Apolipoproteine and KLOTHO Gene Variants Do Not Affect the Penetrance of Fragile X-Associated Tremor/Ataxia Syndrome
Abstract
1. Introduction
2. Results
2.1. FXTAS Stage by APOε4 and KLOTHO Variant Genotypes
2.2. FXTAS Diagnosis by APOε4 and KLOTHO Variant Genotype
2.3. FXTAS Stage and Diagnosis by APOε2 and ε4 Alleles
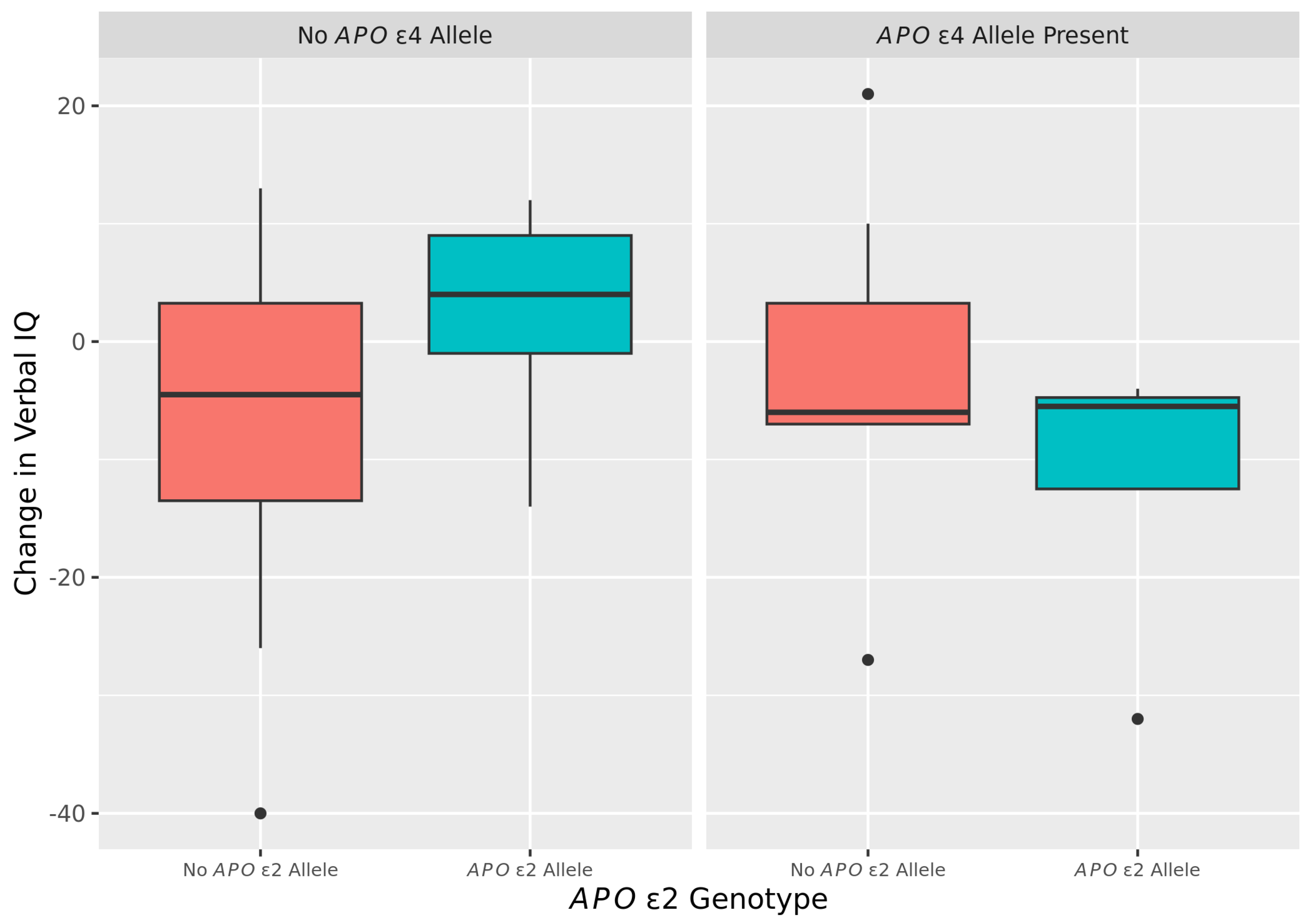
2.4. Changes Overtime in VIQ by APOε2 and ε4 Among FXTAS Subjects
2.5. No Changes Overtime in PIQ and FSIQ by APOε2 and ε4 Alleles among FXTAS Subjects
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Molecular Measures
4.3. Clinical Measures
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tassone, F.; Protic, D.; Allen, E.G.; Archibald, A.D.; Baud, A.; Brown, T.W.; Budimirovic, D.B.; Cohen, J.; Dufour, B.; Eiges, R.; et al. Insight and Recommendations for Fragile X-Premutation-Associated Conditions from the Fifth International Conference on FMR1 Premutation. Cells 2023, 12, 2330. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, R.J.; Protic, D.; Rajaratnam, A.; Salcedo-Arellano, M.J.; Aydin, E.Y.; Schneider, A. Fragile X-Associated Neuropsychiatric Disorders (FXAND). Front. Psychiatry 2018, 9, 564. [Google Scholar] [CrossRef] [PubMed]
- Jacquemont, S.; Hagerman, R.J.; Leehey, M.A.; Hall, D.A.; Levine, R.A.; Brunberg, J.A.; Zhang, L.; Jardini, T.; Gane, L.W.; Harris, S.W.; et al. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA 2004, 291, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, R.J.; Leehey, M.; Heinrichs, W.; Tassone, F.; Wilson, R.; Hills, J.; Grigsby, J.; Gage, B.; Hagerman, P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 2001, 57, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.T.; Han, Y.D.; Cong, L.; Liu, C.C.; Liang, X.Y.; Xue, F.Z.; Du, Y.F. Apolipoprotein E Facilitates Amyloid-β Oligomer-Induced Tau Phosphorylation. J. Alzheimer’s Dis. 2020, 74, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Strittmatter, W.J.; Saunders, A.M.; Schmechel, D.; Pericak-Vance, M.; Enghild, J.; Salvesen, G.S.; Roses, A.D. Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA 1993, 90, 1977–1981. [Google Scholar] [CrossRef] [PubMed]
- Insel, P.S.; Hansson, O.; Mattsson-Carlgren, N. Association Between Apolipoprotein E ε2 vs ε4, Age, and β-Amyloid in Adults without Cognitive Impairment. JAMA Neurol. 2021, 78, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, T.E.; Huey, E.D.; Devanand, D.P. Association of APOE e2 genotype with Alzheimer’s and non-Alzheimer’s neurodegenerative pathologies. Nat. Commun. 2020, 11, 4727. [Google Scholar] [CrossRef]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; van Duijn, C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef]
- ALZGENE. Meta-Analysis of All Published AD Association Studies (Case-Control Only) APOE_e2/3/4. 2010. Available online: http://www.alzgene.org/meta.asp?geneID=83 (accessed on 21 October 2023).
- van der Flier, W.M.; Pijnenburg, Y.A.; Schoonenboom, S.N.; Dik, M.G.; Blankenstein, M.A.; Scheltens, P. Distribution of APOE genotypes in a memory clinic cohort. Dement. Geriatr. Cogn. Disord. 2008, 25, 433–438. [Google Scholar] [CrossRef]
- Kuro-o, M. Klotho and the aging process. Korean J. Intern. Med. 2011, 26, 113–122. [Google Scholar] [CrossRef]
- Kuro-o, M.; Matsumura, Y.; Aizawa, H.; Kawaguchi, H.; Suga, T.; Utsugi, T.; Ohyama, Y.; Kurabayashi, M.; Kaname, T.; Kume, E.; et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 1997, 390, 45–51. [Google Scholar] [CrossRef]
- Neitzel, J.; Franzmeier, N.; Rubinski, A.; Dichgans, M.; Brendel, M.; Weiner, M.; Aisen, P.; Petersen, R.; Jack, C.R.; Jagust, W.; et al. KL-VS heterozygosity is associated with lower amyloid-dependent tau accumulation and memory impairment in Alzheimer’s disease. Nat. Commun. 2021, 12, 3825. [Google Scholar] [CrossRef]
- Belloy, M.E.; Napolioni, V.; Han, S.S.; Le Guen, Y.; Greicius, M.D.; Alzheimer’s Disease Neuroimaging Initiative. Association of Klotho-VS Heterozygosity with Risk of Alzheimer Disease in Individuals Who Carry APOE4. JAMA Neurol. 2020, 77, 849–862. [Google Scholar] [CrossRef]
- Silva, F.; Rodriguez-Revenga, L.; Madrigal, I.; Alvarez-Mora, M.I.; Oliva, R.; Milà, M. High apolipoprotein E4 allele frequency in FXTAS patients. Genet. Med. 2013, 15, 639–642. [Google Scholar] [CrossRef]
- Cabal-Herrera, A.M.; Tassanakijpanich, N.; Salcedo-Arellano, M.J.; Hagerman, R.J. Fragile X-Associated Tremor/Ataxia Syndrome (FXTAS): Pathophysiology and Clinical Implications. Int. J. Mol. Sci. 2020, 21, 4391. [Google Scholar] [CrossRef]
- Seritan, A.L.; Nguyen, D.V.; Farias, S.T.; Hinton, L.; Grigsby, J.; Bourgeois, J.A.; Hagerman, R.J. Dementia in fragile X-associated tremor/ataxia syndrome (FXTAS): Comparison with Alzheimer’s disease. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2008, 147B, 1138–1144. [Google Scholar] [CrossRef]
- Grigsby, J.; Cornish, K.; Hocking, D.; Kraan, C.; Olichney, J.M.; Rivera, S.M.; Schneider, A.; Sherman, S.; Wang, J.Y.; Yang, J.-C. The cognitive neuropsychological phenotype of carriers of the FMR1 premutation. J. Neurodev. Disord. 2014, 6, 28. [Google Scholar] [CrossRef]
- Butler, C.; Zeman, A.Z. Neurological syndromes which can be mistaken for psychiatric conditions. J. Neurol. Neurosurg. Psychiatry 2005, 76 (Suppl. S1), i31–i38. [Google Scholar] [CrossRef]
- Kumar, A.; Sidhu, J.; Goyal, A.; Tsao, J.W. Alzheimer Disease. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2023. [Google Scholar]
- Arking, D.E.; Atzmon, G.; Arking, A.; Barzilai, N.; Dietz, H.C. Association between a functional variant of the KLOTHO gene and high-density lipoprotein cholesterol, blood pressure, stroke, and longevity. Circ. Res. 2005, 96, 412–418. [Google Scholar] [CrossRef]
- Zhao, Y.; Zeng, C.-Y.; Li, X.-H.; Yang, T.-T.; Kuang, X.; Du, J.-R. Klotho overexpression improves amyloid-β clearance and cognition in the APP/PS1 mouse model of Alzheimer’s disease. Aging Cell 2020, 19, e13239. [Google Scholar] [CrossRef]
- Faber, J.; Fonseca, L.M. How sample size influences research outcomes. Dent. Press J. Orthod. 2014, 19, 27–29. [Google Scholar] [CrossRef]
- Tipton, E.; Hallberg, K.; Hedges, L.V.; Chan, W. Implications of Small Samples for Generalization: Adjustments and Rules of Thumb. Eval. Rev. 2017, 41, 472–505. [Google Scholar] [CrossRef]
- Schneider, A.; Summers, S.; Tassone, F.; Seritan, A.; Hessl, D.; Hagerman, P.; Hagerman, R. Women with Fragile X-associated Tremor/Ataxia Syndrome. Mov. Disord. Clin. Pr. 2020, 7, 910–919. [Google Scholar] [CrossRef]
- Jacquemont, S.; Hagerman, R.J.; Leehey, M.; Grigsby, J.; Zhang, L.; Brunberg, J.A.; Greco, C.; Des Portes, V.; Jardini, T.; Levine, R.; et al. Fragile X premutation tremor/ataxia syndrome: Molecular, clinical, and neuroimaging correlates. Am. J. Hum. Genet. 2003, 72, 869–878. [Google Scholar] [CrossRef]
- Bourgeois, J.A.; Cogswell, J.B.; Hessl, D.; Zhang, L.; Ono, M.Y.; Tassone, F.; Farzin, F.; Brunberg, J.A.; Grigsby, J.; Hagerman, R.J. Cognitive, anxiety and mood disorders in the fragile X-associated tremor/ataxia syndrome. Gen. Hosp. Psychiatry 2007, 29, 349–356. [Google Scholar] [CrossRef][Green Version]
- Manly, J.J.; Jones, R.N.; Langa, K.M.; Ryan, L.H.; Levine, D.A.; McCammon, R.; Heeringa, S.G.; Weir, D. Estimating the Prevalence of Dementia and Mild Cognitive Impairment in the US: The 2016 Health and Retirement Study Harmonized Cognitive Assessment Protocol Project. JAMA Neurol. 2022, 79, 1242–1249. [Google Scholar] [CrossRef]
- Błaszczyk, J.W. Pathogenesis of Dementia. Int. J. Mol. Sci. 2023, 24, 543. [Google Scholar] [CrossRef]
- Paroni, G.; Bisceglia, P.; Seripa, D. Understanding the Amyloid Hypothesis in Alzheimer’s Disease. J. Alzheimer’s Dis. 2019, 68, 493–510. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, L.; Jiao, B. Cognitive Dysfunction in Repeat Expansion Diseases: A Review. Front. Aging Neurosci. 2022, 14, 841711. [Google Scholar] [CrossRef]
- Leehey, M.A.; Berry-Kravis, E.; Goetz, C.G.; Zhang, L.; Hall, D.A.; Li, L.; Rice, C.D.; Lara, R.; Cogswell, J.; Reynolds, A.; et al. FMR1 CGG repeat length predicts motor dysfunction in premutation carriers. Neurology 2008, 70, 1397–1402. [Google Scholar] [CrossRef]
- Tassone, F.; Adams, J.; Berry-Kravis, E.M.; Cohen, S.S.; Brusco, A.; Leehey, M.A.; Li, L.; Hagerman, R.J.; Hagerman, P.J. CGG repeat length correlates with age of onset of motor signs of the fragile X-associated tremor/ataxia syndrome (FXTAS). Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2007, 144B, 566–569. [Google Scholar] [CrossRef]
- Greco, C.M.; Berman, R.F.; Martin, R.M.; Tassone, F.; Schwartz, P.H.; Chang, A.; Trapp, B.D.; Iwahashi, C.; Brunberg, J.; Grigsby, J.; et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS). Brain 2006, 129, 243–255. [Google Scholar] [CrossRef]
- Sevin, M.; Kutalik, Z.; Bergman, S.; Vercelletto, M.; Renou, P.; Lamy, E.; Vingerhoets, F.J.; Di Virgilio, G.; Boisseau, P.; Bezieau, S.; et al. Penetrance of marked cognitive impairment in older male carriers of the FMR1 gene premutation. J. Med. Genet. 2009, 46, 818–824. [Google Scholar] [CrossRef]
- Seritan, A.L.; Kim, K.; Benjamin, I.; Seritan, I.; Hagerman, R.J. Risk Factors for Cognitive Impairment in Fragile X-Associated Tremor/Ataxia Syndrome. J. Geriatr. Psychiatry Neurol. 2016, 29, 328–337. [Google Scholar] [CrossRef]
- Gane, L.W.; Iosif, A.-M.; Flynn-Wilson, L.; Venturino, M.; Hagerman, R.J.; Sertian, A.L. Assessment of patient and caregiver needs in fragile X-associaetd tremor/ataxia syndrome (FXTAS) by utilizing Q-sort methodology. Aging Ment. Health 2010, 14, 1000–1007. [Google Scholar] [CrossRef]
- Almeida, O.P.; Hankey, G.J.; Yeap, B.B.; Golledge, J.; Flicker, L. Depression as a modifiable factor to decrease the risk of dementia. Transl. Psychiatry 2017, 7, e1117. [Google Scholar] [CrossRef]
- Bourgeois, J.A.; Seritan, A.L.; Casillas, E.M.; Hessl, D.; Schneider, A.; Yang, Y.; Kaur, I.; Cogswell, J.B.; Nguyen, D.V.; Hagerman, R.J. Lifetime prevalence of mood and anxiety disorders in fragile X premutation carriers. J. Clin. Psychiatry 2011, 72, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, R.; Hagerman, P. Fragile X-associated tremor/ataxia syndrome: Pathophysiology and management. Curr. Opin. Neurol. 2021, 34, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Winarni, T.I.; Chonchaiya, W.; Sumekar, T.A.; Ashwood, P.; Morales, G.M.; Tassone, F.; Nguyen, D.V.; Faradz, S.M.; Van de Water, J.; Cook, K.; et al. Immune-mediated disorders among women carriers of fragile X premutation alleles. Am. J. Med. Genet. Part A 2012, 158A, 2473–2481. [Google Scholar] [CrossRef] [PubMed]
- Hamlin, A.A.; Sukharev, D.; Campos, L.; Mu, Y.; Tassone, F.; Hessl, D.; Nguyen, D.V.; Loesch, D.; Hagerman, R.J. Hypertension in FMR1 premutation males with and without fragile X-associated tremor/ataxia syndrome (FXTAS). Am. J. Med. Genet. Part A 2012, 158A, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Knopman, D.S. Cerebrovascular disease and dementia. Br. J. Radiol. 2007, 80, S121–S127. [Google Scholar] [CrossRef] [PubMed]
- Schmechel, D.E.; Saunders, A.M.; Strittmatter, W.J.; Crain, B.J.; Hulette, C.M.; Joo, S.H.; Pericak-Vance, M.A.; Goldgaber, D.; Roses, A.D. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc. Natl. Acad. Sci. USA 1993, 90, 9649–9653. [Google Scholar] [CrossRef] [PubMed]
- Sweigart, B.; Andersen, S.L.; Gurinovich, A.; Cosentino, S.; Schupf, N.; Perls, T.T.; Sebastiani, P. APOE E2/E2 Is Associated with Slower Rate of Cognitive Decline with Age. J. Alzheimer’s Dis. 2021, 83, 853–860. [Google Scholar] [CrossRef]
- Goedert, M.; Ghetti, B.; Spillantini, M.G. Frontotemporal dementia: Implications for understanding Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006254. [Google Scholar] [CrossRef] [PubMed]
- Bacalman, S.; Farzin, F.; Bourgeois, J.A.; Cogswell, J.; Goodlin-Jones, B.L.; Gane, L.W.; Grigsby, J.; Leehey, M.A.; Tassone, F.; Hagerman, R.J. Psychiatric phenotype of the fragile X-associated tremor/ataxia syndrome (FXTAS) in males: Newly described fronto-subcortical dementia. J. Clin. Psychiatry 2006, 67, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Filley, C.M.; Brown, M.S.; Onderko, K.; Ray, M.; Bennett, R.E.; Berry-Kravis, E.; Grigsby, J. White matter disease and cognitive impairment in FMR1 premutation carriers. Neurology 2015, 84, 2146–2152. [Google Scholar] [CrossRef] [PubMed]
- Caixeta, L. What matters in white matter dementia? Dement. Neuropsychol. 2007, 1, 131–139. [Google Scholar] [CrossRef]
- Tassone, F.; Pan, R.; Amiri, K.; Taylor, A.K.; Hagerman, P.J. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J. Mol. Diagn. JMD 2008, 10, 43–49. [Google Scholar] [CrossRef]
- Filipovic-Sadic, S.; Sah, S.; Chen, L.; Krosting, J.; Sekinger, E.; Zhang, W.; Hagerman, P.J.; Stenzel, T.T.; Hadd, A.G.; Latham, G.J.; et al. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin. Chem. 2010, 56, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Yrigollen, C.M.; Durbin-Johnson, B.; Gane, L.; Nelson, D.L.; Hagerman, R.; Hagerman, P.J.; Tassone, F. AGG interruptions within the maternal FMR1 gene reduce the risk of offspring with fragile X syndrome. Genet. Med. Off. J. Am. Coll. Med. Genet. 2012, 14, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Drozdick, L.W.; Wahlstrom, D.; Zhu, J.; Weiss, L.G. The Wechsler Adult Intelligence Scale—Fourth Edition. In Contemporary Intellectual Assessment: Theories, Tests, and Issues; Flanagan, D.P., McDonough, E.M., Eds.; The Guilford Press: New York, NY, USA, 2018; pp. 486–511. [Google Scholar]
- McCullagh, P. Regression Models for Ordinal Data. J. R. Stat. Soc. Ser. B (Methodol.) 1980, 42, 109–142. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2022; Available online: https://www.R-project.org (accessed on 23 June 2022).
| Characteristics | Overall (n = 245) |
|---|---|
| Age | |
| Mean (SD) | 66.1 (7.93) |
| Median [Min, Max] | 66.0 [50.0, 89.0] |
| Race | |
| American Indian/Alaska Native | 3 (1.2%) |
| Asian | 1 (0.4%) |
| More Than One Race | 1 (0.4%) |
| Unknown | 34 (13.9%) |
| White | 206 (84.1%) |
| Ethnicity | |
| Hispanic or Latino | 10 (4.1%) |
| Non-Hispanic or Latino | 180 (73.5%) |
| Unknown | 55 (22.4%) |
| CGG repeats | |
| Mean (SD) | 86.9 (18.8) |
| Median [Min, Max] | 84.0 [52.0, 183] |
| Missing | 1 (0.4%) |
| AGG Interruptions | |
| 0 | 122 (49.8%) |
| 1 | 76 (31.0%) |
| 2 | 47 (19.2%) |
| APOε | |
| ε2,ε2 | 3 (1.2%) |
| ε2,ε3 | 25 (10.2%) |
| ε2,ε4 | 6 (2.4%) |
| ε3,ε3 | 163 (66.5%) |
| ε3,ε4 | 47 (19.2%) |
| ε4,ε4 | 1 (0.4%) |
| KLOTHO | |
| KL-VShet− | 166 (67.8%) |
| KL-VShet+ | 69 (28.2%) |
| KL-hom+ | 10 (4.1%) |
| FXTAS Stage | |
| 0 | 14 (5.7%) |
| 1 | 20 (8.2%) |
| 2 | 44 (18.0%) |
| 3 | 64 (26.1%) |
| 4 | 39 (15.9%) |
| 5 | 18 (7.3%) |
| Missing | 46 (18.8%) |
| FXTAS Diagnosis | |
| No | 25 (10.2%) |
| Possible | 34 (13.9%) |
| Probable | 37 (15.1%) |
| Definite | 103 (42.0%) |
| Missing | 46 (18.8%) |
| FXTAS (n = 165) | No FXTAS (n = 34) | No Diagnosis (n = 46) | Overall (n = 245) | |
|---|---|---|---|---|
| Age | ||||
| Mean (SD) | 66.2 (7.44) | 63.3 (6.40) | 67.8 (10.0) | 66.1 (7.93) |
| Median [Min, Max] | 66.0 [51.0, 85.0] | 63.0 [50.0, 77.0] | 68.0 [50.0, 89.0] | 66.0 [50.0, 89.0] |
| Race | ||||
| American Indian/Alaska Native | 2 (1.2%) | 1 (2.9%) | 0 (0%) | 3 (1.2%) |
| Asian | 1 (0.6%) | 0 (0%) | 0 (0%) | 1 (0.4%) |
| More Than One Race | 1 (0.6%) | 0 (0%) | 0 (0%) | 1 (0.4%) |
| Unknown | 14 (8.5%) | 1 (2.9%) | 19 (41.3%) | 34 (13.9%) |
| White | 147 (89.1%) | 32 (94.1%) | 27 (58.7%) | 206 (84.1%) |
| Ethnicity | ||||
| Hispanic or Latino | 7 (4.2%) | 0 (0%) | 3 (6.5%) | 10 (4.1%) |
| Non-Hispanic or Latino | 127 (77.0%) | 29 (85.3%) | 24 (52.2%) | 180 (73.5%) |
| Unknown | 31 (18.8%) | 5 (14.7%) | 19 (41.3%) | 55 (22.4%) |
| CGG | ||||
| Mean (SD) | 89.4 (18.6) | 77.5 (17.0) | 84.8 (18.4) | 86.9 (18.8) |
| Median [Min, Max] | 87.0 [52.0, 183] | 76.0 [53.0, 135] | 83.0 [53.0, 130] | 84.0 [52.0, 183] |
| Missing | 0 (0%) | 1 (2.9%) | 0 (0%) | 1 (0.4%) |
| AGG Interruptions | ||||
| 0 | 83 (50.3%) | 20 (57.1%) | 19 (42.2%) | 122 (49.8%) |
| 1 | 50 (30.3%) | 11 (31.4%) | 15 (33.3%) | 76 (31.0%) |
| 2 | 32 (19.4%) | 4 (11.4%) | 11 (24.4%) | 47 (19.2%) |
| APOε Genotype | ||||
| ε2,ε2 | 3 (1.8%) | 0 (0%) | 0 (0%) | 3 (1.2%) |
| ε2,ε3 | 17 (10.3%) | 5 (14.3%) | 3 (6.7%) | 25 (10.2%) |
| ε2,ε4 | 5 (3.0%) | 0 (0%) | 1 (2.2%) | 6 (2.4%) |
| ε3,ε3 | 111 (67.3%) | 24 (68.6%) | 28 (62.2%) | 163 (66.5%) |
| ε3,ε4 | 29 (17.6%) | 6 (17.1%) | 12 (26.7%) | 47 (19.2%) |
| ε4,ε4 | 0 (0%) | 0 (0%) | 1 (2.2%) | 1 (0.4%) |
| KLOTHO Genotype | ||||
| KL-VShet− | 110 (66.7%) | 25 (71.4%) | 31 (68.9%) | 166 (67.8%) |
| KL-VShet+ | 50 (30.3%) | 10 (28.6%) | 9 (20.0%) | 69 (28.2%) |
| KL-hom+ | 5 (3.0%) | 0 (0%) | 5 (11.1%) | 10 (4.1%) |
| APOε4 Genotype | Comparison | Odds Ratio (95% CI) | p-Value |
| No | (KL-VShet+ + or KL-VShom+) − (KL-VShet−) | 1.123 (0.612, 2.062) | 0.707 |
| Yes | (KL-VShet+ + or KL-VShom+) − (KL-VShet−) | 1.63 (0.348, 7.644) | 0.535 |
| KLOTHO Genotype | Comparison | Odds Ratio (95% CI) | p-Value |
| KL-VShet− | APOε4 allele present—No APOε4 allele | 1.396 (0.644, 3.027) | 0.398 |
| KL-VShet+ + or KL-VShom+ | APOε4 allele present—No APOε4 allele | 2.026 (0.46, 8.92) | 0.351 |
| APOε4 Genotype | Comparison | Odds Ratio (95% CI) | p-Value |
| No | (KL-VShet+ + or KL-VShom+) − (KL-VShet−) | 1.24 (0.616, 2.496) | 0.547 |
| Yes | (KL-VShet+ + or KL-VShom+) − (KL-VShet−) | 1.077 (0.107, 10.854) | 0.950 |
| KLOTHO Genotype | Comparison | Odds Ratio (95% CI) | p-Value |
| KL-VShet− | APOε4 allele present—No APOε4 allele | 1.239 (0.502, 3.058) | 0.642 |
| KL-VShet+ + or KL-VShom+ | APOε4 allele present—No APOε4 allele | 1.076 (0.112, 10.318) | 0.949 |
| APOε4 | Comparison | Odds Ratio (95% CI) | p-Value |
| No | APOε2 allele present—No APOε2 allele | 0.727 (0.311, 1.698) | 0.461 |
| Yes | APOε2 allele present—No APOε2 allele | 0.758 (0.149, 3.855) | 0.738 |
| APOε2 | Comparison | Odds Ratio (95% CI) | p-Value |
| No | APOε4 allele present—No APOε4 allele | 1.477 (0.702, 3.105) | 0.304 |
| Yes | APOε4 allele present—No APOε4 allele | 1.539 (0.288, 8.22) | 0.614 |
| APOε4 | Comparison | Odds Ratio (95% CI) | p-Value |
| No | APOε2 allele present—No APOε2 allele | 0.539 (0.218, 1.329) | 0.179 |
| Yes | APOε2 allele present—No APOε2 allele | 0.61 (0.097, 3.822) | 0.597 |
| APOε2 | Comparison | Odds Ratio (95% CI) | p-Value |
| No | APOε4 allele present—No APOε4 allele | 1.142 (0.451, 2.891) | 0.779 |
| Yes | APOε4 allele present—No APOε4 allele | 1.292 (0.206, 8.118) | 0.785 |
| APOε4 | Comparison | Difference in Means (95% CI) | p-Value |
| No | APOε2 allele present—No APOε2 allele | 8.4 (−11, 181) | 0.071 |
| Yes | APOε2 allele present—No APOε2 allele | −4.6 (−181, 91) | 0.496 |
| APOε2 | Comparison | Difference in Means (95% CI) | p-Value |
| No | APOε4 allele present—No APOε4 allele | 3.7 (−51, 121) | 0.399 |
| Yes | APOε4 allele present—No APOε4 allele | −9.3 (−231, 51) | 0.182 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winarni, T.I.; Hwang, Y.H.; Rivera, S.M.; Hessl, D.; Durbin-Johnson, B.P.; Utari, A.; Hagerman, R.; Tassone, F. Apolipoproteine and KLOTHO Gene Variants Do Not Affect the Penetrance of Fragile X-Associated Tremor/Ataxia Syndrome. Int. J. Mol. Sci. 2024, 25, 8103. https://doi.org/10.3390/ijms25158103
Winarni TI, Hwang YH, Rivera SM, Hessl D, Durbin-Johnson BP, Utari A, Hagerman R, Tassone F. Apolipoproteine and KLOTHO Gene Variants Do Not Affect the Penetrance of Fragile X-Associated Tremor/Ataxia Syndrome. International Journal of Molecular Sciences. 2024; 25(15):8103. https://doi.org/10.3390/ijms25158103
Chicago/Turabian StyleWinarni, Tri Indah, Ye Hyun Hwang, Susan M. Rivera, David Hessl, Blythe P. Durbin-Johnson, Agustini Utari, Randi Hagerman, and Flora Tassone. 2024. "Apolipoproteine and KLOTHO Gene Variants Do Not Affect the Penetrance of Fragile X-Associated Tremor/Ataxia Syndrome" International Journal of Molecular Sciences 25, no. 15: 8103. https://doi.org/10.3390/ijms25158103
APA StyleWinarni, T. I., Hwang, Y. H., Rivera, S. M., Hessl, D., Durbin-Johnson, B. P., Utari, A., Hagerman, R., & Tassone, F. (2024). Apolipoproteine and KLOTHO Gene Variants Do Not Affect the Penetrance of Fragile X-Associated Tremor/Ataxia Syndrome. International Journal of Molecular Sciences, 25(15), 8103. https://doi.org/10.3390/ijms25158103







