Genomic Analysis of Cronobacter condimenti s37: Identification of Resistance and Virulence Genes and Comparison with Other Cronobacter and Closely Related Species
Abstract
1. Introduction
2. Results and Discussion
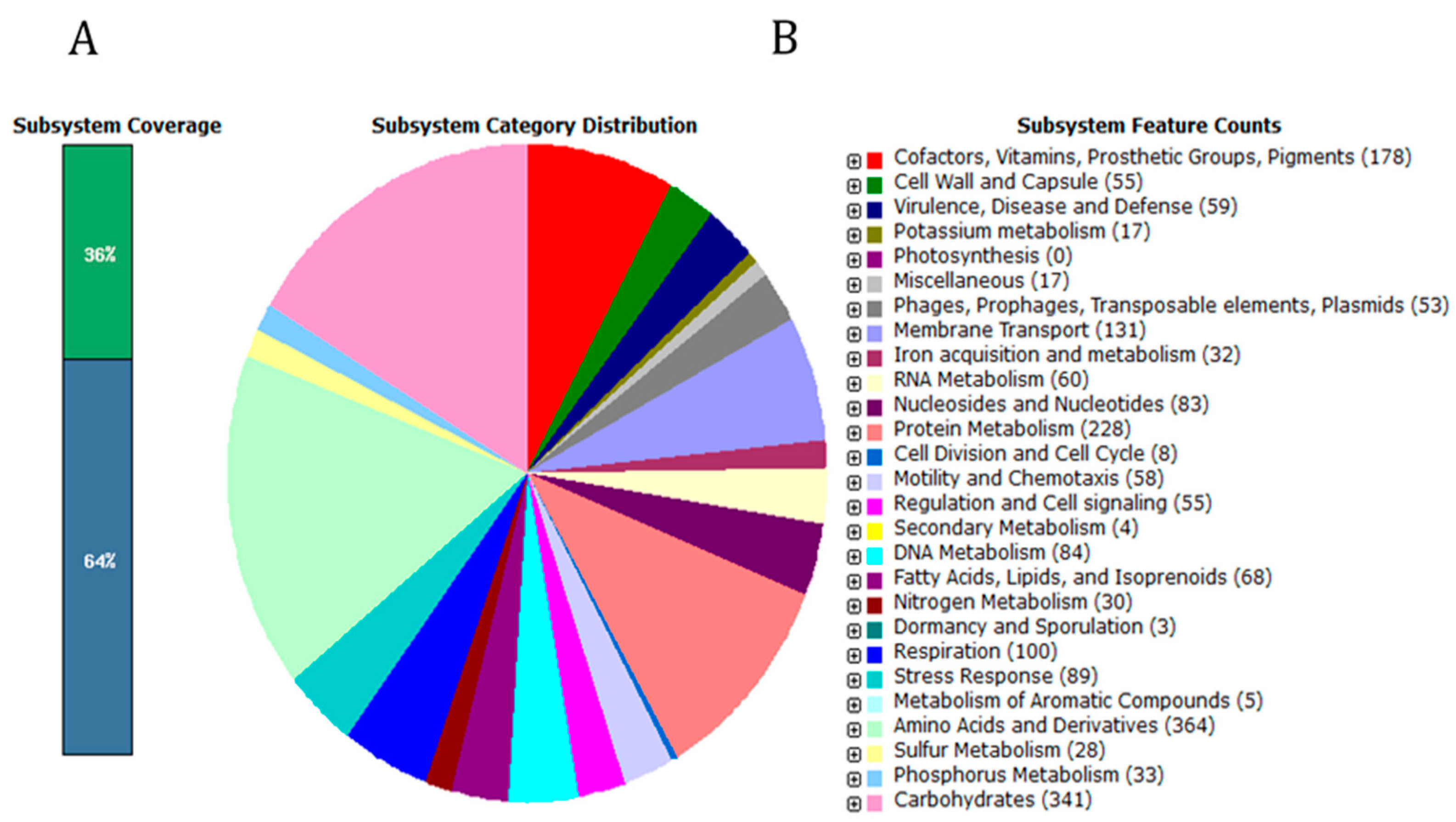
2.1. General Features of C. condimenti s37 Genome
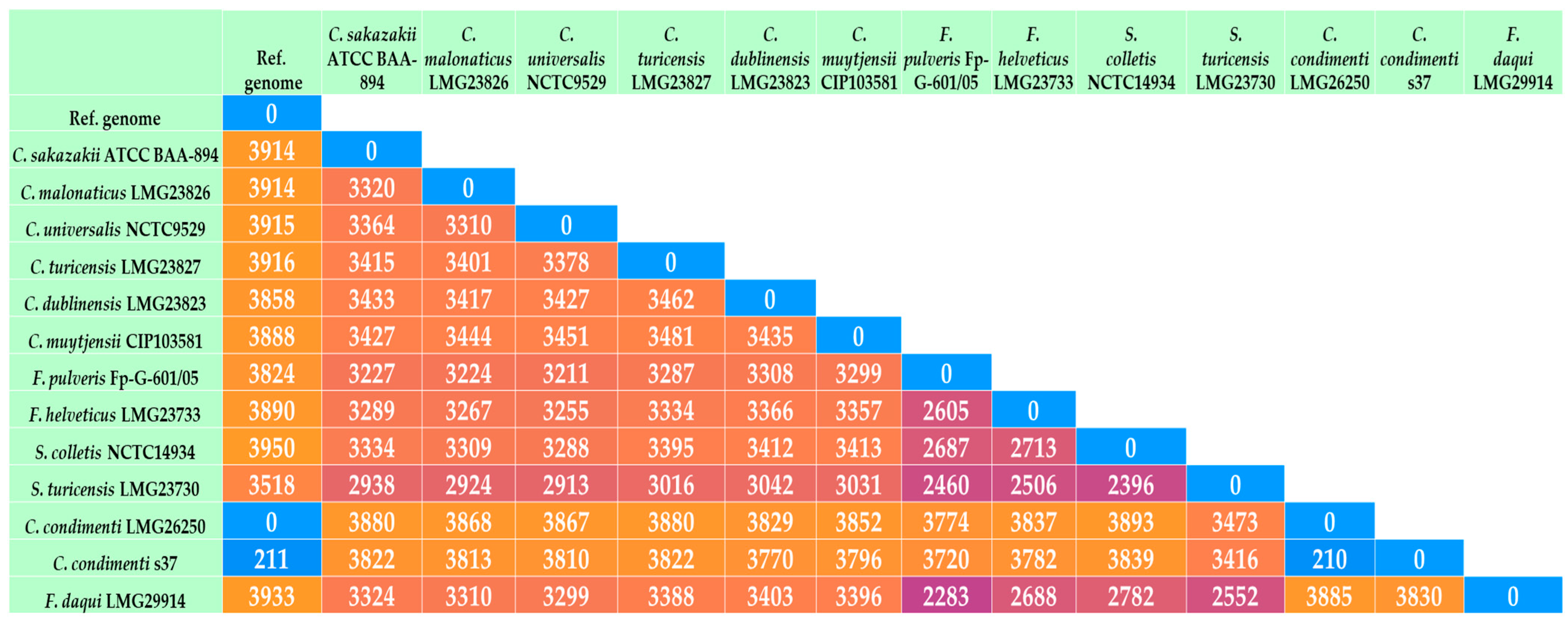
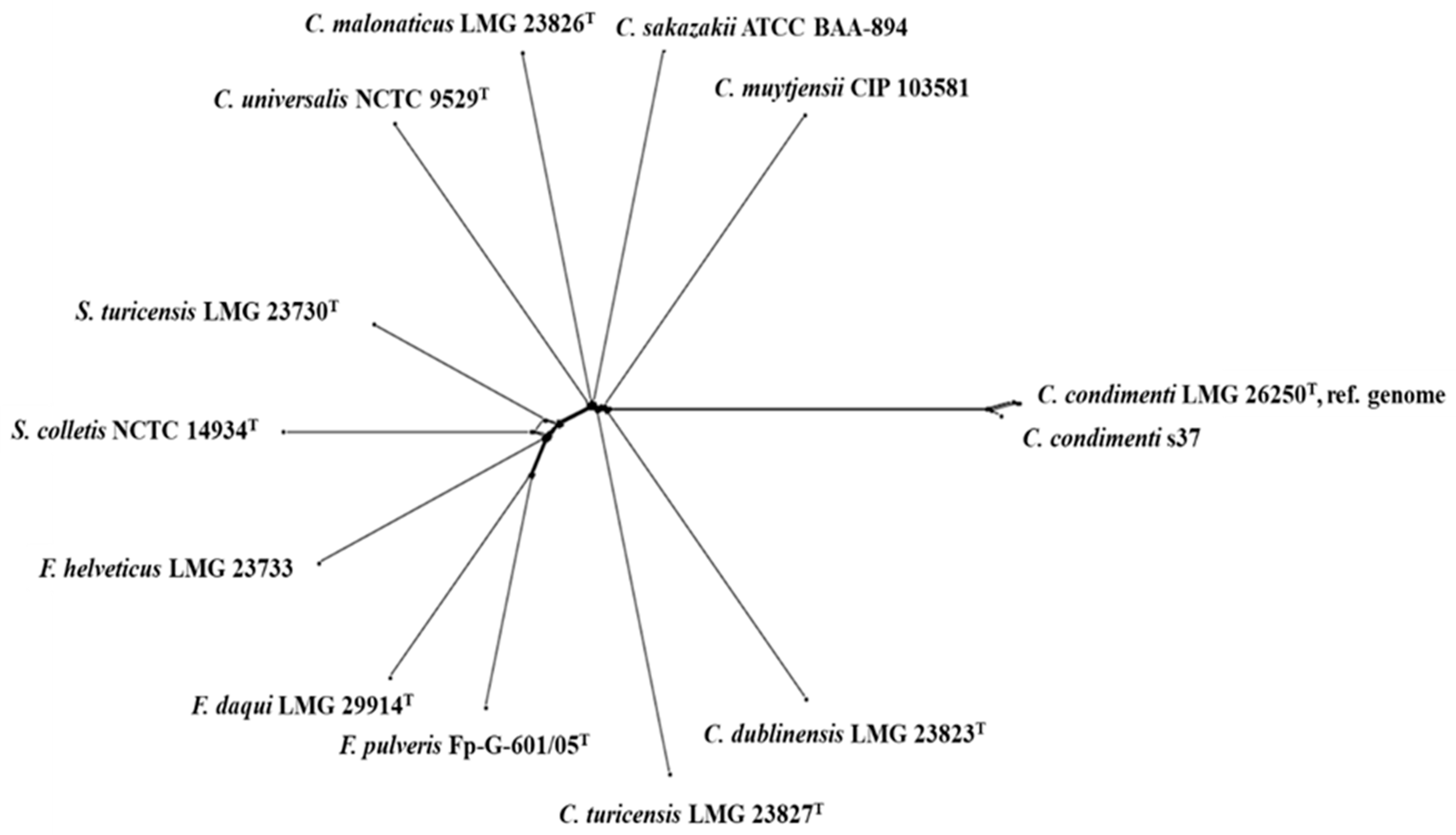
2.2. Genome Comparison of C. condiment s37 with Other Cronobacter and Closely Related Species
2.3. Genes Involved in Resistance to Environmental Stress
2.4. Genes Involved in Antimicrobial Resistance (AMR)
2.5. Genes Involved in Pathogenicity
3. Materials and Methods
3.1. Materials
3.2. Draft Genome Sequence of C. condimenti s37
3.3. Core Genome-MLST Analysis
3.4. Antimicrobial Resistance Genes and Virulence Factors of C. condimenti s37
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, H.; Fu, S.; Song, D.; Qin, X.; Zhang, W.; Man, C.; Yang, X.; Jiang, Y. Identification, Typing and Drug Resistance of Cronobacter spp. in Powdered Infant Formula and Processing Environment. Foods 2023, 12, 1084. [Google Scholar] [CrossRef] [PubMed]
- CDC. FDA Investigation of Cronobacter Infections: Powdered Infant Formula (February 2022). FDA, 2022. Available online: https://www.fda.gov/food/outbreaks-foodborne-illness/fda-investigation-Cronobacter-infections-powdered-infant-formula-february-2022 (accessed on 2 August 2024).
- Cechin, C.F.; Carvalho, G.G.; Bastos, C.P.; Kabuki, D.Y. Cronobacter spp. in foods of plant origin: Occurrence, contamination routes, and pathogenic potential. Crit. Rev. Food Sci. Nutr. 2023, 63, 12398–12412. [Google Scholar] [CrossRef] [PubMed]
- Cechin, C.F.; Carvalho, G.G.; Kabuki, D.Y. Occurrence, genetic characterization, and antibiotic susceptibility of Cronobacter spp. isolated from low water activity functional foods in Brazil. Food Microbiol. 2024, 122, 104570. [Google Scholar] [CrossRef] [PubMed]
- Phair, K.; Pereira, S.G.; Kealey, C.; Fanning, S.; Brady, D.B. Insights into the mechanisms of Cronobacter sakazakii virulence. Microb Pathog. 2022, 169, 105643. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Cetinkaya, E.; Drahovska, H.; Levican, A.; Fiqueras, M.J.; Forsythe, S.J. Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. Genomospecies 1, recovered from a leg infection, water and food ingredients. Int. J. Syst. Evol. Microbiol. 2012, 62, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, S.J. Updates on the Cronobacter genus. Ann. Rev. Food Sci. Technol. 2018, 9, 23–44. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Woo, J.; Lee, Y.; Negrete, F.; Finkelstein, S.; Chase, H.R.; Addy, N.; Ewing, L.; Gilles Beaubrun, J.J.; Patel, I.; et al. Draft genomes of Cronobacter sakazakii strains isolated from dried spices bring unique insights into the diversity of plant-associated strains. Stand. Genom. Sci. 2018, 13, 35. [Google Scholar] [CrossRef]
- Ling, N.; Jiang, X.; Forsythe, S.; Zhang, D.; Shen, Y.; Ding, Y.; Wang, J.; Zhang, J.; Wu, Q.; Ye, Y. Food Safety Risks and Contributing Factors of Cronobacter spp. Engineering 2022, 12, 128–138. [Google Scholar] [CrossRef]
- Stephan, R.; Grim, C.J.; Gopinath, G.R.; Mammel, M.K.; Sathyamoorthy, V.; Trach, L.H.; Chase, H.R.; Fanning, S.; Tall, B.D. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov, respectively. Int. J. Syst. Evol. Microbiol. 2014, 64, 3402–3410. [Google Scholar] [CrossRef]
- Gao, Z.; Su, C.; Yang, X.; Sun, D.; Zeng, C.; Chen, M.; Hu, W.; Zhang, C. Franconibacter daqui sp. nov., a facultatively alkaliphilic species isolated from a Daqu sample. Int. J. Syst. Evol. Microbiol. 2017, 67, 4962–4966. [Google Scholar] [CrossRef]
- Jackson, E.E.; Masood, N.; Ibrahim, K.; Urvoy, N.; Hariri, S.; Forsythe, S.J. Description of Siccibacter colletis sp. nov., a novel species isolated from plant material, and emended description of Siccibacter turicensis. Int. J. Syst. Evol. Microbiol. 2015, 65 Pt 4, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Svobodová, B.; Vlach, J.; Junková, P.; Karamonová, L.; Blažková, M.; Fukal, L. Novel method for reliable identification of Siccibacter and Franconibacter strains: From “pseudo-Cronobacter” to new Enterobacteriaceae genera. Appl. Environ. Microbiol. 2017, 83, e00234-17. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.E.; Forsythe, S.J. Comparative study of Cronobacter identification according to phenotyping methods. BMC Microbiol. 2016, 16, 146. [Google Scholar] [CrossRef] [PubMed]
- Lepuschitz, S.; Pekard-Amenitsch, S.; Haunold, R.; Schill, S.; Schriebl, A.; Mach, R.; Allerberger, F.; Ruppitsch, W.; Forsythe, S.J. Draft genome sequence of the first documented clinical Siccibacter turicensis isolate in Austria. Genome Announc. 2018, 6, e00380-18. [Google Scholar] [CrossRef] [PubMed]
- Aly, M.A.; Domig, K.J.; Kneifel, W.; Reimhult, E. Whole Genome Sequencing-Based Comparison of Food Isolates of Cronobacter sakazakii. Front. Microbiol. 2019, 10, 1464. [Google Scholar] [CrossRef] [PubMed]
- Berthold-Pluta, A.; Garbowska, M.; Stefańska, I.; Pluta, A. Microbiological quality of selected ready-to-eat leaf vegetables, sprouts and non-pasteurized fresh fruit-vegetable juices including the presence of Cronobacter spp. Food Microbiol. 2017, 65, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Hamby, S.E.; Joseph, S.; Forsythe, S.J.; Chuzhanova, N. In Silico identification of pathogenic strains of Cronobacter from biochemical reveals association of inositol fermentation with pathogenicity. BMC Microbiol. 2011, 11, 204. [Google Scholar] [CrossRef] [PubMed]
- Grim, C.J.; Kotewicz, M.L.; Power, K.A.; Gopinath, G.; Franco, A.A.; Jarvis, K.G.; Yan, Q.; Jackson, S.A.; Sathyamoorthy, V.; Hu, L.; et al. Pan-genome analysis of the emerging foodborne pathogen Cronobacter spp. suggests a species-level bidirectional divergence driven by niche adaptation. BMC Genom. 2013, 14, 366. [Google Scholar] [CrossRef] [PubMed]
- Ikryannikova, L.N.; Kurbatov, L.K.; Gorokhovets, N.V.; Zamyatnin, A.A. Contact-Dependent Growth Inhibition in Bacteria: Do Not Get Too Close! Int. J. Mol. Sci. 2020, 21, 7990. [Google Scholar] [CrossRef]
- Mariano, G.; Faba-Rodriguez, R.; Bui, S.; Zhao, W.; Ross, J.; Tzokov, S.B.; Bergeron, J.R.C. Oligomerization of the FliF Domains Suggests Coordinated Assembly of the Bacterial Flagellum MS Ring. Front. Microbiol. 2022, 12, 781960. [Google Scholar] [CrossRef]
- Rogov, V.V.; Rogova, N.Y.; Bernhard, F.; Koglin, A.; Löhr, F.; Dötsch, V. A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains. J. Mol. Biol. 2006, 364, 68–79. [Google Scholar] [CrossRef]
- Yin, R.; Cheng, J.; Lin, J. The role of the type VI secretion system in the stress resistance of plant-associated bacteria. Stress Biol. 2024, 4, 16. [Google Scholar] [CrossRef]
- Koskiniemi, S.; Lamoureux, J.G.; Nikolakakis, K.C.; t’Kint de Roodenbeke, C.; Kaplan, M.D.; Low, D.A.; Hayes, C.S. Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. USA 2013, 110, 7032–7037. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, D.K.; Morby, A.P. Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 291–311. [Google Scholar] [CrossRef]
- Rensing, C.; Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003, 27, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Du, X.-J.; Wang, X.-Y.; Dong, X.; Li, P.; Wang, S. Characterization of the Desiccation Tolerance of Cronobacter sakazakii Strains. Front. Microbiol. 2018, 9, 2867. [Google Scholar] [CrossRef]
- Wang, L.; Forsythe, S.J.; Yang, X.; Fu, S.; Man, C.; Jiang, Y. Stress resistance of Cronobacter spp. affecting control of its growth during food production. J. Dairy Sci. 2021, 104, 11348–11367. [Google Scholar] [CrossRef]
- Srikumar, S.; Cao, Y.; Yan, Q.; Van Hoorde, K.; Nguyen, S.; Cooney, S.; Gopinath, G.R.; Tall, B.D.; Sivasankaran, S.K.; Lehner, A.; et al. RNA sequencing-based transcriptional overview of xerotolerance in Cronobacter sakazakii SP291. Appl. Environ. Microbiol. 2019, 85, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yu, Y.; Wu, X.; Xia, X.; Xiao, X.; Wu, H. Comparative proteomic analysis of Cronobacter sakazakii by iTRAQ provides insights into response to desiccation. Food Res. Int. 2017, 100, 631–639. [Google Scholar] [CrossRef]
- Iversen, C.; Forsythe, S. Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci. Technol. 2003, 14, 443–454. [Google Scholar] [CrossRef]
- Chase, H.R.; Gopinath, G.R.; Eshwar, A.K.; Stoller, A.; Fricker-Feer, C.; Gangiredla, J.; Patel, I.R.; Cinar, H.N.; Jeong, H.; Lee, C.; et al. Comparative genomic characterization of the highly persistent and potentially virulent Cronobacter sakazakii ST83, CC65 strain H322 and other ST83 strains. Front. Microbiol. 2017, 8, 1136. [Google Scholar] [CrossRef]
- Jang, H.; Addy, N.; Ewing, L.; Beaubrun, J.; Lee, Y.; Woo, J.; Negrete, F.; Finkelstein, S.; Tall, B.D.; Lehner, A.; et al. Whole-genome sequences of Cronobacter sakazakii isolates obtained from foods of plant origin and dried-food manufacturing environments. Genome Announc. 2018, 6, e00223-18. [Google Scholar] [CrossRef]
- Joseph, S.; Desai, P.; Ji, Y.; Cummings, C.A.; Shih, R.; Degoricija, L.; Rico, A.; Brzoska, P.; Hamby, S.E.; Masood, N.; et al. Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS ONE 2012, 7, e49455. [Google Scholar] [CrossRef]
- Stevens, M.J.A.; Cernela, N.; Stephan, R.; Lehner, A. Comparative genomics of Cronobacter sakazakii strains from a powdered infant formula plant reveals evolving populations. LWT 2023, 184, 115034. [Google Scholar] [CrossRef]
- Joseph, S.; Hariri, S.; Masood, N.; Forsythe, S.J. Sialic acid utilization by Cronobacter sakazakii. Microb. Inform. Exp. 2013, 3, 3. [Google Scholar] [CrossRef]
- Singh, N.; Goel, G.; Raghav, M. Insights into virulence factors determining the pathogenicity of Cronobacter sakazakii. Virulence 2015, 6, 433–440. [Google Scholar] [CrossRef]
- Gan, X.; Li, M.; Xu, J.; Yan, S.; Wang, W.; Li, F. Emerging of Multidrug-Resistant Cronobacter sakazakii Isolated from Infant Supplementary Food in China. Microbiol. Spectr. 2022, 10, e01197-22. [Google Scholar] [CrossRef] [PubMed]
- Severi, E.; Hood, D.W.; Thomas, G.H. Sialic acid utilization by bacterial pathogens. Microbiology 2007, 153, 2817–2822. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.; Lee, C.-R. Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front. Microbiol. 2019, 10, 953. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, K.-P.; Choi, J.; Lim, J.-A.; Lee, J.; Hwang, S.; Ryu, S. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 2010, 76, 5188–5198. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, J.; Ling, N.; Zeng, H.; Tong, L.; Zhang, M.; Zhang, J.; Wu, Q.; Ye, Y. Short communication: Roles of outer membrane protein W on survival, cellular morphology, and biofilm formation of Cronobacter sakazakii in response to oxidative stress. J. Dairy Sci. 2019, 102, 2017–2021. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ling, N.; Gao, J.; Zhang, X.; Zhang, M.; Tong, L.; Zeng, H.; Zhang, J.; Wu, Q. Roles of outer membrane protein W (OmpW) on survival, morphology, and biofilm formation under NaCl stresses in Cronobacter sakazakii. J. Dairy Sci. 2018, 101, 3844–3850. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tao, Y.; Yu, H.; Ni, J.; Zeng, L.; Teng, Q.; Kim, K.S.; Zhao, G.P.; Guo, X.; Yao, Y. Hcp Family Proteins Secreted via the Type VI Secretion System Coordinately Regulate Escherichia coli K1 Interaction with Human Brain Microvascular Endothelial Cells. Inf. Immun. 2012, 80, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, J.; Zhou, Y.; Tang, X. Prospecting the plant growth—promoting activities of endophytic bacteria Franconibacter sp. YSD YN2 isolated from Cyperus esculentus L. var. sativus leaves. Ann. Microbiol. 2022, 72, 1. [Google Scholar] [CrossRef]
- Aly, M.A.; Reimhult, E.; Kneifel, W.; Domig, K.J. Characterization of biofilm formation by Cronobacter spp. isolates of different food origin under model conditions. J. Food Prot. 2019, 82, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, F.Y.; Darcan, C.; Kariptaş, E. The Determination, Monitoring, Molecular Mechanisms and Formation of Biofilm in E. coli. Braz. J. Microbiol. 2023, 54, 259–277. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 2 August 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Maiden, M.C.J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. Available online: https://www.biomedcentral.com/1471-2105/11/595 (accessed on 2 August 2024). [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
| Feature | C. condimenti s37 |
|---|---|
| Genome size (bp) | 4,590,991 |
| GC content (%) | 55.7 |
| Number of genes (total) | 4384 |
| Total number of RNA | 96 |
| Locus | Product | Isolates | Isolate Count |
|---|---|---|---|
| AFK62_RS01850 | efflux RND transporter permease subunit | Cronobacter sakazakii a Cronobacter malonaticus b Cronobacter turicensis c Cronobacter muytjensii d Cronobacter dublinensis e Cronobacter condimenti f Cronobacter condimenti s37 Cronobacter universalis g Franconibacter helveticus h Franconibacter pulveris Franconibacter daqui i Siccibacter colletis j Siccibacter turicensis k | 13 |
| AFK62_RS03000 | organic hydroperoxide resistance protein | ||
| AFK62_RS05310 | multidrug efflux RND transporter permease subunit AcrB | ||
| AFK62_RS09870 | general stress protein | ||
| AFK62_RS11360 | L-serine ammonia-lyase | ||
| AFK62_RS11405 | transcription antiterminator/RNA stability regulator CspE | ||
| AFK62_RS15790 | L-serine ammonia-lyase | ||
| AFK62_RS16240 | transketolase | ||
| AFK62_RS17895 | maltose/maltodextrin ABC transporter ATP-binding protein MalK | ||
| AFK62_RS08905 | PTS sugar transporter subunit IIC | Cronobacter sakazakii a Cronobacter malonaticus b Cronobacter turicensis c Cronobacter muytjensii d Cronobacter dublinensis e Cronobacter condimenti f Cronobacter condimenti s37 Cronobacter universalis g Siccibacter colletis j Siccibacter turicensis k | 10 |
| AFK62_RS11020 | contact-dependent growth inhibition system immunity protein | Cronobacter dublinensis e Cronobacter condimenti f Cronobacter condimenti s37 | 3 |
| AFK62_RS11030 | contact-dependent growth inhibition system immunity protein | Cronobacter dublinensis e Cronobacter condimenti f Cronobacter condimenti s37 | 3 |
| AFK62_RS22640 | hypothetical protein | Cronobacter dublinensis e Cronobacter condimenti f Cronobacter condimenti s37 | 3 |
| AFK62_RS05535 | hypothetical protein | Cronobacter condimenti f Cronobacter condimenti s37 | 2 |
| AFK62_RS05550 | hypothetical protein | Cronobacter condimenti f Cronobacter condimenti s37 | 2 |
| Antimicrobial Resistance Gene (AMR Gene) | AMR Gene Family | Drug Class | Resistance Mechanism | Identity of Matching Region (%) | Length of Reference Sequence (%) |
|---|---|---|---|---|---|
| vanG | glycopeptide resistance gene cluster, D-alanine--D-alanine ligase | glycopeptide antibiotic | antibiotic target alteration | 36.59 | 104.87 |
| msbA | ATP-binding cassette (ABC) antibiotic efflux pump | nitroimidazole antibiotic | antibiotic efflux | 92.27 | 100.00 |
| qacJ | small multidrug resistance (SMR) antibiotic efflux pump | disinfecting agents and antiseptics | antibiotic efflux | 45.63 | 101.87 |
| marA | resistance-nodulation-cell division (RND) antibiotic efflux pump, General Bacterial Porin with reduced permeability to beta-lactams | fluoroquinolone antibiotic, monobactam, carbapenem, cephalosporin, glycylcycline, cephamycin, penam, tetracycline antibiotic, rifamycin antibiotic, phenicol antibiotic, penem, disinfecting agents, and antiseptics | antibiotic efflux, reduced permeability to antibiotic | 89.52 | 97.64 |
| Klebsiella pneumoniae kpnF | small multidrug resistance (SMR) antibiotic efflux pump | macrolide antibiotic, aminoglycoside antibiotic, cephalosporin, tetracycline antibiotic, peptide antibiotic, rifamycin antibiotic, disinfecting agents, and antiseptics | antibiotic efflux | 73.39 | 100.00 |
| Klebsiella pneumoniae kpnE | small multidrug resistance (SMR) antibiotic efflux pump | macrolide antibiotic, aminoglycoside antibiotic, cephalosporin, tetracycline antibiotic, peptide antibiotic, rifamycin antibiotic, disinfecting agents, and antiseptics | antibiotic efflux | 73.11 | 99.17 |
| fosA8 | fosfomycin thiol transferase | phosphonic acid antibiotic | antibiotic inactivation | 58.82 | 95.74 |
| H-NS | major facilitator superfamily (MFS) antibiotic efflux pump, resistance-nodulation-cell division (RND) antibiotic efflux pump | macrolide antibiotic, fluoroquinolone antibiotic, cephalosporin, cephamycin, penam, tetracycline antibiotic | antibiotic efflux | 91.91 | 100.00 |
| adeF | resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, tetracycline antibiotic | antibiotic efflux | 41.7 | 98.02 |
| emrR | major facilitator superfamily (MFS) antibiotic efflux pump | fluoroquinolone antibiotic | antibiotic efflux | 91.43 | 100.00 |
| Klebsiella pneumoniae kpnH | major facilitator superfamily (MFS) antibiotic efflux pump | macrolide antibiotic, fluoroquinolone antibiotic, aminoglycoside antibiotic, carbapenem, cephalosporin, penam, peptide antibiotic, penem | antibiotic efflux | 92.19 | 100.00 |
| rsmA | resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, diaminopyrimidine antibiotic, phenicol antibiotic | antibiotic efflux | 85.25 | 100.00 |
| CRP | resistance-nodulation-cell division (RND) antibiotic efflux pump | macrolide antibiotic, fluoroquinolone antibiotic, penam | antibiotic efflux | 99.05 | 100.00 |
| Escherichia coli EF-Tu mutants conferring resistance to Pulvomycin | elfamycin resistant EF-Tu | elfamycin antibiotic | antibiotic target alteration | 98.22 | 96.33 |
| Haemophilus influenzae PBP3 conferring resistance to beta-lactam antibiotics | Penicillin-binding protein mutations conferring resistance to beta-lactam antibiotics | cephalosporin, cephamycin, penam | antibiotic target alteration | 52.04 | 96.39 |
| Escherichia coli glpT with mutation conferring resistance to fosfomycin | antibiotic-resistant GlpT | phosphonic acid antibiotic | antibiotic target alteration | 92.43 | 99.56 |
| Escherichia coli AcrAB-TolC with MarR mutations conferring resistance to ciprofloxacin and tetracycline | resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, cephalosporin, glycylcycline, penam, tetracycline antibiotic, rifamycin antibiotic, phenicol antibiotic, disinfecting agents, and antiseptics | antibiotic target alteration, antibiotic efflux | 84.72 | 113.19 |
| Species | Strain | Source | MLST Sequence Type | PubMLST ID |
|---|---|---|---|---|
| Cronobacter sakazakii | ATCC BAA-894 | Powdered formula | 1 | 5 |
| Cronobacter malonaticus | LMG 23826T | Clinical | 7 | 61 |
| Cronobacter turicensis | LMG23827T | Clinical | 19 | 110 |
| Cronobacter muytjensii | CIP 103581 | Clinical | 81 | 598 |
| Cronobacter dublinensis | LMG 23823T | Environmental | 106 | 146 |
| Cronobacter condimenti | LMG 26250T | Food | 98 | 1554 |
| Cronobacter condimenti | s37 | Small radish sprouts | 98 | 1896 |
| Cronobacter universalis | NCTC 9529T | Water | 54 | 84 |
| Franconibacter helveticus | LMG 23733 | Fruit powder | 217 | 643 |
| Franconibacter pulveris | Fp-G-601/05T | Fruit powder | 231 | 611 |
| Franconibacter daqui | LMG 29914T | Food ingredient | 766 | 3273 |
| Siccibacter colletis | NCTC 14934T | Food ingredient | 227 | 1165 |
| Siccibacter turicensis | LMG 23730T | Fruit powder | 216 | 1179 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berthold-Pluta, A.; Stefańska, I.; Forsythe, S.; Aleksandrzak-Piekarczyk, T.; Stasiak-Różańska, L.; Garbowska, M. Genomic Analysis of Cronobacter condimenti s37: Identification of Resistance and Virulence Genes and Comparison with Other Cronobacter and Closely Related Species. Int. J. Mol. Sci. 2024, 25, 8622. https://doi.org/10.3390/ijms25168622
Berthold-Pluta A, Stefańska I, Forsythe S, Aleksandrzak-Piekarczyk T, Stasiak-Różańska L, Garbowska M. Genomic Analysis of Cronobacter condimenti s37: Identification of Resistance and Virulence Genes and Comparison with Other Cronobacter and Closely Related Species. International Journal of Molecular Sciences. 2024; 25(16):8622. https://doi.org/10.3390/ijms25168622
Chicago/Turabian StyleBerthold-Pluta, Anna, Ilona Stefańska, Stephen Forsythe, Tamara Aleksandrzak-Piekarczyk, Lidia Stasiak-Różańska, and Monika Garbowska. 2024. "Genomic Analysis of Cronobacter condimenti s37: Identification of Resistance and Virulence Genes and Comparison with Other Cronobacter and Closely Related Species" International Journal of Molecular Sciences 25, no. 16: 8622. https://doi.org/10.3390/ijms25168622
APA StyleBerthold-Pluta, A., Stefańska, I., Forsythe, S., Aleksandrzak-Piekarczyk, T., Stasiak-Różańska, L., & Garbowska, M. (2024). Genomic Analysis of Cronobacter condimenti s37: Identification of Resistance and Virulence Genes and Comparison with Other Cronobacter and Closely Related Species. International Journal of Molecular Sciences, 25(16), 8622. https://doi.org/10.3390/ijms25168622






