EPAD1 Orthologs Play a Conserved Role in Pollen Exine Patterning
Abstract
1. Introduction
2. Results
2.1. Poaceae Pollen Morphology Is Conserved but Surface Ornamentation Is Diverse
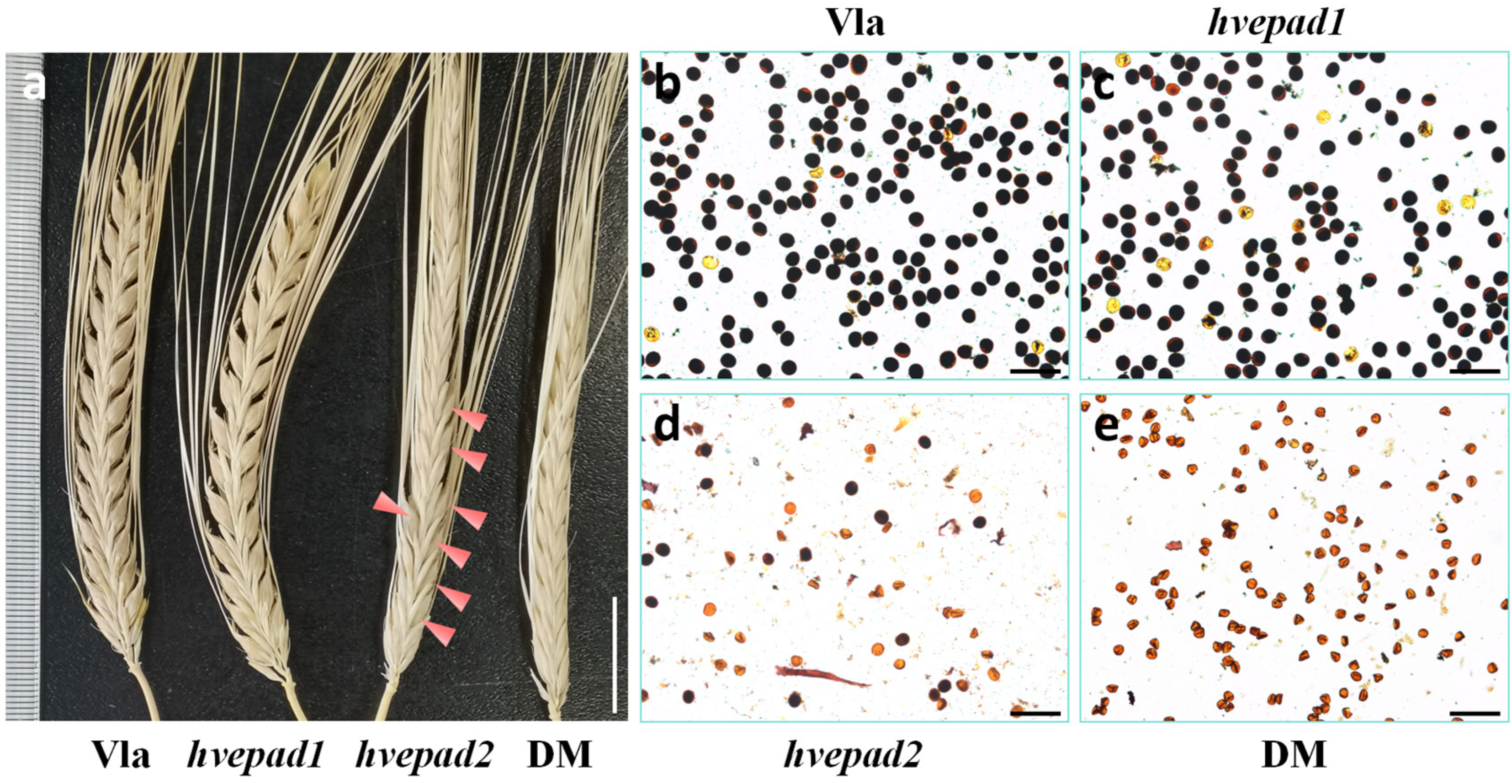
2.2. Mutations of BOP Clade HvEPAD1 and HvEPAD2 Affect Pollen Development
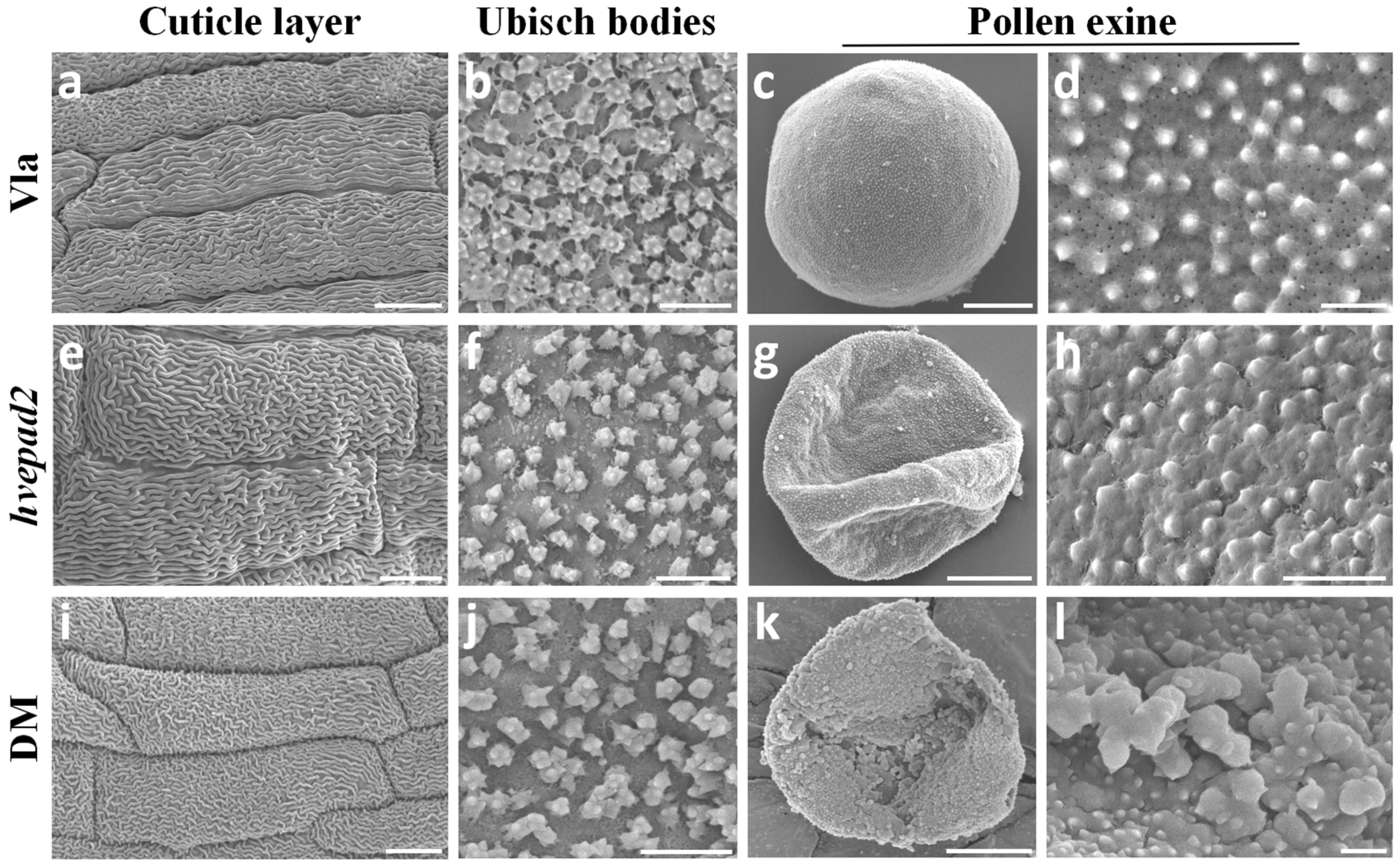
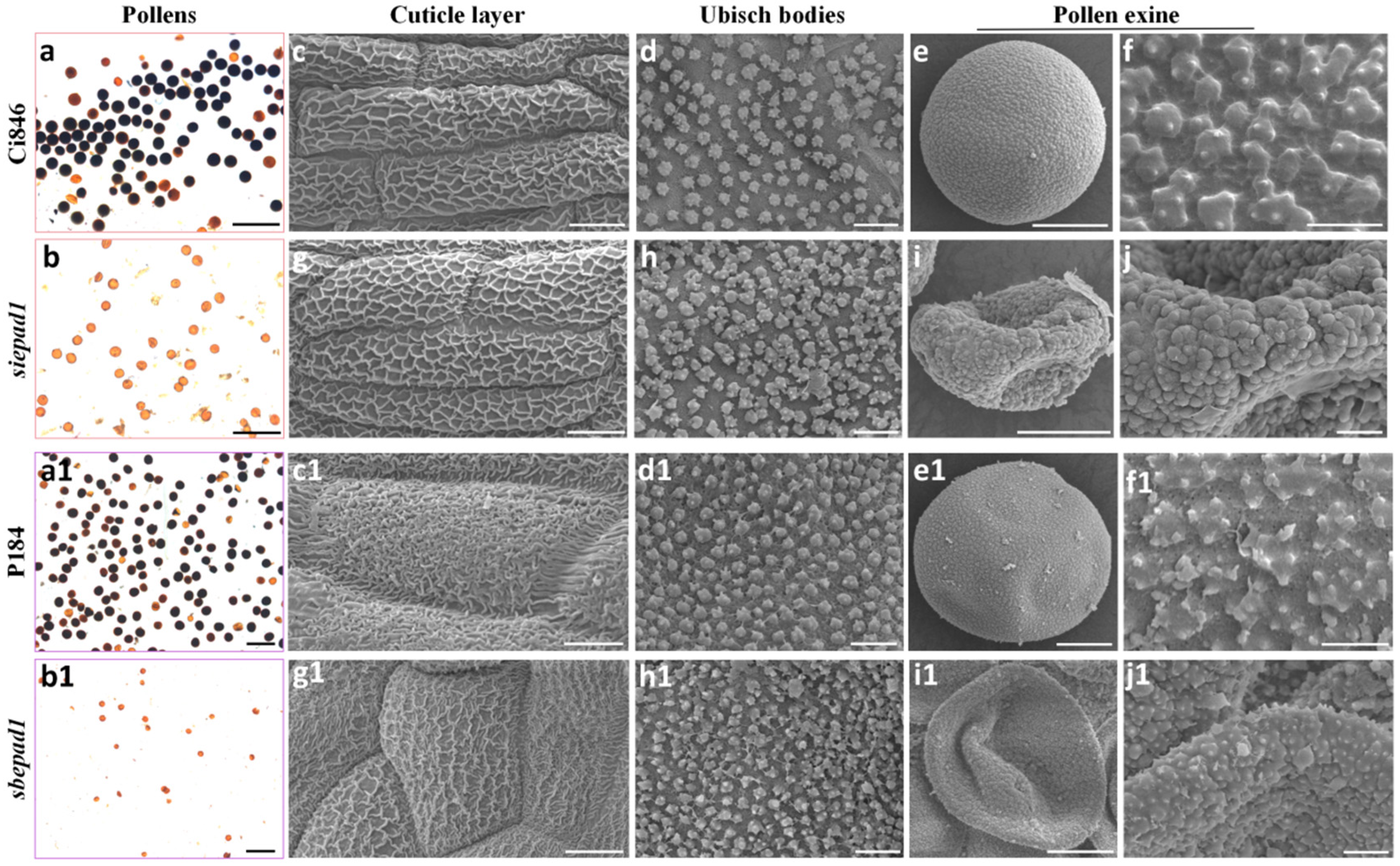
2.3. Pollen Exine Patterns Are Defective in BOP Clade Orthologs Mutants
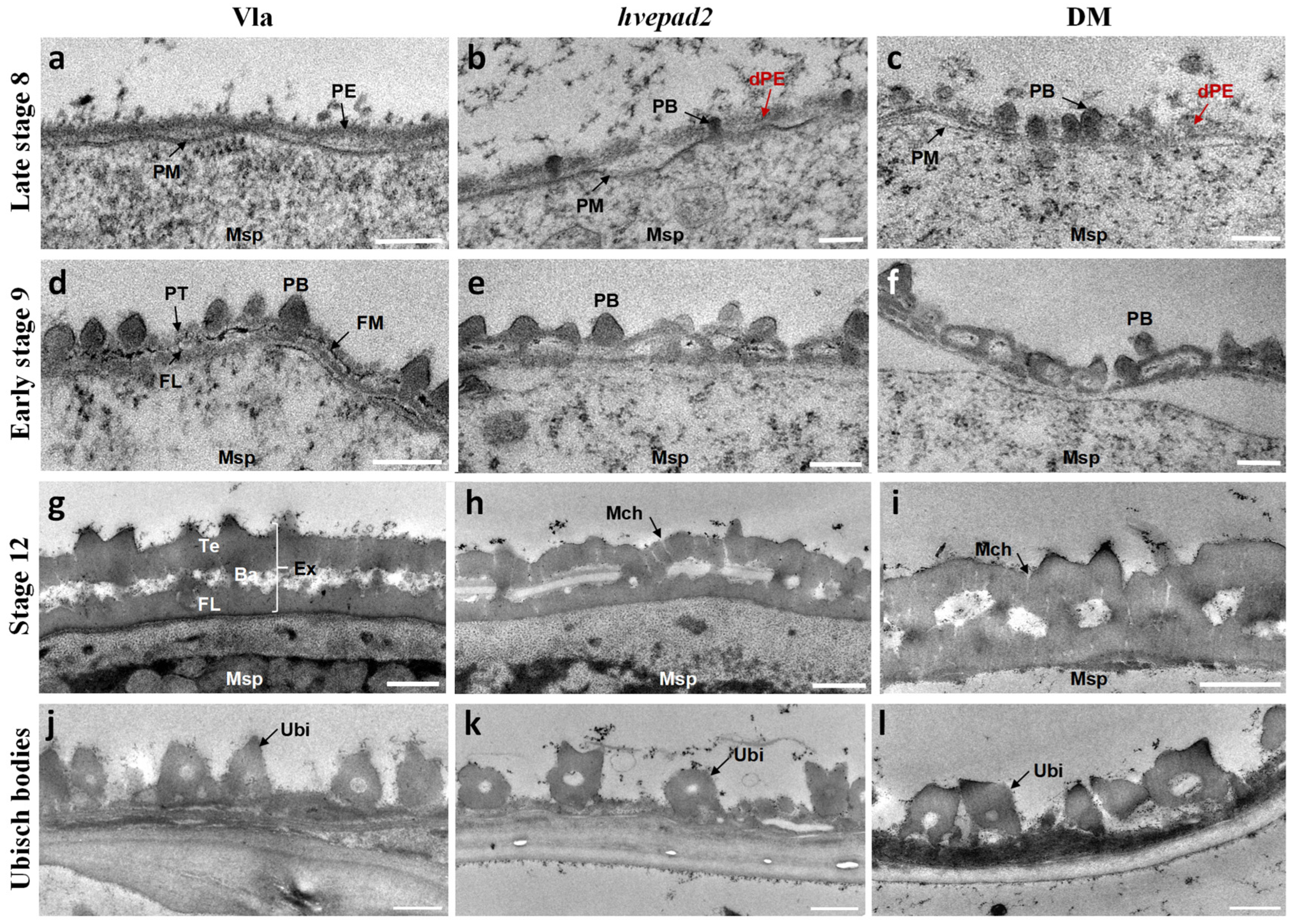
2.4. The Primexine Integrity Was Disrupted in BOP Clade Mutants
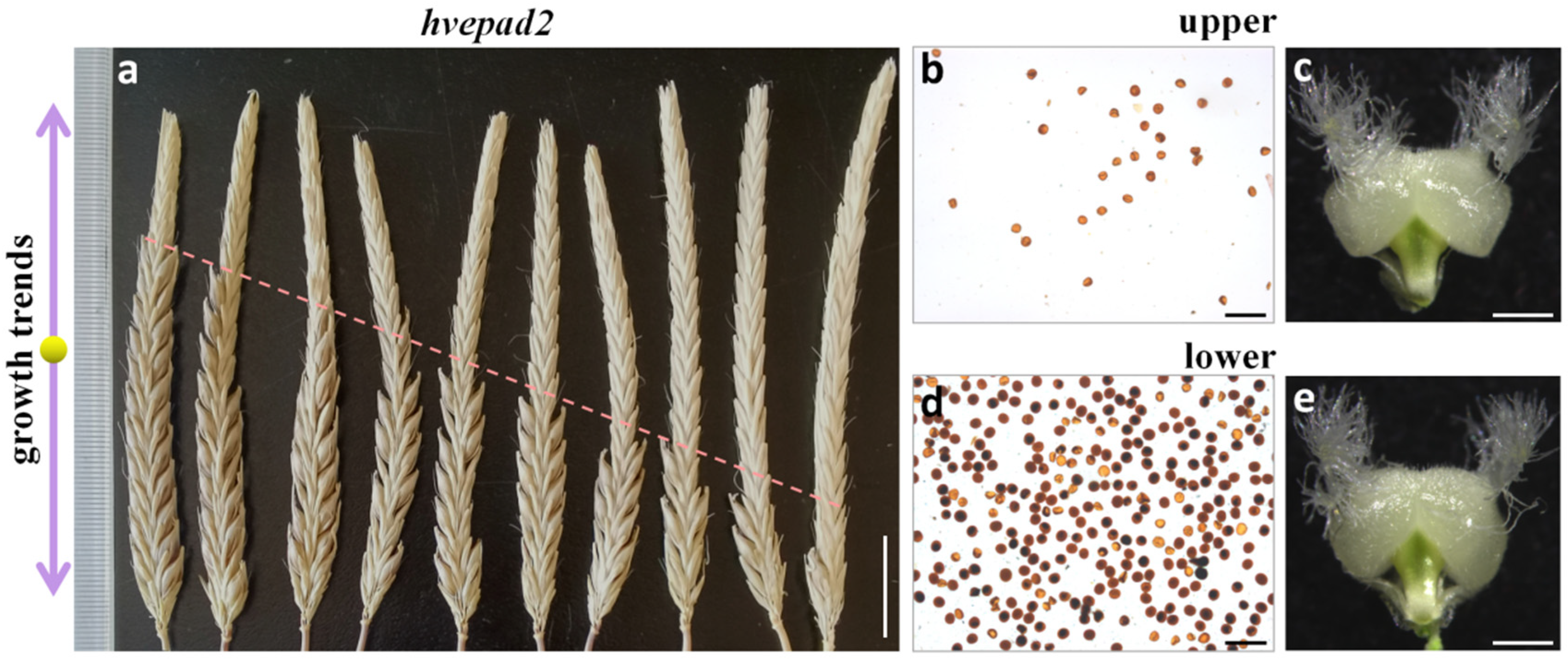
2.5. HvEPAD2 Exerts an Impact on the Male Fertility of Apical Spikelets
2.6. Mutants of PACMAD Clade Genes Showed Similar Pollen Exine Defects as BOP Clade Mutants
3. Discussion
3.1. Poaceae Pollen Exine Stereostructure Is Conserved but Surface Ornamentation Is Diverse
3.2. EPAD1 and Orthologs Function Conserved in Pollen Exine Patterning
3.3. The Gradient of Gene Expression Contributes to the Abortion of Apical Florets
4. Material and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phylogenetic Tree Analysis
4.3. Online Pollen Palynological Data
4.4. Characterization of Mutant Phenotypes
4.5. Transcriptome Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, R.; Dobritsa, A.A. Exine and Aperture Patterns on the Pollen Surface: Their Formation and Roles in Plant Reproduction. Annu. Plant Rev. 2018, 1, 589–628. [Google Scholar]
- Hesse, M. Pollen wall stratification and pollination. Plant Syst. Evol. 2000, 222, 1–17. [Google Scholar] [CrossRef]
- Wallace, S.; Fleming, A.; Wellman, C.H.; Beerling, D.J. Evolutionary development of the plant and spore wall. AoB Plants 2011, 2011, plr027. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, H.G.; Heslop-Harrison, J. Common mode of deposition for the sporopollenin of sexine and nexine. Nature 1968, 220, 926–927. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Liu, Y.; Douglas, C.J. Role of glycosyltransferases in pollen wall primexine formation and exine patterning. Plant Physiol. 2017, 173, 167–182. [Google Scholar] [CrossRef]
- Suzuki, T.; Narciso, J.O.; Zeng, W.; van de Meene, A.; Yasutomi, M.; Takemura, S.; Lampugnani, E.R.; Doblin, M.S.; Bacic, A.; Ishiguro, S. KNS4/UPEX1: A type II arabinogalactan b-(1,3)-galactosyltransferase required for pollen exine development. Plant Physiol. 2017, 173, 183–205. [Google Scholar] [CrossRef]
- Shi, J.; Cui, M.; Yang, L.; Kim, Y.J.; Zhang, D. Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 2015, 20, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 2011, 62, 437–460. [Google Scholar] [CrossRef]
- Li, H.; Kim, Y.J.; Yang, L.; Liu, Z.; Zhang, J.; Shi, H.; Huang, G.; Persson, S.; Zhang, D.; Liang, W. Grass-Specific EPAD1 Is Essential for Pollen Exine Patterning in Rice. Plant Cell. 2020, 32, 3961–3977. [Google Scholar] [CrossRef]
- Wei, C. Morphometrics of Modern and Fossil Poaceae Pollen from South America. Ph.D. Thesis, Fully Internal, University of Amsterdam, Amsterdam, The Netherlands, 2023. [Google Scholar]
- Radaeski, J.N.; Evaldt, A.C.P.; Bauermann, S.G. Anthropic pollen indicators: Poaceae pollen of non-native species in Southern Brazil. Open Access J. Sci. 2018, 2, 137–144. [Google Scholar] [CrossRef][Green Version]
- Tucker, E.J.; Baumann, U.; Kouidri, A.; Suchecki, R.; Baes, M.; Garcia, M.; Okada, T.; Dong, C.; Wu, Y.; Sandhu, A.; et al. Molecular identification of the wheat male fertility gene Ms1 and its prospects for hybrid breeding. Nat. Commun. 2017, 8, 869. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, J.; Chen, S.; Heng, Y.; Chen, Z.; Yang, J.; Zhou, K.; Pei, J.; He, H.; Deng, X.W.; et al. Poaceae-specific MS1 encodes a phospholipid-binding protein for male fertility in bread wheat. Proc. Natl. Acad. Sci. USA 2017, 114, 12614–12619. [Google Scholar] [CrossRef] [PubMed]
- Heng, Y.; Li, J.; Wang, Z.; Chen, Z.; He, H.; Deng, X.; Ma, L. Cloning, expression and functional analysis of a male fertility gene ThMsl in bread wheat. Sci. Agric. Sin. 2020, 53, 4727–4742. [Google Scholar]
- Li, J.; Wang, Z.; Chang, Z.; He, H.; Tang, X.; Ma, L.; Deng, X.W. A functional characterization of TaMs1 orthologs in Poaceae plants. Crop J. 2021, 9, 1291–1300. [Google Scholar] [CrossRef]
- Zhang, T.; Huang, W.; Zhang, L.; Li, D.Z.; Qi, J.; Ma, H. Phylogenomic profiles of whole-genome duplications in Poaceae and landscape of differential duplicate retention and losses among major Poaceae lineages. Nat. Commun. 2024, 15, 3305. [Google Scholar] [CrossRef] [PubMed]
- Grass Phylogeny Working Group II. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 2012, 193, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhang, L.; Columbus, J.T.; Hu, Y.; Zhao, Y.; Tang, L.; Guo, Z.; Chen, W.; McKain, M.; Bartlett, M.; et al. A well-supported nuclear phylogeny of Poaceae and implications for the evolution of C4 photosynthesis. Mol. Plant 2022, 15, 755–777. [Google Scholar] [CrossRef] [PubMed]
- Gallaher, T.J.; Peterson, P.M.; Soreng, R.J.; Zuloaga, F.O.; Li, D.-Z.; Clark, L.G.; Tyrrell, C.D.; Welker, C.A.D.; Kellogg, E.A.; Teisher, J.K. Grasses through space and time: An overview of the biogeographical and macroevolutionary history of Poaceae. J. Syst. Evol. 2022, 60, 522–569. [Google Scholar] [CrossRef]
- Huysmans, S.; El-Ghazaly, G.; Smets, E. Orbicules in angiosperms: Morphology, function, distribution, and relation with tapetum types. Bot. Rev. 1998, 64, 240–272. [Google Scholar] [CrossRef]
- Radja, A.; Horsley, E.M.; Lavrentovich, M.O.; Sweeney, A.M. Pollen Cell Wall Patterns Form from Modulated Phases. Cell 2019, 176, 856–868. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Tan, K.; Huang, W.; Shi, J.; Li, T.; Hu, J.; Wang, K.; Wang, C.; Xin, B.; et al. A complete telomere-to-telomere assembly of the maize genome. Nat. Genet. 2023, 55, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Shanmugaraj, N.; Rajaraman, J.; Kale, S.; Kamal, R.; Huang, Y.; Thirulogachandar, V.; Garibay-Hernández, A.; Budhagatapalli, N.; Tandron Moya, Y.A.; Hajirezaei, M.R.; et al. Multilayered regulation of developmentally programmed pre-anthesis tip degeneration of the barley inflorescence. Plant Cell 2023, 35, 3973–4001. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liu, Y.; Zhang, L.; Sun, Z.; Wang, Z.; Jiao, Y.; Shen, K.; Guo, Z. A transcriptional atlas identifies key regulators and networks for the development of spike tissues in barley. Cell Rep. 2023, 42, 113441. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.M.; Sharma, R.; Börner, A. Insight into the genetic contribution of maximum yield potential, spikelet development and abortion in barley. Plants People Planet 2021, 3, 721–736. [Google Scholar] [CrossRef]
- Meng, Z.-D.; Zhang, F.-J.; Ding, Z.-H.; Sun, Q.; Wang, L.; Guo, Q.-F.; Wang, H.-G. Inheritance of Ear Tip-Barrenness Trait in Maize. Agric. Sci. China 2007, 6, 628–633. [Google Scholar] [CrossRef]
- Sun, W.; Lu, C.; Wen, L.; Liu, Y.; Zhou, X.; Xiao, X.; Guo, X.; Wang, Z.; Sun, Z.; Zhang, Z.; et al. Low sucrose availability reduces basal spikelet fertility by inducing abscisic acid and jasmonic acid synthesis in wheat. J. Exp. Bot. 2024, 75, 1967–1981. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.L.; Sun, A.Z.; Chen, S.T.; Chen, L.S.; Guo, F.Q. SPL6 represses signalling outputs of ER stress in control of panicle cell death in rice. Nat. Plants 2018, 4, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Shen, W.; Cui, Y.; Liu, Y.; Zheng, X.; Li, Y.; Wu, M.; Fang, S.; Liu, C.; Tang, M.; et al. APICAL SPIKELET ABORTION (ASA) Controls Apical Panicle Development in Rice by Regulating Salicylic Acid Biosynthesis. Front Plant Sci. 2021, 12, 636877. [Google Scholar] [CrossRef]
- Jiang, G.; Koppolu, R.; Rutten, T.; Hensel, G.; Lundqvist, U.; Tandron Moya, Y.A.; Huang, Y.; Rajaraman, J.; Poursarebani, N.; von Wirén, N.; et al. Non-cell-autonomous signaling associated with barley ALOG1 specifies spikelet meristem determinacy. Curr. Biol. 2024, 34, 2344–2358. [Google Scholar] [CrossRef]
- Bai, J.; Zhu, X.; Wang, Q.; Zhang, J.; Chen, H.; Dong, G.; Zhu, L.; Zheng, H.; Xie, Q.; Nian, J.; et al. Rice TUTOU1 Encodes a Suppressor of cAMP Receptor-Like Protein That Is Important for Actin Organization and Panicle Development. Plant Physiol. 2015, 169, 1179–1191. [Google Scholar] [CrossRef]
- Zafar, S.A.; Patil, S.B.; Uzair, M.; Fang, J.; Zhao, J.; Guo, T.; Yuan, S.; Uzair, M.; Luo, Q.; Shi, J.; et al. DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol. 2020, 225, 356–375. [Google Scholar] [CrossRef] [PubMed]
- Bretani, G.; Rossini, L.; Ferrandi, C.; Russell, J.; Waugh, R.; Kilian, B.; Bagnaresi, P.; Cattivelli, L.; Fricano, A. Segmental duplications are hot spots of copy number variants affecting barley gene content. Plant J. 2020, 103, 1073–1088. [Google Scholar] [CrossRef]
- Sutton, T.; Baumann, U.; Hayes, J.; Collins, N.C.; Shi, B.J.; Schnurbusch, T.; Hay, A.; Mayo, G.; Pallotta, M.; Tester, M.; et al. Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 2007, 318, 1446–1449. [Google Scholar] [CrossRef] [PubMed]
- Swanson-Wagner, R.A.; Eichten, S.R.; Kumari, S.; Tiffin, P.; Stein, J.C.; Ware, D.; Springer, N.M. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 2010, 20, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Díaz, A.; Zikhali, M.; Turner, A.S.; Isaac, P.; Laurie, D.A. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS ONE. 2012, 7, e33234. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Hua, M.; Yin, W.; Fernández Gómez, J.; Tidy, A.; Xing, G.; Zong, J.; Shi, S.; Wilson, Z.A. Barley TAPETAL DEVELOPMENT and FUNCTION1 (HvTDF1) gene reveals conserved and unique roles in controlling anther tapetum development in dicot and monocot plants. New Phytol. 2023, 240, 173–190. [Google Scholar] [CrossRef]
- Fernández Gómez, J.; Wilson, Z.A. Non-destructive staging of barley reproductive development for molecular analysis based upon external morphology. J. Exp. Bot. 2012, 63, 4085–4094. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Hua, M.; Tariq, N.; Li, X.; Zhang, Y.; Zhang, D.; Liang, W. EPAD1 Orthologs Play a Conserved Role in Pollen Exine Patterning. Int. J. Mol. Sci. 2024, 25, 8914. https://doi.org/10.3390/ijms25168914
Li H, Hua M, Tariq N, Li X, Zhang Y, Zhang D, Liang W. EPAD1 Orthologs Play a Conserved Role in Pollen Exine Patterning. International Journal of Molecular Sciences. 2024; 25(16):8914. https://doi.org/10.3390/ijms25168914
Chicago/Turabian StyleLi, Huanjun, Miaoyuan Hua, Naveed Tariq, Xian Li, Yushi Zhang, Dabing Zhang, and Wanqi Liang. 2024. "EPAD1 Orthologs Play a Conserved Role in Pollen Exine Patterning" International Journal of Molecular Sciences 25, no. 16: 8914. https://doi.org/10.3390/ijms25168914
APA StyleLi, H., Hua, M., Tariq, N., Li, X., Zhang, Y., Zhang, D., & Liang, W. (2024). EPAD1 Orthologs Play a Conserved Role in Pollen Exine Patterning. International Journal of Molecular Sciences, 25(16), 8914. https://doi.org/10.3390/ijms25168914





