single nucleotide polymorphism |
| SQTS | short QT syndrome |
| SR | sarcoplasmic reticulum |
| SWORD | Survival with oral d-sotalol Trial |
| TdP | torsade de pointes type polymorphic ventricular tachyarrhythmia |
| VEGF | vascular endothelial growth factor |
References
- Kistamás, K.; Müller, A.; Muenthaisong, S.; Lamberto, F.; Zana, M.; Dulac, M.; Leal, F.; Maziz, A.; Costa, P.; Bernotiene, E.; et al. Multifactorial approaches to enhance maturation of human iPSC-derived cardiomyocytes. J. Mol. Liq. 2023, 387, 122668. [Google Scholar] [CrossRef]
- Echt, D.S.; Liebson, P.R.; Mitchell, L.B.; Peters, R.W.; Obias-Manno, D.; Barker, A.H.; Arensberg, D.; Baker, A.; Friedman, L.; Greene, H.L.; et al. Mortality and morbidity in patients receiving encainide, flecainide, or placebo. The Cardiac Arrhythmia Suppression Trial. N. Engl. J. Med. 1991, 324, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Cardiac Arrhythmia Suppression Trial, I.I.I. Effect of the antiarrhythmic agent moricizine on survival after myocardial infarction. N. Engl. J. Med. 1992, 327, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Pratt, C.M.; Camm, A.J.; Cooper, W.; Friedman, P.L.; MacNeil, D.J.; Moulton, K.M.; Pitt, B.; Schwartz, P.J.; Veltri, E.P.; Waldo, A.L. Mortality in the Survival with ORal D-sotalol (SWORD) trial: Why did patients die? Am. J. Cardiol. 1998, 81, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Waldo, A.L.; Camm, A.J.; deRuyter, H.; Freidman, P.L.; MacNeil, D.J.; Pitt, B.; Pratt, C.M.; Rodda, B.E.; Schwartz, P.J. Survival with oral d-sotalol in patients with left ventricular dysfunction after myocardial infarction: Rationale, design, and methods (the SWORD trial). Am. J. Cardiol. 1995, 75, 1023–1027. [Google Scholar] [CrossRef] [PubMed]
- Ferdinandy, P.; Baczko, I.; Bencsik, P.; Giricz, Z.; Gorbe, A.; Pacher, P.; Varga, Z.V.; Varro, A.; Schulz, R. Definition of hidden drug cardiotoxicity: Paradigm change in cardiac safety testing and its clinical implications. Eur. Heart J. 2019, 40, 1771–1777. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- James, S.K.; Roe, M.T.; Cannon, C.P.; Cornel, J.H.; Horrow, J.; Husted, S.; Katus, H.; Morais, J.; Steg, P.G.; Storey, R.F.; et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes intended for non-invasive management: Substudy from prospective randomised PLATelet inhibition and patient Outcomes (PLATO) trial. BMJ 2011, 342, d3527. [Google Scholar] [CrossRef] [PubMed]
- Bonaca, M.P.; Creager, M.A.; Olin, J.; Scirica, B.M.; Gilchrist, I.C., Jr.; Murphy, S.A.; Goodrich, E.L.; Braunwald, E.; Morrow, D.A. Peripheral Revascularization in Patients with Peripheral Artery Disease with Vorapaxar: Insights from the TRA 2 degrees P-TIMI 50 Trial. JACC. Cardiovasc. Interv. 2016, 9, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Swedberg, K.; Komajda, M.; Bohm, M.; Borer, J.S.; Ford, I.; Dubost-Brama, A.; Lerebours, G.; Tavazzi, L.; Investigators, S. Ivabradine and outcomes in chronic heart failure (SHIFT): A randomised placebo-controlled study. Lancet 2010, 376, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Sauer, A.J.; Cole, R.; Jensen, B.C.; Pal, J.; Sharma, N.; Yehya, A.; Vader, J. Practical guidance on the use of sacubitril/valsartan for heart failure. Heart Fail. Rev. 2019, 24, 167–176. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Damy, T.; Garcia-Pavia, P.; Hanna, M.; Judge, D.P.; Merlini, G.; Gundapaneni, B.; Patterson, T.A.; Riley, S.; Schwartz, J.H.; Sultan, M.B.; et al. Efficacy and safety of tafamidis doses in the Tafamidis in Transthyretin Cardiomyopathy Clinical Trial (ATTR-ACT) and long-term extension study. Eur. J. Heart Fail. 2021, 23, 277–285. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ezekowitz, J.A.; O’Connor, C.M.; Troughton, R.W.; Alemayehu, W.G.; Westerhout, C.M.; Voors, A.A.; Butler, J.; Lam, C.S.P.; Ponikowski, P.; Emdin, M.; et al. N-Terminal Pro-B-Type Natriuretic Peptide and Clinical Outcomes: Vericiguat Heart Failure with Reduced Ejection Fraction Study. JACC. Heart Fail. 2020, 8, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Olivotto, I.; Oreziak, A.; Barriales-Villa, R.; Abraham, T.P.; Masri, A.; Garcia-Pavia, P.; Saberi, S.; Lakdawala, N.K.; Wheeler, M.T.; Owens, A.; et al. Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER-HCM): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2020, 396, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, D.L.; Szarek, M.; Steg, P.G.; Cannon, C.P.; Leiter, L.A.; McGuire, D.K.; Lewis, J.B.; Riddle, M.C.; Voors, A.A.; Metra, M.; et al. Sotagliflozin in Patients with Diabetes and Recent Worsening Heart Failure. N. Engl. J. Med. 2021, 384, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Schlaich, M.P.; Bellet, M.; Weber, M.A.; Danaietash, P.; Bakris, G.L.; Flack, J.M.; Dreier, R.F.; Sassi-Sayadi, M.; Haskell, L.P.; Narkiewicz, K.; et al. Dual endothelin antagonist aprocitentan for resistant hypertension (PRECISION): A multicentre, blinded, randomised, parallel-group, phase 3 trial. Lancet 2022, 400, 1927–1937. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, S.; Kainuma, S.; Kawamura, T.; Suzuki, K.; Ito, Y.; Iseoka, H.; Ito, E.; Takeda, M.; Sasai, M.; Mochizuki-Oda, N.; et al. Case report: Transplantation of human induced pluripotent stem cell-derived cardiomyocyte patches for ischemic cardiomyopathy. Front. Cardiovasc. Med. 2022, 9, 950829. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- A Study of iPS Cell-derived Cardiomyocyte Spheroids (HS-001) in Patients with Heart Failure (LAPiS Study); Heartseed Inc.: Tokyo, Japan, 2022. Available online: https://clinicaltrials.gov/study/NCT04945018 (accessed on 30 July 2024).
- Funakoshi, S.; Yoshida, Y. Recent progress of iPSC technology in cardiac diseases. Arch. Toxicol. 2021, 95, 3633–3650. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Lyra-Leite, D.M.; Gutierrez-Gutierrez, O.; Wang, M.; Zhou, Y.; Cyganek, L.; Burridge, P.W. A review of protocols for human iPSC culture, cardiac differentiation, subtype-specification, maturation, and direct reprogramming. STAR Protoc. 2022, 3, 101560. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Clinical Trial of Human (Allogeneic) iPS Cell-Derived Cardiomyocytes Sheet for Ischemic Cardiomyopathy. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04696328 (accessed on 30 July 2024).
- Safety and Efficacy of Induced Pluripotent Stem Cell-derived Engineered Human Myocardium as Biological Ventricular Assist Tissue in Terminal Heart Failure (BioVAT-HF). 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT04396899 (accessed on 30 July 2024).
- Treating Heart Failure with hPSC-CMs (HEAL-CHF). 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT03763136 (accessed on 30 July 2024).
- Treating Congestive HF with hiPSC-CMs through Endocardial Injection. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04982081 (accessed on 30 July 2024).
- Modeling and Pharmacological Targeting of Genetic Cardiomyopathy in Children via Cardiomyocytes Derived from Induced Pluripotent Stem Cells (DMDstem). 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT03696628 (accessed on 30 July 2024).
- Allogeneic iPSC-Derived Cardiomyocyte Therapy in Patients with Worsening Ischemic Heart Failure. 2022. Available online: https://clinicaltrials.gov/ct2/show/NCT05566600 (accessed on 30 July 2024).
- Vreeker, A.; van Stuijvenberg, L.; Hund, T.J.; Mohler, P.J.; Nikkels, P.G.; van Veen, T.A. Assembly of the cardiac intercalated disk during pre- and postnatal development of the human heart. PLoS ONE 2014, 9, e94722. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shiba, Y.; Fernandes, S.; Zhu, W.Z.; Filice, D.; Muskheli, V.; Kim, J.; Palpant, N.J.; Gantz, J.; Moyes, K.W.; Reinecke, H.; et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature 2012, 489, 322–325. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shiba, Y.; Gomibuchi, T.; Seto, T.; Wada, Y.; Ichimura, H.; Tanaka, Y.; Ogasawara, T.; Okada, K.; Shiba, N.; Sakamoto, K.; et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 2016, 538, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.J.; Yang, X.; Don, C.W.; Minami, E.; Liu, Y.W.; Weyers, J.J.; Mahoney, W.M.; Van Biber, B.; Cook, S.M.; Palpant, N.J.; et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 2014, 510, 273–277. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rodgers, J.L.; Jones, J.; Bolleddu, S.I.; Vanthenapalli, S.; Rodgers, L.E.; Shah, K.; Karia, K.; Panguluri, S.K. Cardiovascular Risks Associated with Gender and Aging. J. Cardiovasc. Dev. Dis. 2019, 6, 19. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Elkhoury, K.; Kodeih, S.; Enciso-Martinez, E.; Maziz, A.; Bergaud, C. Advancing Cardiomyocyte Maturation: Current Strategies and Promising Conductive Polymer-Based Approaches. Adv. Healthc. Mater. 2024, 13, e2303288. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lami, B.; Ikonomou, L.; Gu, M. Unlocking the potential of induced pluripotent stem cells for neonatal disease modeling and drug development. Semin. Perinatol. 2023, 47, 151729. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lamberto, F.; Shashikadze, B.; Elkhateib, R.; Lombardo, S.D.; Horanszky, A.; Balogh, A.; Kistamas, K.; Zana, M.; Menche, J.; Frohlich, T.; et al. Low-dose Bisphenol A exposure alters the functionality and cellular environment in a human cardiomyocyte model. Environ. Pollut. 2023, 335, 122359. [Google Scholar] [CrossRef] [PubMed]
- Lamberto, F.; Peral-Sanchez, I.; Muenthaisong, S.; Zana, M.; Willaime-Morawek, S.; Dinnyes, A. Environmental Alterations during Embryonic Development: Studying the Impact of Stressors on Pluripotent Stem Cell-Derived Cardiomyocytes. Genes 2021, 12, 1564. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Burridge, P.W.; Keller, G.; Gold, J.D.; Wu, J.C. Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 2012, 10, 16–28. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lin, Y.; Zou, J. Differentiation of Cardiomyocytes from Human Pluripotent Stem Cells in Fully Chemically Defined Conditions. STAR Protoc. 2020, 1, 100015. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cofino-Fabres, C.; Passier, R.; Schwach, V. Towards Improved Human In Vitro Models for Cardiac Arrhythmia: Disease Mechanisms, Treatment, and Models of Atrial Fibrillation. Biomedicines 2023, 11, 2355. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Veerman, C.C.; Kosmidis, G.; Mummery, C.L.; Casini, S.; Verkerk, A.O.; Bellin, M. Immaturity of human stem-cell-derived cardiomyocytes in culture: Fatal flaw or soluble problem? Stem Cells Dev. 2015, 24, 1035–1052. [Google Scholar] [CrossRef] [PubMed]
- Souidi, M.; Sleiman, Y.; Acimovic, I.; Pribyl, J.; Charrabi, A.; Baecker, V.; Scheuermann, V.; Pesl, M.; Jelinkova, S.; Skladal, P.; et al. Oxygen Is an Ambivalent Factor for the Differentiation of Human Pluripotent Stem Cells in Cardiac 2D Monolayer and 3D Cardiac Spheroids. Int. J. Mol. Sci. 2021, 22, 662. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leitolis, A.; Robert, A.W.; Pereira, I.T.; Correa, A.; Stimamiglio, M.A. Cardiomyogenesis Modeling Using Pluripotent Stem Cells: The Role of Microenvironmental Signaling. Front. Cell Dev. Biol. 2019, 7, 164. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, J.; Klos, M.; Wilson, G.F.; Herman, A.M.; Lian, X.; Raval, K.K.; Barron, M.R.; Hou, L.; Soerens, A.G.; Yu, J.; et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: The matrix sandwich method. Circ. Res. 2012, 111, 1125–1136. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, J.; Wilson, G.F.; Soerens, A.G.; Koonce, C.H.; Yu, J.; Palecek, S.P.; Thomson, J.A.; Kamp, T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009, 104, e30–e41. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kalkunte, N.G.; Delambre, T.E.; Sohn, S.; Pickett, M.; Parekh, S.; Zoldan, J. Engineering Alignment Has Mixed Effects on Human Induced Pluripotent Stem Cell Differentiated Cardiomyocyte Maturation. Tissue Engineering. Part A 2023, 29, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Garcia, N.A.; Ontoria-Oviedo, I.; Gonzalez-King, H.; Diez-Juan, A.; Sepulveda, P. Glucose Starvation in Cardiomyocytes Enhances Exosome Secretion and Promotes Angiogenesis in Endothelial Cells. PLoS ONE 2015, 10, e0138849. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rupert, C.E.; Irofuala, C.; Coulombe, K.L.K. Practical adoption of state-of-the-art hiPSC-cardiomyocyte differentiation techniques. PLoS ONE 2020, 15, e0230001. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Takaki, T.; Inagaki, A.; Chonabayashi, K.; Inoue, K.; Miki, K.; Ohno, S.; Makiyama, T.; Horie, M.; Yoshida, Y. Optical Recording of Action Potentials in Human Induced Pluripotent Stem Cell-Derived Cardiac Single Cells and Monolayers Generated from Long QT Syndrome Type 1 Patients. Stem Cells Int. 2019, 2019, 7532657. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Giacomelli, E.; Meraviglia, V.; Campostrini, G.; Cochrane, A.; Cao, X.; van Helden, R.W.J.; Krotenberg Garcia, A.; Mircea, M.; Kostidis, S.; Davis, R.P.; et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 2020, 26, 862–879.e11. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Reilly, L.; Munawar, S.; Zhang, J.; Crone, W.C.; Eckhardt, L.L. Challenges and innovation: Disease modeling using human-induced pluripotent stem cell-derived cardiomyocytes. Front. Cardiovasc. Med. 2022, 9, 966094. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Parrotta, E.I.; Lucchino, V.; Scaramuzzino, L.; Scalise, S.; Cuda, G. Modeling Cardiac Disease Mechanisms Using Induced Pluripotent Stem Cell-Derived Cardiomyocytes: Progress, Promises and Challenges. Int. J. Mol. Sci. 2020, 21, 4354. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kleinsorge, M.; Cyganek, L. Subtype-Directed Differentiation of Human iPSCs into Atrial and Ventricular Cardiomyocytes. STAR Protoc. 2020, 1, 100026. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vuckovic, S.; Dinani, R.; Nollet, E.E.; Kuster, D.W.D.; Buikema, J.W.; Houtkooper, R.H.; Nabben, M.; van der Velden, J.; Goversen, B. Characterization of cardiac metabolism in iPSC-derived cardiomyocytes: Lessons from maturation and disease modeling. Stem Cell Res. Ther. 2022, 13, 332. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, J.; Laksman, Z.; Backx, P.H. The electrophysiological development of cardiomyocytes. Adv. Drug Deliv. Rev. 2016, 96, 253–273. [Google Scholar] [CrossRef] [PubMed]
- Chirikian, O.; Goodyer, W.R.; Dzilic, E.; Serpooshan, V.; Buikema, J.W.; McKeithan, W.; Wu, H.; Li, G.; Lee, S.; Merk, M.; et al. CRISPR/Cas9-based targeting of fluorescent reporters to human iPSCs to isolate atrial and ventricular-specific cardiomyocytes. Sci. Rep. 2021, 11, 3026. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mehta, A.; Ramachandra, C.J.A.; Singh, P.; Chitre, A.; Lua, C.H.; Mura, M.; Crotti, L.; Wong, P.; Schwartz, P.J.; Gnecchi, M.; et al. Identification of a targeted and testable antiarrhythmic therapy for long-QT syndrome type 2 using a patient-specific cellular model. Eur. Heart J. 2018, 39, 1446–1455. [Google Scholar] [CrossRef] [PubMed]
- James, E.C.; Tomaskovic-Crook, E.; Crook, J.M. Bioengineering Clinically Relevant Cardiomyocytes and Cardiac Tissues from Pluripotent Stem Cells. Int. J. Mol. Sci. 2021, 22, 3005. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Knollmann, B.C. Induced pluripotent stem cell-derived cardiomyocytes: Boutique science or valuable arrhythmia model? Circ. Res. 2013, 112, 969–976; discussion 976. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cyganek, L.; Tiburcy, M.; Sekeres, K.; Gerstenberg, K.; Bohnenberger, H.; Lenz, C.; Henze, S.; Stauske, M.; Salinas, G.; Zimmermann, W.H.; et al. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight 2018, 3, e99941. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhu, K.; Bao, X.; Wang, Y.; Lu, T.; Zhang, L. Human induced pluripotent stem cell (hiPSC)-derived cardiomyocyte modelling of cardiovascular diseases for natural compound discovery. Biomed. Pharmacother. 2023, 157, 113970. [Google Scholar] [CrossRef] [PubMed]
- Hanses, U.; Kleinsorge, M.; Roos, L.; Yigit, G.; Li, Y.; Barbarics, B.; El-Battrawy, I.; Lan, H.; Tiburcy, M.; Hindmarsh, R.; et al. Intronic CRISPR Repair in a Preclinical Model of Noonan Syndrome-Associated Cardiomyopathy. Circulation 2020, 142, 1059–1076. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Zhang, J.Z.; Itzhaki, I.; Zhang, S.L.; Chen, H.; Haddad, F.; Kitani, T.; Wilson, K.D.; Tian, L.; Shrestha, R.; et al. Determining the Pathogenicity of a Genomic Variant of Uncertain Significance Using CRISPR/Cas9 and Human-Induced Pluripotent Stem Cells. Circulation 2018, 138, 2666–2681. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kapplinger, J.D.; Giudicessi, J.R.; Ye, D.; Tester, D.J.; Callis, T.E.; Valdivia, C.R.; Makielski, J.C.; Wilde, A.A.; Ackerman, M.J. Enhanced Classification of Brugada Syndrome-Associated and Long-QT Syndrome-Associated Genetic Variants in the SCN5A-Encoded Na(v)1.5 Cardiac Sodium Channel. Circ. Cardiovasc. Genet. 2015, 8, 582–595. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hancox, J.C.; Stuart, A.G.; Harmer, S.C. Functional evaluation of gene mutations in Long QT Syndrome: Strength of evidence from in vitro assays for deciphering variants of uncertain significance. J. Congenit. Cardiol. 2020, 4, 6. [Google Scholar] [CrossRef]
- Kamga, M.V.K.; Reppel, M.; Hescheler, J.; Nguemo, F. Modeling genetic cardiac channelopathies using induced pluripotent stem cells—Status quo from an electrophysiological perspective. Biochem. Pharmacol. 2021, 192, 114746. [Google Scholar] [CrossRef] [PubMed]
- Altomare, C.; Pianezzi, E.; Cervio, E.; Bolis, S.; Biemmi, V.; Benzoni, P.; Camici, G.G.; Moccetti, T.; Barile, L.; Vassalli, G. Human-induced pluripotent stem cell-derived cardiomyocytes from cardiac progenitor cells: Effects of selective ion channel blockade. EP Eur. 2016, 18, iv67–iv76. [Google Scholar] [CrossRef] [PubMed]
- Maurissen, T.L.; Kawatou, M.; Lopez-Davila, V.; Minatoya, K.; Yamashita, J.K.; Woltjen, K. Modeling mutation-specific arrhythmogenic phenotypes in isogenic human iPSC-derived cardiac tissues. Sci. Rep. 2024, 14, 2586. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mamoshina, P.; Rodriguez, B.; Bueno-Orovio, A. Toward a broader view of mechanisms of drug cardiotoxicity. Cell Rep. Med. 2021, 2, 100216. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mulla, W.; Murninkas, M.; Levi, O.; Etzion, Y. Incorrectly corrected? QT interval analysis in rats and mice. Front. Physiol. 2022, 13, 1002203. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chang, Y.; Li, Y.N.; Bai, R.; Wu, F.; Ma, S.; Saleem, A.; Zhang, S.; Jiang, Y.; Dong, T.; Guo, T.; et al. hERG-deficient human embryonic stem cell-derived cardiomyocytes for modelling QT prolongation. Stem Cell Res. Ther. 2021, 12, 278. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vargas, H.M.; Bass, A.S.; Koerner, J.; Matis-Mitchell, S.; Pugsley, M.K.; Skinner, M.; Burnham, M.; Bridgland-Taylor, M.; Pettit, S.; Valentin, J.P. Evaluation of drug-induced QT interval prolongation in animal and human studies: A literature review of concordance. Br. J. Pharmacol. 2015, 172, 4002–4011. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Colatsky, T.; Fermini, B.; Gintant, G.; Pierson, J.B.; Sager, P.; Sekino, Y.; Strauss, D.G.; Stockbridge, N. The Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative—Update on progress. J. Pharmacol. Toxicol. Methods 2016, 81, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.; Zusterzeel, R.; Johannesen, L.; Ochoa-Jimenez, R.; Mason, J.W.; Sanabria, C.; Kemp, S.; Sager, P.T.; Patel, V.; Matta, M.K.; et al. Assessment of Multi-Ion Channel Block in a Phase I Randomized Study Design: Results of the CiPA Phase I ECG Biomarker Validation Study. Clin. Pharmacol. Ther. 2019, 105, 943–953. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Blinova, K.; Dang, Q.; Millard, D.; Smith, G.; Pierson, J.; Guo, L.; Brock, M.; Lu, H.R.; Kraushaar, U.; Zeng, H.; et al. International Multisite Study of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Proarrhythmic Potential Assessment. Cell Rep. 2018, 24, 3582–3592. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, Z.; Ridder, B.J.; Han, X.; Wu, W.W.; Sheng, J.; Tran, P.N.; Wu, M.; Randolph, A.; Johnstone, R.H.; Mirams, G.R.; et al. Assessment of an In Silico Mechanistic Model for Proarrhythmia Risk Prediction under the CiPA Initiative. Clin. Pharmacol. Ther. 2019, 105, 466–475. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Crumb, W.J., Jr.; Vicente, J.; Johannesen, L.; Strauss, D.G. An evaluation of 30 clinical drugs against the comprehensive in vitro proarrhythmia assay (CiPA) proposed ion channel panel. J. Pharmacol. Toxicol. Methods 2016, 81, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Rafatian, N.; Wang, E.Y.; Wu, Q.; Lai, B.F.L.; Lu, R.X.; Savoji, H.; Radisic, M. Towards chamber specific heart-on-a-chip for drug testing applications. Adv. Drug Deliv. Rev. 2020, 165–166, 60–76. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pognan, F.; Beilmann, M.; Boonen, H.C.M.; Czich, A.; Dear, G.; Hewitt, P.; Mow, T.; Oinonen, T.; Roth, A.; Steger-Hartmann, T.; et al. The evolving role of investigative toxicology in the pharmaceutical industry. Nat. Rev. Drug Discov. 2023, 22, 317–335. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abulaiti, M.; Yalikun, Y.; Murata, K.; Sato, A.; Sami, M.M.; Sasaki, Y.; Fujiwara, Y.; Minatoya, K.; Shiba, Y.; Tanaka, Y.; et al. Establishment of a heart-on-a-chip microdevice based on human iPS cells for the evaluation of human heart tissue function. Sci. Rep. 2020, 10, 19201. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Xia, S.; Li, Y.; Qi, Y.; Wang, Y.; Zhang, D.; Hua, Y.; Luo, S. Application of hiPSC as a Drug Tester Via Mimicking a Personalized Mini Heart. Front. Genet. 2022, 13, 891159. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pang, L. Toxicity testing in the era of induced pluripotent stem cells: A perspective regarding the use of patient-specific induced pluripotent stem cell–derived cardiomyocytes for cardiac safety evaluation. Curr. Opin. Toxicol. 2020, 23–24, 50–55. [Google Scholar] [CrossRef]
- Mathur, A.; Loskill, P.; Shao, K.; Huebsch, N.; Hong, S.; Marcus, S.G.; Marks, N.; Mandegar, M.; Conklin, B.R.; Lee, L.P.; et al. Human iPSC-based Cardiac Microphysiological System For Drug Screening Applications. Sci. Rep. 2015, 5, 8883. [Google Scholar] [CrossRef]
- Tisdale, J.E.; Chung, M.K.; Campbell, K.B.; Hammadah, M.; Joglar, J.A.; Leclerc, J.; Rajagopalan, B.; American Heart Association Clinical Pharmacology Committee of the Council on Clinical Cardiology and Council on Cardiovascular and Stroke Nursing. Drug-Induced Arrhythmias: A Scientific Statement from the American Heart Association. Circulation 2020, 142, e214–e233. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, M.L.; Pettersson, M.; Meyboom, R.H.; Hoes, A.W.; Leufkens, H.G. Anti-HERG activity and the risk of drug-induced arrhythmias and sudden death. Eur. Heart J. 2005, 26, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.V.; Ferdinandy, P.; Liaudet, L.; Pacher, P. Drug-induced mitochondrial dysfunction and cardiotoxicity. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1453–H1467. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, A.; Gao, M.; Liu, B.; Qin, Y.; Chen, L.; Liu, H.; Wu, H.; Gong, G. Mitochondrial autophagy: Molecular mechanisms and implications for cardiovascular disease. Cell Death Dis. 2022, 13, 444. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Russo, M.; Della Sala, A.; Tocchetti, C.G.; Porporato, P.E.; Ghigo, A. Metabolic Aspects of Anthracycline Cardiotoxicity. Curr. Treat. Options Oncol. 2021, 22, 18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- L’Ecuyer, T.; Sanjeev, S.; Thomas, R.; Novak, R.; Das, L.; Campbell, W.; Heide, R.V. DNA damage is an early event in doxorubicin-induced cardiac myocyte death. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H1273–H1280. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yang, Y.; Chen, T.; Wang, Y.; Guo, T.; Liu, Y.; Li, H.; Yang, L. Cell death regulation in myocardial toxicity induced by antineoplastic drugs. Front. Cell Dev. Biol. 2023, 11, 1075917. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Doherty, K.R.; Talbert, D.R.; Trusk, P.B.; Moran, D.M.; Shell, S.A.; Bacus, S. Structural and functional screening in human induced-pluripotent stem cell-derived cardiomyocytes accurately identifies cardiotoxicity of multiple drug types. Toxicol. Appl. Pharmacol. 2015, 285, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; McKeithan, W.L.; Serrano, R.; Kitani, T.; Burridge, P.W.; Del Alamo, J.C.; Mercola, M.; Wu, J.C. Use of human induced pluripotent stem cell-derived cardiomyocytes to assess drug cardiotoxicity. Nat. Protoc. 2018, 13, 3018–3041. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shi, M.; Tien, N.T.; de Haan, L.; Louisse, J.; Rietjens, I.; Bouwmeester, H. Evaluation of in vitro models of stem cell-derived cardiomyocytes to screen for potential cardiotoxicity of chemicals. Toxicol. Vitr. Int. J. Publ. Assoc. BIBRA 2020, 67, 104891. [Google Scholar] [CrossRef] [PubMed]
- Draghici, A.E.; Taylor, J.A. The physiological basis and measurement of heart rate variability in humans. J. Physiol. Anthropol. 2016, 35, 22. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pan, D.; Li, B.; Wang, S. Establishment and validation of a torsade de pointes prediction model based on human iPSC-derived cardiomyocytes. Exp. Ther. Med. 2023, 25, 61. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Faulkner-Jones, A.; Zamora, V.; Hortigon-Vinagre, M.P.; Wang, W.; Ardron, M.; Smith, G.L.; Shu, W. A Bioprinted Heart-on-a-Chip with Human Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Evaluation. Bioengineering 2022, 9, 32. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, Z.; Li, X.; El-Battrawy, I.; Lan, H.; Zhong, R.; Xu, Q.; Huang, M.; Liao, Z.; Lang, S.; Zimmermann, W.H.; et al. Drug Testing in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes from a Patient with Short QT Syndrome Type 1. Clin. Pharmacol. Ther. 2019, 106, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Horvath, B.; Kiss, D.; Dienes, C.; Hezso, T.; Kovacs, Z.; Szentandrassy, N.; Almassy, J.; Magyar, J.; Banyasz, T.; Nanasi, P.P. Ion current profiles in canine ventricular myocytes obtained by the “onion peeling” technique. J. Mol. Cell. Cardiol. 2021, 158, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Szentandrassy, N.; Kistamas, K.; Hegyi, B.; Horvath, B.; Ruzsnavszky, F.; Vaczi, K.; Magyar, J.; Banyasz, T.; Varro, A.; Nanasi, P.P. Contribution of ion currents to beat-to-beat variability of action potential duration in canine ventricular myocytes. Pflug. Arch. Eur. J. Physiol. 2015, 467, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.L.; Zlochiver, V.; Conrad, D.B.; Vaidyanathan, R.; Valiquette, A.M.; Joshi-Mukherjee, R. A Multiwell Cardiac muGMEA Platform for Action Potential Recordings from Human iPSC-Derived Cardiomyocyte Constructs. Stem Cell Rep. 2018, 11, 522–536. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Honda, Y.; Li, J.; Hino, A.; Tsujimoto, S.; Lee, J.K. High-Throughput Drug Screening System Based on Human Induced Pluripotent Stem Cell-Derived Atrial Myocytes approximately A Novel Platform to Detect Cardiac Toxicity for Atrial Arrhythmias. Front. Pharmacol. 2021, 12, 680618. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Eisner, D.A.; Caldwell, J.L.; Kistamas, K.; Trafford, A.W. Calcium and Excitation-Contraction Coupling in the Heart. Circ. Res. 2017, 121, 181–195. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sankaranarayanan, R.; Kistamas, K.; Greensmith, D.J.; Venetucci, L.A.; Eisner, D.A. Systolic [Ca2+]i regulates diastolic levels in rat ventricular myocytes. J. Physiol. 2017, 595, 5545–5555. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kistamas, K.; Szentandrassy, N.; Hegyi, B.; Vaczi, K.; Ruzsnavszky, F.; Horvath, B.; Banyasz, T.; Nanasi, P.P.; Magyar, J. Changes in intracellular calcium concentration influence beat-to-beat variability of action potential duration in canine ventricular myocytes. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2015, 66, 73–81. [Google Scholar] [PubMed]
- Watanabe, H.; Honda, Y.; Deguchi, J.; Yamada, T.; Bando, K. Usefulness of cardiotoxicity assessment using calcium transient in human induced pluripotent stem cell-derived cardiomyocytes. J. Toxicol. Sci. 2017, 42, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Tadano, K.; Miyagawa, S.; Takeda, M.; Tsukamoto, Y.; Kazusa, K.; Takamatsu, K.; Akashi, M.; Sawa, Y. Cardiotoxicity assessment using 3D vascularized cardiac tissue consisting of human iPSC-derived cardiomyocytes and fibroblasts. Mol. Therapy. Methods Clin. Dev. 2021, 22, 338–349. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, R.; Li, D.; Sun, F.; Rampoldi, A.; Maxwell, J.T.; Wu, R.; Fischbach, P.; Castellino, S.M.; Du, Y.; Fu, H.; et al. Melphalan induces cardiotoxicity through oxidative stress in cardiomyocytes derived from human induced pluripotent stem cells. Stem Cell Res. Ther. 2020, 11, 470. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stein, J.M.; Mummery, C.L.; Bellin, M. Engineered models of the human heart: Directions and challenges. Stem Cell Rep. 2021, 16, 2049–2057. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kitsara, M.; Agbulut, O.; Kontziampasis, D.; Chen, Y.; Menasche, P. Fibers for hearts: A critical review on electrospinning for cardiac tissue engineering. Acta Biomater. 2017, 48, 20–40. [Google Scholar] [CrossRef] [PubMed]
- Campostrini, G.; Windt, L.M.; van Meer, B.J.; Bellin, M.; Mummery, C.L. Cardiac Tissues from Stem Cells: New Routes to Maturation and Cardiac Regeneration. Circ. Res. 2021, 128, 775–801. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lock, R.I.; Graney, P.L.; Tavakol, D.N.; Nash, T.R.; Kim, Y.; Sanchez, E., Jr.; Morsink, M.; Ning, D.; Chen, C.; Fleischer, S.; et al. Macrophages enhance contractile force in iPSC-derived human engineered cardiac tissue. Cell Rep. 2024, 43, 114302. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ronaldson-Bouchard, K.; Yeager, K.; Teles, D.; Chen, T.; Ma, S.; Song, L.; Morikawa, K.; Wobma, H.M.; Vasciaveo, A.; Ruiz, E.C.; et al. Engineering of human cardiac muscle electromechanically matured to an adult-like phenotype. Nat. Protoc. 2019, 14, 2781–2817. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Seguret, M.; Vermersch, E.; Jouve, C.; Hulot, J.S. Cardiac Organoids to Model and Heal Heart Failure and Cardiomyopathies. Biomedicines 2021, 9, 563. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rogozinski, N.; Yanez, A.; Bhoi, R.; Lee, M.Y.; Yang, H. Current methods for fabricating 3D cardiac engineered constructs. iScience 2022, 25, 104330. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, M.; Wu, H.; Yuan, Y.; Hu, B.; Gu, N. Recent fabrications and applications of cardiac patch in myocardial infarction treatment. VIEW 2022, 3, 20200153. [Google Scholar] [CrossRef]
- Cho, S.; Discher, D.E.; Leong, K.A.-O.; Vunjak-Novakovic, G.A.-O.; Wu, J.A.-O. Challenges and opportunities for the next generation of cardiovascular tissue engineering. Nat. Methods 2022, 19, 1064–1071. [Google Scholar] [CrossRef]
- Murata, K.; Masumoto, H. Systems for the Functional Evaluation of Human Heart Tissues Derived from Pluripotent Stem Cells. Stem Cells 2022, 40, 537–545. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, Y.; Lin, L.; Qiao, L.A.-O. Recent developments in organ-on-a-chip technology for cardiovascular disease research. Anal. Bioanal. Chem. 2023, 415, 3911–3925. [Google Scholar] [CrossRef]
- Christoffersson, J.; Meier, F.; Kempf, H.; Schwanke, K.; Coffee, M.; Beilmann, M.; Zweigerdt, R.; Mandenius, C.F. A Cardiac Cell Outgrowth Assay for Evaluating Drug Compounds Using a Cardiac Spheroid-on-a-Chip Device. Bioengineering 2018, 5, 36. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tang, Y.; Tian, F.; Miao, X.; Wu, D.; Wang, Y.; Wang, H.; You, K.; Li, Q.; Zhao, S.; Wang, W. Heart-on-a-chip using human iPSC-derived cardiomyocytes with an integrated vascular endothelial layer based on a culture patch as a potential platform for drug evaluation. Biofabrication 2022, 15, 015010. [Google Scholar] [CrossRef] [PubMed]
- Crestani, T.; Steichen, C.; Neri, E.; Rodrigues, M.; Fonseca-Alaniz, M.H.; Ormrod, B.; Holt, M.R.; Pandey, P.; Harding, S.; Ehler, E.; et al. Electrical stimulation applied during differentiation drives the hiPSC-CMs towards a mature cardiac conduction-like cells. Biochem. Biophys. Res. Commun. 2020, 533, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Rafatian, N.; Feric, N.T.; Cox, B.J.; Aschar-Sobbi, R.; Wang, E.Y.; Aggarwal, P.; Zhang, B.; Conant, G.; Ronaldson-Bouchard, K.; et al. A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 2019, 176, 913–927.e18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dou, W.; Malhi, M.; Zhao, Q.; Wang, L.; Huang, Z.; Law, J.; Liu, N.; Simmons, C.A.; Maynes, J.T.; Sun, Y. Microengineered platforms for characterizing the contractile function of in vitro cardiac models. Microsyst. Nanoeng. 2022, 8, 26. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qian, F.; Huang, C.; Lin, Y.D.; Ivanovskaya, A.N.; O’Hara, T.J.; Booth, R.H.; Creek, C.J.; Enright, H.A.; Soscia, D.A.; Belle, A.M.; et al. Simultaneous electrical recording of cardiac electrophysiology and contraction on chip. Lab Chip 2017, 17, 1732–1739. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dou, W.; Malhi, M.; Zhu, M.; Liu, H.; Plakhotnik, J.; Xu, Z.; Zhao, Q.; Chen, J.; Chen, S.; et al. Microdevice Platform for Continuous Measurement of Contractility, Beating Rate, and Beating Rhythm of Human-Induced Pluripotent Stem Cell-Cardiomyocytes inside a Controlled Incubator Environment. ACS Appl. Mater. Interfaces 2018, 10, 21173–21183. [Google Scholar] [CrossRef] [PubMed]
- Lind, J.U.; Busbee, T.A.; Valentine, A.D.; Pasqualini, F.S.; Yuan, H.; Yadid, M.; Park, S.J.; Kotikian, A.; Nesmith, A.P.; Campbell, P.H.; et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 2017, 16, 303–308. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, D.S.; Choi, Y.W.; Shanmugasundaram, A.; Jeong, Y.J.; Park, J.; Oyunbaatar, N.E.; Kim, E.S.; Choi, M.; Lee, D.W. Highly durable crack sensor integrated with silicone rubber cantilever for measuring cardiac contractility. Nat. Commun. 2020, 11, 535. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McCain, M.L.; Agarwal, A.; Nesmith, H.W.; Nesmith, A.P.; Parker, K.K. Micromolded gelatin hydrogels for extended culture of engineered cardiac tissues. Biomaterials 2014, 35, 5462–5471. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fu, F.; Shang, L.; Chen, Z.; Yu, Y.; Zhao, Y. Bioinspired living structural color hydrogels. Sci. Robot. 2018, 3, eaar8580. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chen, Z.; Xu, D.; Zhao, Y. Electroconductive and Anisotropic Structural Color Hydrogels for Visual Heart-on-a-Chip Construction. Adv. Sci. 2022, 9, e2105777. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sun, L.; Wang, Y.; Bian, F.; Xu, D.; Zhao, Y. Bioinspired optical and electrical dual-responsive heart-on-a-chip for hormone testing. Sci. Bull. 2023, 68, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, E.Y.; Lai, F.B.L.; Cheung, K.; Radisic, M. Organs-on-a-chip: A union of tissue engineering and microfabrication. Trends Biotechnol. 2023, 41, 410–424. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hodgkin, A.L.; Huxley, A.F. Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J. Physiol. 1952, 116, 449–472. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, H.; Stebbeds, W.; Francis, J.; Pointon, A.; Obrezanova, O.; Beattie, K.A.; Clements, P.; Harvey, J.S.; Smith, G.F.; Bender, A. Deriving waveform parameters from calcium transients in human iPSC-derived cardiomyocytes to predict cardiac activity with machine learning. Stem Cell Rep. 2022, 17, 556–568. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Akwaboah, A.D.; Tsevi, B.; Yamlome, P.; Treat, J.A.; Brucal-Hallare, M.; Cordeiro, J.M.; Deo, M. An in silico hiPSC-Derived Cardiomyocyte Model Built with Genetic Algorithm. Front. Physiol. 2021, 12, 675867. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mohammadzadeh, S.; Lejeune, E. SarcGraph: A Python package for analyzing the contractile behavior of pluripotent stem cell-derived cardiomyocytes. J. Open Source Softw. 2023, 8, 5322. [Google Scholar] [CrossRef]
- Grancharova, T.; Gerbin, K.A.; Rosenberg, A.B.; Roco, C.M.; Arakaki, J.E.; DeLizo, C.M.; Dinh, S.Q.; Donovan-Maiye, R.M.; Hirano, M.; Nelson, A.M.; et al. A comprehensive analysis of gene expression changes in a high replicate and open-source dataset of differentiating hiPSC-derived cardiomyocytes. Sci. Rep. 2021, 11, 15845. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, J.; Morgan, W.; Saini, A.; Liu, T.; Lough, J.; Han, L. Single-cell transcriptomic profiling reveals specific maturation signatures in human cardiomyocytes derived from LMNB2-inactivated induced pluripotent stem cells. Front. Cell Dev. Biol. 2022, 10, 895162. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dunham, C.S.; Mackenzie, M.E.; Nakano, H.; Kim, A.R.; Nakano, A.; Stieg, A.Z.; Gimzewski, J.K. Cardio PyMEA: A user-friendly, open-source Python application for cardiomyocyte microelectrode array analysis. PLoS ONE 2022, 17, e0266647. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, B.; Zhang, K.; Chen, C.S.; Lejeune, E. Sarc-Graph: Automated segmentation, tracking, and analysis of sarcomeres in hiPSC-derived cardiomyocytes. PLoS Comput. Biol. 2021, 17, e1009443. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Toepfer, C.N.; Sharma, A.; Cicconet, M.; Garfinkel, A.C.; Mucke, M.; Neyazi, M.; Willcox, J.A.L.; Agarwal, R.; Schmid, M.; Rao, J.; et al. SarcTrack. Circ. Res. 2019, 124, 1172–1183. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grafton, F.; Ho, J.; Ranjbarvaziri, S.; Farshidfar, F.; Budan, A.; Steltzer, S.; Maddah, M.; Loewke, K.E.; Green, K.; Patel, S.; et al. Deep learning detects cardiotoxicity in a high-content screen with induced pluripotent stem cell-derived cardiomyocytes. eLife 2021, 10, e68714. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pasqualin, C.; Gannier, F.; Yu, A.; Malécot, C.O.; Bredeloux, P.; Maupoil, V. 0426: SarcOptiM, an ImageJ plug-in for cardiomyocyte contractility recording. Arch. Cardiovasc. Dis. Suppl. 2016, 8, 234. [Google Scholar] [CrossRef]
- Maddah, M.; Mandegar, M.A.; Dame, K.; Grafton, F.; Loewke, K.; Ribeiro, A.J.S. Quantifying drug-induced structural toxicity in hepatocytes and cardiomyocytes derived from hiPSCs using a deep learning method. J. Pharmacol. Toxicol. Methods 2020, 105, 106895. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.J.S.; Schwab, O.; Mandegar, M.A.; Ang, Y.S.; Conklin, B.R.; Srivastava, D.; Pruitt, B.L. Multi-Imaging Method to Assay the Contractile Mechanical Output of Micropatterned Human iPSC-Derived Cardiac Myocytes. Circ. Res. 2017, 120, 1572–1583. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Orita, K.; Sawada, K.; Koyama, R.; Ikegaya, Y. Deep learning-based quality control of cultured human-induced pluripotent stem cell-derived cardiomyocytes. J. Pharmacol. Sci. 2019, 140, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Orita, K.; Sawada, K.; Matsumoto, N.; Ikegaya, Y. Machine-learning-based quality control of contractility of cultured human-induced pluripotent stem-cell-derived cardiomyocytes. Biochem. Biophys. Res. Commun. 2020, 526, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Tohyama, S. Human Engineered Heart Tissue Models for Disease Modeling and Drug Discovery. Front. Cell Dev. Biol. 2022, 10, 855763. [Google Scholar] [CrossRef]
- Brennan, T.; Fink, M.; Rodriguez, B. Multiscale modelling of drug-induced effects on cardiac electrophysiological activity. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2009, 36, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.L.; Wang, K.; Clerx, M.; Johnstone, R.H.; Hortigon-Vinagre, M.P.; Zamora, V.; Allan, A.; Smith, G.L.; Gavaghan, D.J.; Mirams, G.R.; et al. Tailoring Mathematical Models to Stem-Cell Derived Cardiomyocyte Lines Can Improve Predictions of Drug-Induced Changes to Their Electrophysiology. Front. Physiol. 2017, 8, 986. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.H.; Edwards, A.G.; Giles, W.R.; Tveito, A. A computational method for identifying an optimal combination of existing drugs to repair the action potentials of SQT1 ventricular myocytes. PLoS Comput. Biol. 2021, 17, e1009233. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jaeger, K.H.; Wall, S.; Tveito, A. Computational prediction of drug response in short QT syndrome type 1 based on measurements of compound effect in stem cell-derived cardiomyocytes. PLoS Comput. Biol. 2021, 17, e1008089. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Johnstone, R.H.; Bardenet, R.; Gavaghan, D.J.; Mirams, G.R. Hierarchical Bayesian inference for ion channel screening dose-response data. Wellcome Open Res. 2016, 1, 6. [Google Scholar] [CrossRef]
- Aghasafari, P.; Yang, P.C.; Kernik, D.C.; Sakamoto, K.; Kanda, Y.; Kurokawa, J.; Vorobyov, I.; Clancy, C.E. A deep learning algorithm to translate and classify cardiac electrophysiology. eLife 2021, 10, e68335. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Juhola, M.; Penttinen, K.; Joutsijoki, H.; Aalto-Setälä, K. Analysis of Drug Effects on iPSC Cardiomyocytes with Machine Learning. Ann. Biomed. Eng. 2021, 49, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Pourrier, M.; Fedida, D. The Emergence of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes (hiPSC-CMs) as a Platform to Model Arrhythmogenic Diseases. Int. J. Mol. Sci. 2020, 21, 657. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, K.C.; Breitbart, A.; De Lange, W.J.; Hofsteen, P.; Futakuchi-Tsuchida, A.; Xu, J.; Schopf, C.; Razumova, M.V.; Jiao, A.; Boucek, R.; et al. Novel Adult-Onset Systolic Cardiomyopathy Due to MYH7 E848G Mutation in Patient-Derived Induced Pluripotent Stem Cells. JACC. Basic Transl. Sci. 2018, 3, 728–740. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hwang, H.; Liu, R.; Maxwell, J.T.; Yang, J.; Xu, C. Machine learning identifies abnormal Ca2+ transients in human induced pluripotent stem cell-derived cardiomyocytes. Sci. Rep. 2020, 10, 16977. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Seibertz, F.; Sutanto, H.; Dulk, R.; Pronto, J.R.D.; Springer, R.; Rapedius, M.; Liutkute, A.; Ritter, M.; Jung, P.; Stelzer, L.; et al. Electrophysiological and calcium-handling development during long-term culture of human-induced pluripotent stem cell-derived cardiomyocytes. Basic Res. Cardiol. 2023, 118, 14. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kernik, D.C.; Morotti, S.; Wu, H.; Garg, P.; Duff, H.J.; Kurokawa, J.; Jalife, J.; Wu, J.C.; Grandi, E.; Clancy, C.E. A computational model of induced pluripotent stem-cell derived cardiomyocytes incorporating experimental variability from multiple data sources. J. Physiol. 2019, 597, 4533–4564. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sacchetto, C.; Vitiello, L.; de Windt, L.J.; Rampazzo, A.; Calore, M. Modeling Cardiovascular Diseases with hiPSC-Derived Cardiomyocytes in 2D and 3D Cultures. Int. J. Mol. Sci. 2020, 21, 3404. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kannan, S.; Miyamoto, M.; Zhu, R.; Lynott, M.; Guo, J.; Chen, E.Z.; Colas, A.R.; Lin, B.L.; Kwon, C. Trajectory reconstruction identifies dysregulation of perinatal maturation programs in pluripotent stem cell-derived cardiomyocytes. Cell Rep. 2023, 42, 112330. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Paci, M.; Hyttinen, J.; Rodriguez, B.; Severi, S. Human induced pluripotent stem cell-derived versus adult cardiomyocytes: An in silico electrophysiological study on effects of ionic current block. Br. J. Pharmacol. 2015, 172, 5147–5160. [Google Scholar] [CrossRef]
- Pir, P.; Le Novere, N. Mathematical Models of Pluripotent Stem Cells: At the Dawn of Predictive Regenerative Medicine. Methods Mol. Biol. 2016, 1386, 331–350. [Google Scholar] [CrossRef] [PubMed]
- Wadkin, L.E.; Orozco-Fuentes, S.; Neganova, I.; Lako, M.; Shukurov, A.; Parker, N.G. The recent advances in the mathematical modelling of human pluripotent stem cells. SN Appl. Sci. 2020, 2, 276. [Google Scholar] [CrossRef]
- Hashizume, T.; Ying, B.W. Challenges in developing cell culture media using machine learning. Biotechnol. Adv. 2024, 70, 108293. [Google Scholar] [CrossRef] [PubMed]
- Asadi-Eydivand, M.; Solati-Hashjin, M.; Fathi, A.; Padashi, M.; Abu Osman, N.A. Optimal design of a 3D-printed scaffold using intelligent evolutionary algorithms. Appl. Soft Comput. 2016, 39, 36–47. [Google Scholar] [CrossRef]
- Pueyo, E.; Dangerfield, C.E.; Britton, O.J.; Virag, L.; Kistamas, K.; Szentandrassy, N.; Jost, N.; Varro, A.; Nanasi, P.P.; Burrage, K.; et al. Experimentally-Based Computational Investigation into Beat-To-Beat Variability in Ventricular Repolarization and Its Response to Ionic Current Inhibition. PLoS ONE 2016, 11, e0151461. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zang, Y.; Dai, L.; Zhan, H.; Dou, J.; Xia, L.; Zhang, H. Theoretical investigation of the mechanism of heart failure using a canine ventricular cell model: Especially the role of up-regulated CaMKII and SR Ca2+ leak. J. Mol. Cell. Cardiol. 2013, 56, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhao, X.; Gong, Y.; Zhang, J.; Zang, Y.; Xia, L. Exploring Impaired SERCA Pump-Caused Alternation Occurrence in Ischemia. Comput. Math. Methods Med. 2019, 2019, 8237071. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ghosh, S.; Nehme, R.; Barrett, L.E. Greater genetic diversity is needed in human pluripotent stem cell models. Nat. Commun. 2022, 13, 7301. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Varshneya, M.; Mei, X.; Sobie, E.A. Prediction of arrhythmia susceptibility through mathematical modeling and machine learning. Proc. Natl. Acad. Sci. USA 2021, 118, e2104019118. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990-2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kornej, J.; Borschel, C.S.; Benjamin, E.J.; Schnabel, R.B. Epidemiology of Atrial Fibrillation in the 21st Century: Novel Methods and New Insights. Circ. Res. 2020, 127, 4–20. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hindricks, G.; Potpara, T.; Dagres, N.; Arbelo, E.; Bax, J.J.; Blomstrom-Lundqvist, C.; Boriani, G.; Castella, M.; Dan, G.A.; Dilaveris, P.E.; et al. 2020 ESC Guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS): The Task Force for the diagnosis and management of atrial fibrillation of the European Society of Cardiology (ESC) Developed with the special contribution of the European Heart Rhythm Association (EHRA) of the ESC. Eur. Heart J. 2021, 42, 373–498. [Google Scholar] [CrossRef] [PubMed]
- Goette, A.; Auricchio, A.; Boriani, G.; Braunschweig, F.; Terradellas, J.B.; Burri, H.; Camm, A.J.; Crijns, H.; Dagres, N.; Deharo, J.C.; et al. EHRA White Paper: Knowledge gaps in arrhythmia management-status 2019. EP Eur. 2019, 21, 993–994. [Google Scholar] [CrossRef] [PubMed]
- Heijman, J.; Voigt, N.; Dobrev, D. New directions in antiarrhythmic drug therapy for atrial fibrillation. Future Cardiol. 2013, 9, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.E.; Han, Y.D.; Michell, A.R.; Ly, O.T.; Vanoye, C.G.; Spanghero, E.; George, A.L., Jr.; Darbar, D.; Khetani, S.R. Engineered cocultures of iPSC-derived atrial cardiomyocytes and atrial fibroblasts for modeling atrial fibrillation. Sci. Adv. 2024, 10, eadg1222. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- He, J.Q.; Ma, Y.; Lee, Y.; Thomson, J.A.; Kamp, T.J. Human embryonic stem cells develop into multiple types of cardiac myocytes: Action potential characterization. Circ. Res. 2003, 93, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Yechikov, S.; Kao, H.K.J.; Chang, C.W.; Pretto, D.; Zhang, X.D.; Sun, Y.H.; Smithers, R.; Sirish, P.; Nolta, J.A.; Chan, J.W.; et al. NODAL inhibition promotes differentiation of pacemaker-like cardiomyocytes from human induced pluripotent stem cells. Stem Cell Res. 2020, 49, 102043. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Goldfracht, I.; Protze, S.; Shiti, A.; Setter, N.; Gruber, A.; Shaheen, N.; Nartiss, Y.; Keller, G.; Gepstein, L. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 2020, 11, 75. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van Gorp, P.R.R.; Trines, S.A.; Pijnappels, D.A.; de Vries, A.A.F. Multicellular In vitro Models of Cardiac Arrhythmias: Focus on Atrial Fibrillation. Front. Cardiovasc. Med. 2020, 7, 43. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, F.; Long, D.; Huang, W.; Peng, W.; Lan, H.; Zhou, Y.; Dang, X.; Zhou, R. The Biphasic Effect of Retinoic Acid Signaling Pathway on the Biased Differentiation of Atrial-like and Sinoatrial Node-like Cells from hiPSC. Int. J. Stem Cells 2022, 15, 247–257. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, M.T.; Shao, N.Y.; Garg, V. Subtype-specific cardiomyocytes for precision medicine: Where are we now? Stem Cells 2020, 38, 822–833. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thorpe, J.; Perry, M.D.; Contreras, O.; Hurley, E.; Parker, G.; Harvey, R.P.; Hill, A.P.; Vandenberg, J.I. Development of a robust induced pluripotent stem cell atrial cardiomyocyte differentiation protocol to model atrial arrhythmia. Stem Cell Res. Ther. 2023, 14, 183. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schulz, C.; Sonmez, M.; Krause, J.; Schwedhelm, E.; Bangfen, P.; Alihodzic, D.; Hansen, A.; Eschenhagen, T.; Christ, T. A critical role of retinoic acid concentration for the induction of a fully human-like atrial action potential phenotype in hiPSC-CM. Stem Cell Rep. 2023, 18, 2096–2107. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Worth, C.L.; Lindberg, E.L.; Kanda, M.; Polanski, K.; Heinig, M.; Lee, M.; et al. Cells of the adult human heart. Nature 2020, 588, 466–472. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Baena-Montes, J.M.; Krasny, M.J.; O’Halloran, M.; Dunne, E.; Quinlan, L.R. In Vitro Models for Improved Therapeutic Interventions in Atrial Fibrillation. J. Pers. Med. 2023, 13, 1237. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lemme, M.; Ulmer, B.M.; Lemoine, M.D.; Zech, A.T.L.; Flenner, F.; Ravens, U.; Reichenspurner, H.; Rol-Garcia, M.; Smith, G.; Hansen, A.; et al. Atrial-like Engineered Heart Tissue: An In Vitro Model of the Human Atrium. Stem Cell Rep. 2018, 11, 1378–1390. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sagris, M.; Vardas, E.P.; Theofilis, P.; Antonopoulos, A.S.; Oikonomou, E.; Tousoulis, D. Atrial Fibrillation: Pathogenesis, Predisposing Factors, and Genetics. Int. J. Mol. Sci. 2021, 23, 6. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ragab, A.A.Y.; Sitorus, G.D.S.; Brundel, B.; de Groot, N.M.S. The Genetic Puzzle of Familial Atrial Fibrillation. Front. Cardiovasc. Med. 2020, 7, 14. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Moore, O.M.; Ho, K.S.; Copeland, J.S.; Parthasarathy, V.; Wehrens, X.H.T. Genome Editing and Cardiac Arrhythmias. Cells 2023, 12, 1363. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Benzoni, P.; Campostrini, G.; Landi, S.; Bertini, V.; Marchina, E.; Iascone, M.; Ahlberg, G.; Olesen, M.S.; Crescini, E.; Mora, C.; et al. Human iPSC modelling of a familial form of atrial fibrillation reveals a gain of function of If and ICaL in patient-derived cardiomyocytes. Cardiovasc. Res. 2020, 116, 1147–1160. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hong, L.; Zhang, M.; Ly, O.T.; Chen, H.; Sridhar, A.; Lambers, E.; Chalazan, B.; Youn, S.W.; Maienschein-Cline, M.; Feferman, L.; et al. Human induced pluripotent stem cell-derived atrial cardiomyocytes carrying an SCN5A mutation identify nitric oxide signaling as a mediator of atrial fibrillation. Stem Cell Rep. 2021, 16, 1542–1554. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sumer, S.A.; Hoffmann, S.; Laue, S.; Campbell, B.; Raedecke, K.; Frajs, V.; Clauss, S.; Kaab, S.; Janssen, J.W.G.; Jauch, A.; et al. Precise Correction of Heterozygous SHOX2 Mutations in hiPSCs Derived from Patients with Atrial Fibrillation via Genome Editing and Sib Selection. Stem Cell Rep. 2020, 15, 999–1013. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Babini, H.; Jimenez-Sabado, V.; Stogova, E.; Arslanova, A.; Butt, M.; Dababneh, S.; Asghari, P.; Moore, E.D.W.; Claydon, T.W.; Chiamvimonvat, N.; et al. hiPSC-derived cardiomyocytes as a model to study the role of small-conductance Ca2+-activated K+ (SK) ion channel variants associated with atrial fibrillation. Front. Cell Dev. Biol. 2024, 12, 1298007. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nakamura, M.; Sadoshima, J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018, 15, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, I.; Minamino, T. Physiological and pathological cardiac hypertrophy. J. Mol. Cell. Cardiol. 2016, 97, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Kastner, N.; Zlabinger, K.; Spannbauer, A.; Traxler, D.; Mester-Tonczar, J.; Hasimbegovic, E.; Gyongyosi, M. New Insights and Current Approaches in Cardiac Hypertrophy Cell Culture, Tissue Engineering Models, and Novel Pathways Involving Non-Coding RNA. Front. Pharmacol. 2020, 11, 1314. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Knight, W.E.; Cao, Y.; Lin, Y.H.; Chi, C.; Bai, B.; Sparagna, G.C.; Zhao, Y.; Du, Y.; Londono, P.; Reisz, J.A.; et al. Maturation of Pluripotent Stem Cell-Derived Cardiomyocytes Enables Modeling of Human Hypertrophic Cardiomyopathy. Stem Cell Rep. 2021, 16, 519–533. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Aggarwal, P.; Turner, A.; Matter, A.; Kattman, S.J.; Stoddard, A.; Lorier, R.; Swanson, B.J.; Arnett, D.K.; Broeckel, U. RNA expression profiling of human iPSC-derived cardiomyocytes in a cardiac hypertrophy model. PLoS ONE 2014, 9, e108051. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gerdes, A.M.; Liu, Z.; Zimmer, H.G. Changes in nuclear size of cardiac myocytes during the development and progression of hypertrophy in rats. Cardioscience 1994, 5, 203–208. [Google Scholar] [PubMed]
- Chang, C.C.; Cheng, H.C.; Chou, W.C.; Huang, Y.T.; Hsieh, P.L.; Chu, P.M.; Lee, S.D. Sesamin suppresses angiotensin-II-enhanced oxidative stress and hypertrophic markers in H9c2 cells. Environ. Toxicol. 2023, 38, 2165–2172. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.; Ulfenborg, B.; Andersson, C.X.; Heydarkhan-Hagvall, S.; Jeppsson, A.; Sartipy, P.; Synnergren, J. Cardiac hypertrophy in a dish: A human stem cell based model. Biol. Open 2020, 9, bio052381. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hesse, M.; Welz, A.; Fleischmann, B.K. Heart regeneration and the cardiomyocyte cell cycle. Pflug. Arch. Eur. J. Physiol. 2018, 470, 241–248. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kolwicz, S.C., Jr.; Tian, R. Glucose metabolism and cardiac hypertrophy. Cardiovasc. Res. 2011, 90, 194–201. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pohjolainen, L.; Ruskoaho, H.; Talman, V. Transcriptomics reveal stretched human pluripotent stem cell-derived cardiomyocytes as an advantageous hypertrophy model. J. Mol. Cell. Cardiol. Plus 2022, 2, 100020. [Google Scholar] [CrossRef]
- Johansson, M.; Ulfenborg, B.; Andersson, C.X.; Heydarkhan-Hagvall, S.; Jeppsson, A.; Sartipy, P.; Synnergren, J. Multi-Omics Characterization of a Human Stem Cell-Based Model of Cardiac Hypertrophy. Life 2022, 12, 293. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leonard, A.; Bertero, A.; Powers, J.D.; Beussman, K.M.; Bhandari, S.; Regnier, M.; Murry, C.E.; Sniadecki, N.J. Afterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues. J. Mol. Cell. Cardiol. 2018, 118, 147–158. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhu, Y.; Wu, F.; Yang, Q.; Feng, H.; Xu, D. Resveratrol Inhibits High Glucose-Induced H9c2 Cardiomyocyte Hypertrophy and Damage via RAGE-Dependent Inhibition of the NF-kappaB and TGF-beta1/Smad3 Pathways. Evid.-Based Complement. Altern. Med. Ecam 2022, 2022, 7781910. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Troncoso, M.F.; Pavez, M.; Wilson, C.; Lagos, D.; Duran, J.; Ramos, S.; Barrientos, G.; Silva, P.; Llanos, P.; Basualto-Alarcon, C.; et al. Testosterone activates glucose metabolism through AMPK and androgen signaling in cardiomyocyte hypertrophy. Biol. Res. 2021, 54, 3. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Drawnel, F.M.; Boccardo, S.; Prummer, M.; Delobel, F.; Graff, A.; Weber, M.; Gerard, R.; Badi, L.; Kam-Thong, T.; Bu, L.; et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 2014, 9, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Fandl, H.K.; Garcia, V.P.; Treuth, J.W.; Brewster, L.M.; Greiner, J.J.; Davy, K.P.; Stauffer, B.L.; Desouza, C.A. Endothelial-derived extracellular vesicles from obese/hypertensive adults increase factors associated with hypertrophy and fibrosis in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2023, 324, H675–H685. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, H.; Yang, H.; Rhee, J.W.; Zhang, J.Z.; Lam, C.K.; Sallam, K.; Chang, A.C.Y.; Ma, N.; Lee, J.; Zhang, H.; et al. Modelling diastolic dysfunction in induced pluripotent stem cell-derived cardiomyocytes from hypertrophic cardiomyopathy patients. Eur. Heart J. 2019, 40, 3685–3695. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, J.; Ross, L.; Grube, C.; Wang, H.S. Toxicity of low dose bisphenols in human iPSC-derived cardiomyocytes and human cardiac organoids—Impact on contractile function and hypertrophy. Chemosphere 2024, 353, 141567. [Google Scholar] [CrossRef] [PubMed]
- Garg, P.; Garg, V.; Shrestha, R.; Sanguinetti, M.C.; Kamp, T.J.; Wu, J.C. Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes as Models for Cardiac Channelopathies: A Primer for Non-Electrophysiologists. Circ. Res. 2018, 123, 224–243. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, J.B. Channelopathies. Korean J. Pediatr. 2014, 57, 1–18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cai, D.; Wang, X.; Sun, Y.; Fan, H.; Zhou, J.; Yang, Z.; Qiu, H.; Wang, J.; Su, J.; Gong, T.; et al. Patient-specific iPSC-derived cardiomyocytes reveal aberrant activation of Wnt/beta-catenin signaling in SCN5A-related Brugada syndrome. Stem Cell Res. Ther. 2023, 14, 241. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Egashira, T.; Yuasa, S.; Suzuki, T.; Aizawa, Y.; Yamakawa, H.; Matsuhashi, T.; Ohno, Y.; Tohyama, S.; Okata, S.; Seki, T.; et al. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc. Res. 2012, 95, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Lehnart, S.E.; Ackerman, M.J.; Benson, D.W., Jr.; Brugada, R.; Clancy, C.E.; Donahue, J.K.; George, A.L., Jr.; Grant, A.O.; Groft, S.C.; January, C.T.; et al. Inherited arrhythmias: A National Heart, Lung, and Blood Institute and Office of Rare Diseases workshop consensus report about the diagnosis, phenotyping, molecular mechanisms, and therapeutic approaches for primary cardiomyopathies of gene mutations affecting ion channel function. Circulation 2007, 116, 2325–2345. [Google Scholar] [CrossRef] [PubMed]
- Krahn, A.D.; Behr, E.R.; Hamilton, R.; Probst, V.; Laksman, Z.; Han, H.C. Brugada Syndrome. JACC. Clin. Electrophysiol. 2022, 8, 386–405. [Google Scholar] [CrossRef] [PubMed]
- Tse, G.; Liu, T.; Li, K.H.; Laxton, V.; Chan, Y.W.; Keung, W.; Li, R.A.; Yan, B.P. Electrophysiological Mechanisms of Brugada Syndrome: Insights from Pre-clinical and Clinical Studies. Front. Physiol. 2016, 7, 467. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Belbachir, N.; Portero, V.; Al Sayed, Z.R.; Gourraud, J.B.; Dilasser, F.; Jesel, L.; Guo, H.; Wu, H.; Gaborit, N.; Guilluy, C.; et al. RRAD mutation causes electrical and cytoskeletal defects in cardiomyocytes derived from a familial case of Brugada syndrome. Eur. Heart J. 2019, 40, 3081–3094. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cerrone, M.; Lin, X.; Zhang, M.; Agullo-Pascual, E.; Pfenniger, A.; Chkourko Gusky, H.; Novelli, V.; Kim, C.; Tirasawadichai, T.; Judge, D.P.; et al. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation 2014, 129, 1092–1103. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- El-Battrawy, I.; Lan, H.; Cyganek, L.; Zhao, Z.; Li, X.; Buljubasic, F.; Lang, S.; Yucel, G.; Sattler, K.; Zimmermann, W.H.; et al. Modeling Short QT Syndrome Using Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. J. Am. Heart Assoc. 2018, 7, e007394. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yazawa, M.; Hsueh, B.; Jia, X.; Pasca, A.M.; Bernstein, J.A.; Hallmayer, J.; Dolmetsch, R.E. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 2011, 471, 230–234. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fatima, A.; Xu, G.; Shao, K.; Papadopoulos, S.; Lehmann, M.; Arnaiz-Cot, J.J.; Rosa, A.O.; Nguemo, F.; Matzkies, M.; Dittmann, S.; et al. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell. Physiol. Biochem. 2011, 28, 579–592. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nam, Y.W.; Downey, M.; Rahman, M.A.; Cui, M.; Zhang, M. Channelopathy of small- and intermediate-conductance Ca2+-activated K+ channels. Acta Pharmacol. Sin. 2023, 44, 259–267. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kistamás, K.; Veress, R.; Horvath, B.; Banyasz, T.; Nanasi, P.P.; Eisner, D.A. Calcium Handling Defects and Cardiac Arrhythmia Syndromes. Front. Pharmacol. 2020, 11, 72. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hoekstra, M.; Mummery, C.L.; Wilde, A.A.; Bezzina, C.R.; Verkerk, A.O. Induced pluripotent stem cell derived cardiomyocytes as models for cardiac arrhythmias. Front. Physiol. 2012, 3, 346. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Song, Y.; Zheng, Z.; Lian, J. Deciphering Common Long QT Syndrome Using CRISPR/Cas9 in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Front. Cardiovasc. Med. 2022, 9, 889519. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Feyen, D.A.M.; McKeithan, W.L.; Bruyneel, A.A.N.; Spiering, S.; Hormann, L.; Ulmer, B.; Zhang, H.; Briganti, F.; Schweizer, M.; Hegyi, B.; et al. Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes. Cell Rep. 2020, 32, 107925. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lieu, D.K.; Fu, J.D.; Chiamvimonvat, N.; Tung, K.C.; McNerney, G.P.; Huser, T.; Keller, G.; Kong, C.W.; Li, R.A. Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Arrhythmia Electrophysiol. 2013, 6, 191–201. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meijer van Putten, R.M.; Mengarelli, I.; Guan, K.; Zegers, J.G.; van Ginneken, A.C.; Verkerk, A.O.; Wilders, R. Ion channelopathies in human induced pluripotent stem cell derived cardiomyocytes: A dynamic clamp study with virtual IK1. Front. Physiol. 2015, 6, 7. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gattenlohner, S.; Waller, C.; Ertl, G.; Bultmann, B.D.; Muller-Hermelink, H.K.; Marx, A. NCAM(CD56) and RUNX1(AML1) are up-regulated in human ischemic cardiomyopathy and a rat model of chronic cardiac ischemia. Am. J. Pathol. 2003, 163, 1081–1090. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rodriguez, B.; Trayanova, N.; Noble, D. Modeling cardiac ischemia. Ann. N. Y. Acad. Sci. 2006, 1080, 395–414. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rezende, P.C.; Ribas, F.F.; Serrano, C.V., Jr.; Hueb, W. Clinical significance of chronic myocardial ischemia in coronary artery disease patients. J. Thorac. Dis. 2019, 11, 1005–1015. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, Y.; Liang, Y.; Wang, M.; Wang, C.; Wei, H.; Naruse, K.; Takahashi, K. Model of Ischemic Heart Disease and Video-Based Comparison of Cardiomyocyte Contraction Using hiPSC-Derived Cardiomyocytes. J. Vis. Exp. 2020, 159, e61104. [Google Scholar] [CrossRef] [PubMed]
- Hakli, M.; Kreutzer, J.; Maki, A.J.; Valimaki, H.; Lappi, H.; Huhtala, H.; Kallio, P.; Aalto-Setala, K.; Pekkanen-Mattila, M. Human induced pluripotent stem cell-based platform for modeling cardiac ischemia. Sci. Rep. 2021, 11, 4153. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gaballah, M.; Penttinen, K.; Kreutzer, J.; Maki, A.J.; Kallio, P.; Aalto-Setala, K. Cardiac Ischemia On-a-Chip: Antiarrhythmic Effect of Levosimendan on Ischemic Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Cells 2022, 11, 1045. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Forouzandehmehr, M.; Paci, M.; Hyttinen, J.; Koivumaki, J.T. In silico study of the mechanisms of hypoxia and contractile dysfunction during ischemia and reperfusion of hiPSC cardiomyocytes. Dis. Models Mech. 2024, 17, dmm050365. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mesquita, F.C.P.; King, M.; da Costa Lopez, P.L.; Thevasagayampillai, S.; Gunaratne, P.H.; Hochman-Mendez, C. Laminin Alpha 2 Enhances the Protective Effect of Exosomes on Human iPSC-Derived Cardiomyocytes in an In Vitro Ischemia-Reoxygenation Model. Int. J. Mol. Sci. 2024, 25, 3773. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xiao, Y.; Li, J.; Qiu, L.; Jiang, C.; Huang, Y.; Liu, J.; Sun, Q.; Hong, H.; Ye, L. Dexmedetomidine Protects Human Cardiomyocytes Against Ischemia-Reperfusion Injury Through alpha2-Adrenergic Receptor/AMPK-Dependent Autophagy. Front. Pharmacol. 2021, 12, 615424. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Onodi, Z.; Visnovitz, T.; Kiss, B.; Hambalko, S.; Koncz, A.; Agg, B.; Varadi, B.; Toth, V.E.; Nagy, R.N.; Gergely, T.G.; et al. Systematic transcriptomic and phenotypic characterization of human and murine cardiac myocyte cell lines and primary cardiomyocytes reveals serious limitations and low resemblances to adult cardiac phenotype. J. Mol. Cell. Cardiol. 2022, 165, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Hakli, M.; Kreutzer, J.; Maki, A.J.; Valimaki, H.; Cherian, R.M.; Kallio, P.; Aalto-Setala, K.; Pekkanen-Mattila, M. Electrophysiological Changes of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes during Acute Hypoxia and Reoxygenation. Stem Cells Int. 2022, 2022, 9438281. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Davis, J.; Chouman, A.; Creech, J.; Monteiro da Rocha, A.; Ponce-Balbuena, D.; Jimenez Vazquez, E.N.; Nichols, R.; Lozhkin, A.; Madamanchi, N.R.; Campbell, K.F.; et al. In vitro model of ischemic heart failure using human induced pluripotent stem cell-derived cardiomyocytes. JCI Insight 2021, 6, e134368. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Peters, M.C.; Maas, R.G.C.; van Adrichem, I.; Doevendans, P.A.M.; Mercola, M.; Saric, T.; Buikema, J.W.; van Mil, A.; Chamuleau, S.A.J.; Sluijter, J.P.G.; et al. Metabolic Maturation Increases Susceptibility to Hypoxia-induced Damage in Human iPSC-derived Cardiomyocytes. Stem Cells Transl. Med. 2022, 11, 1040–1051. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).