Genome-Wide Identification and Expression Analysis of ADK Gene Family Members in Cotton under Abiotic Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Sequence Analysis of Cotton ADK Genes
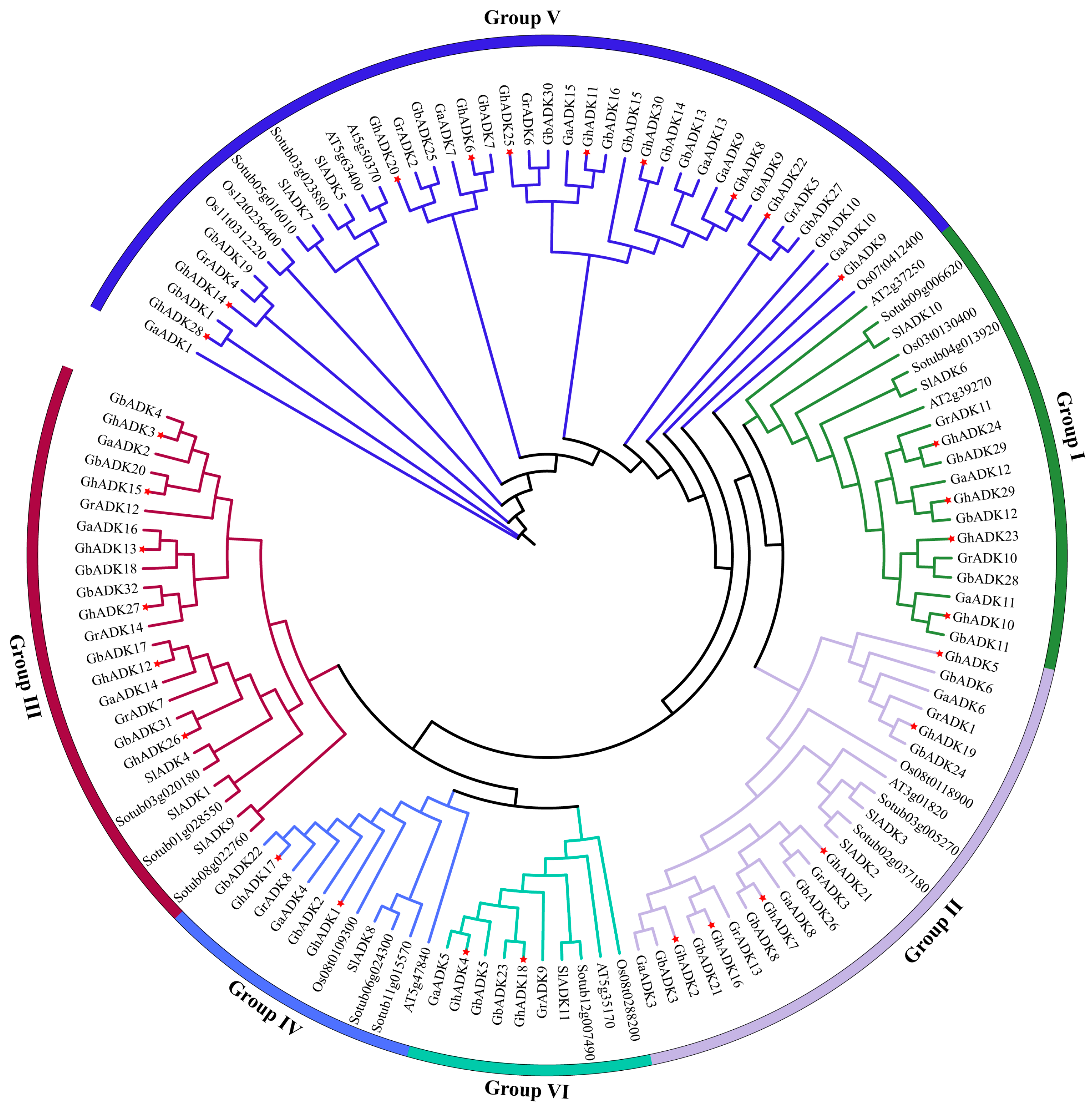
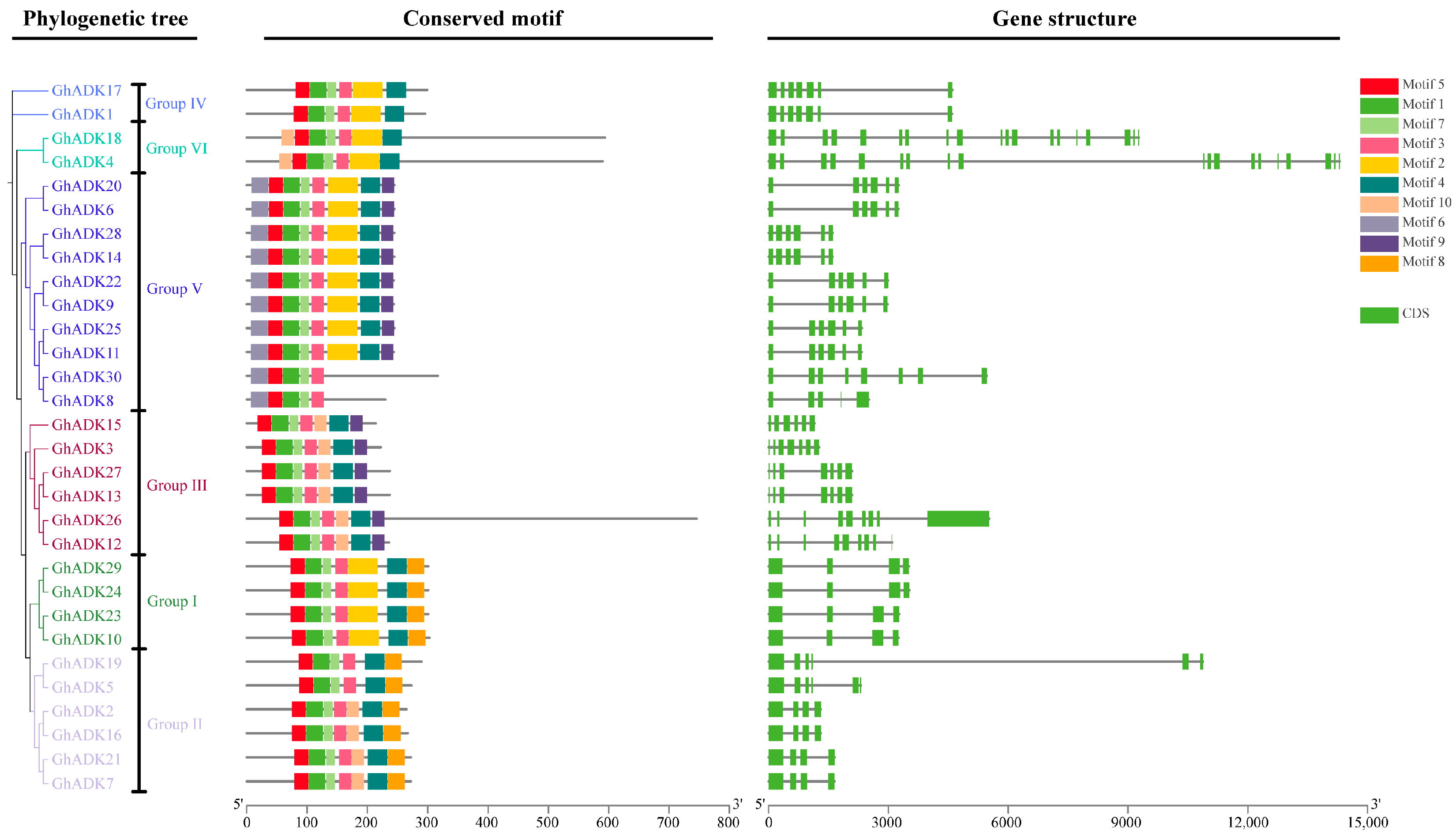
2.2. Phylogenetic Analysis and Classification of the GhADK Gene Family
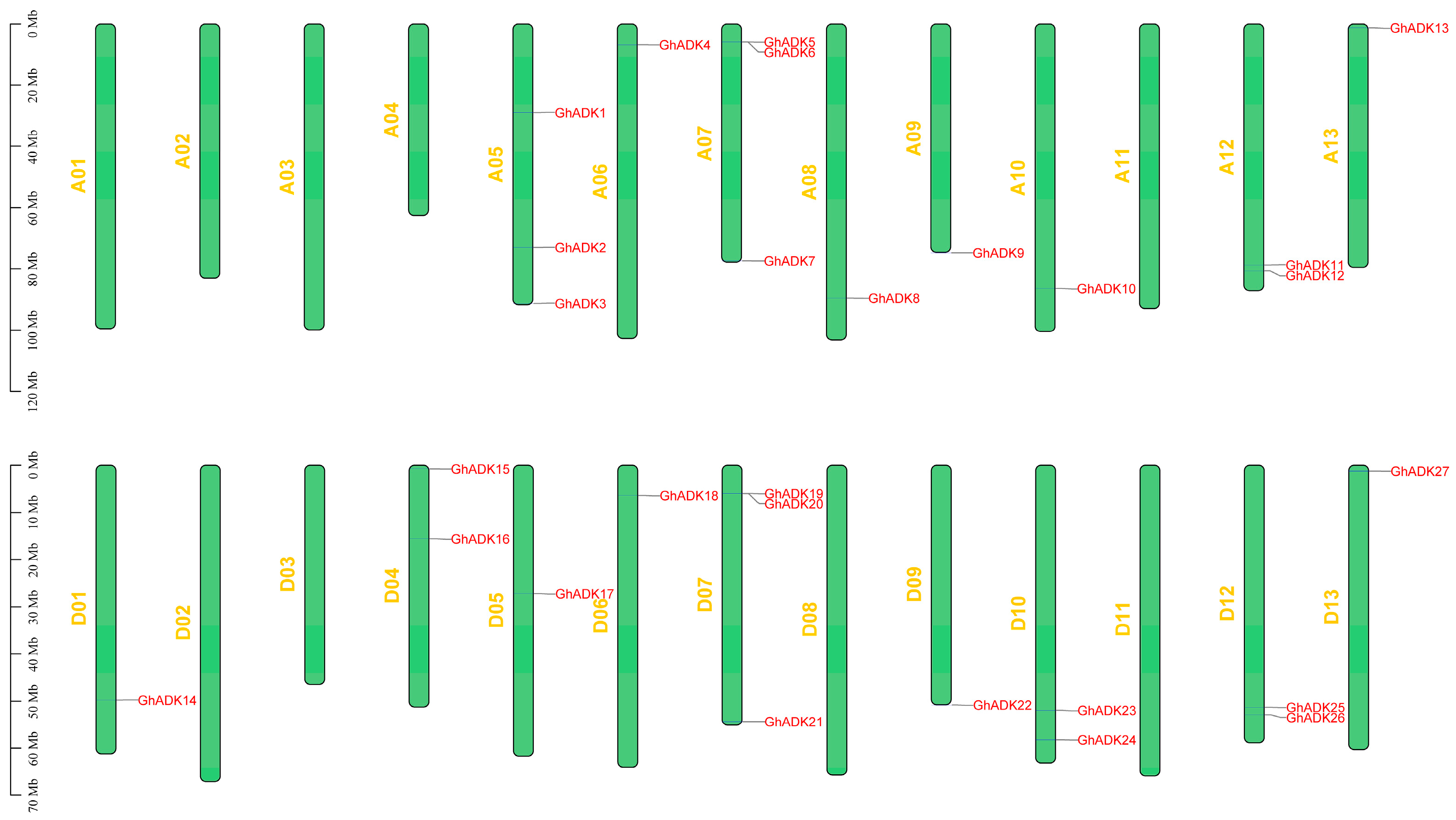
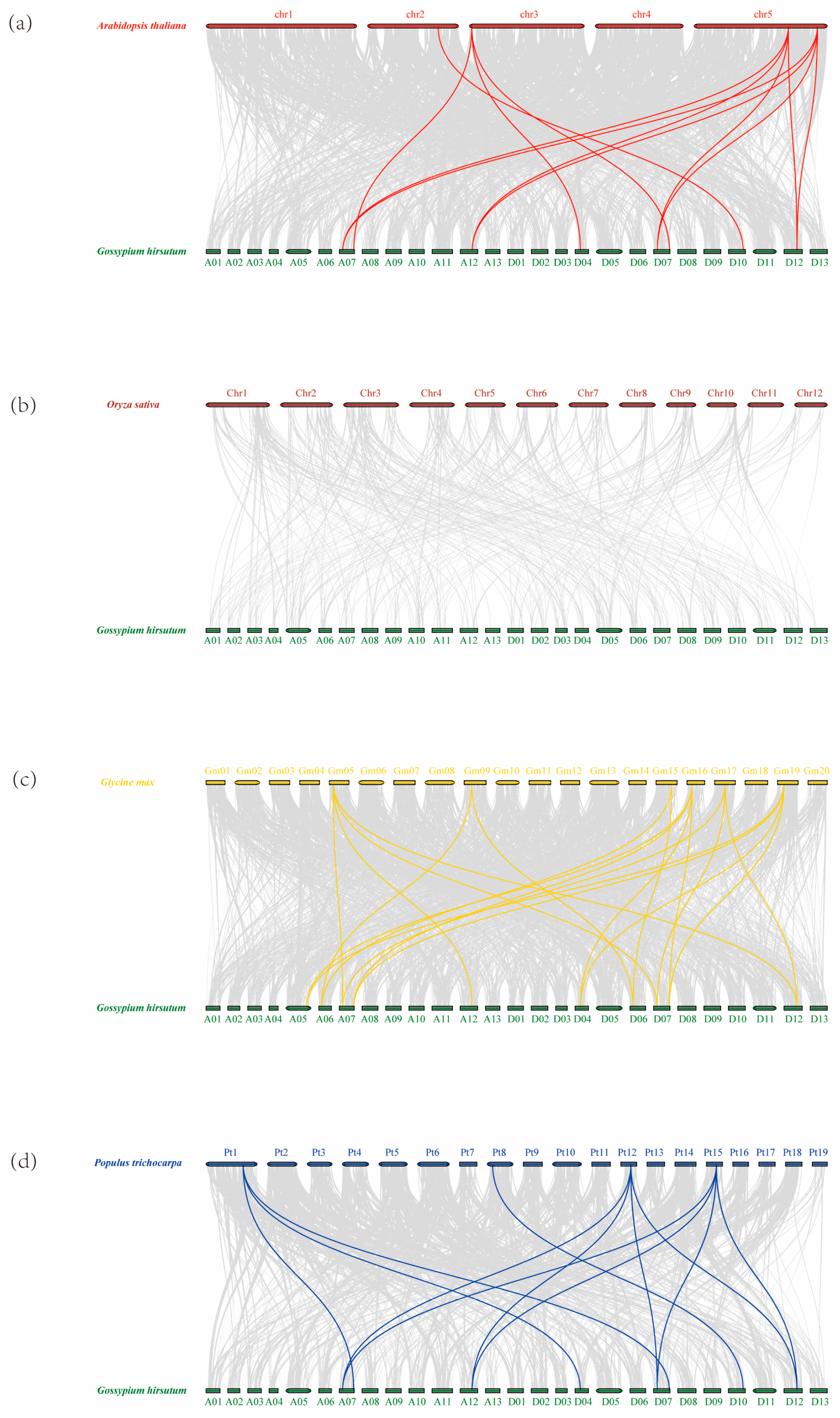
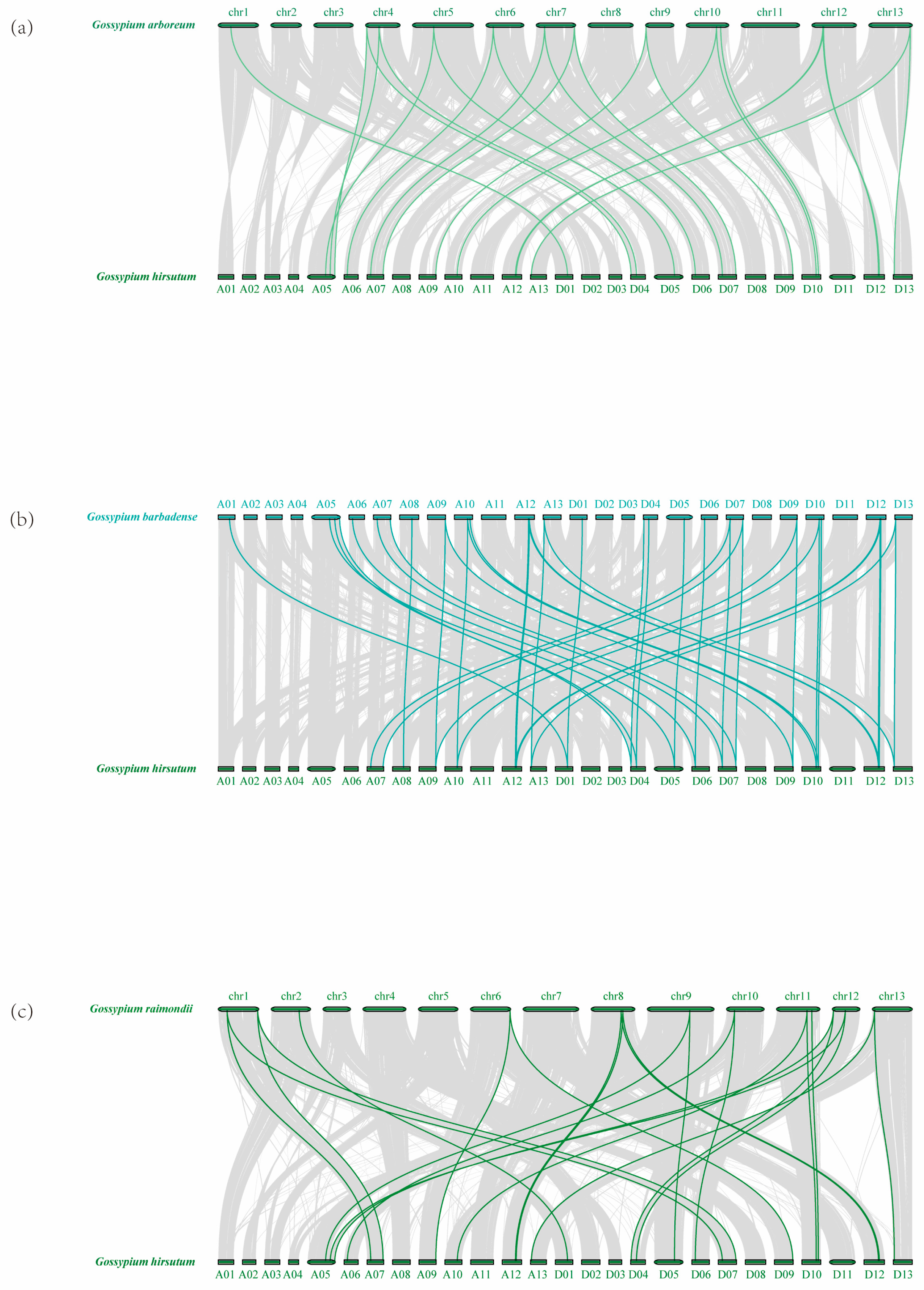
2.3. Chromosomal Mapping and Gene Duplication of the ADK Gene Family in G. hirsutum
2.4. Gene Structure Analysis and Conserved Motif Detection of Upland Cotton ADK Genes
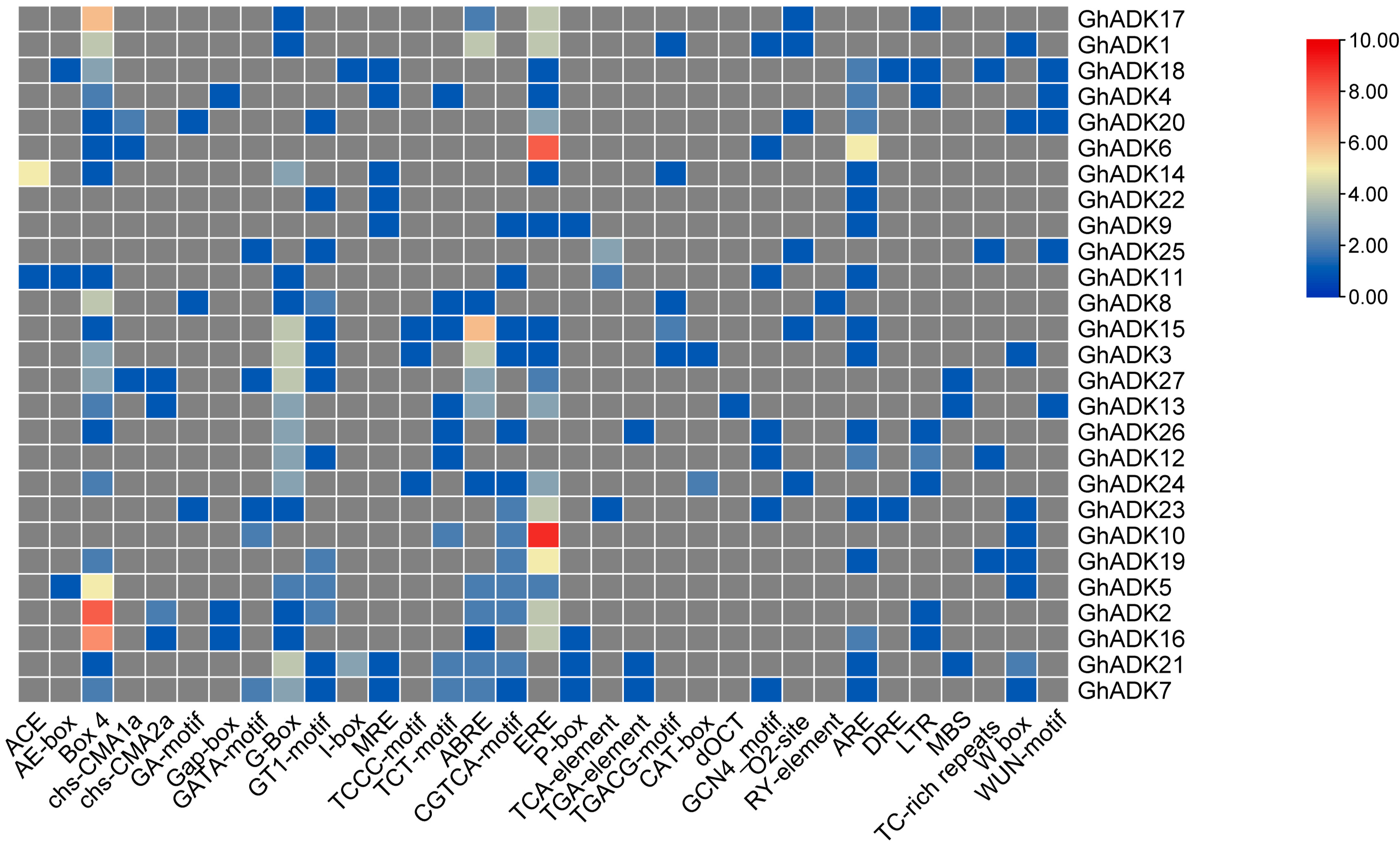
2.5. Analysis of Cis-Acting Elements of the G. hirsutum ADK Gene Promoter
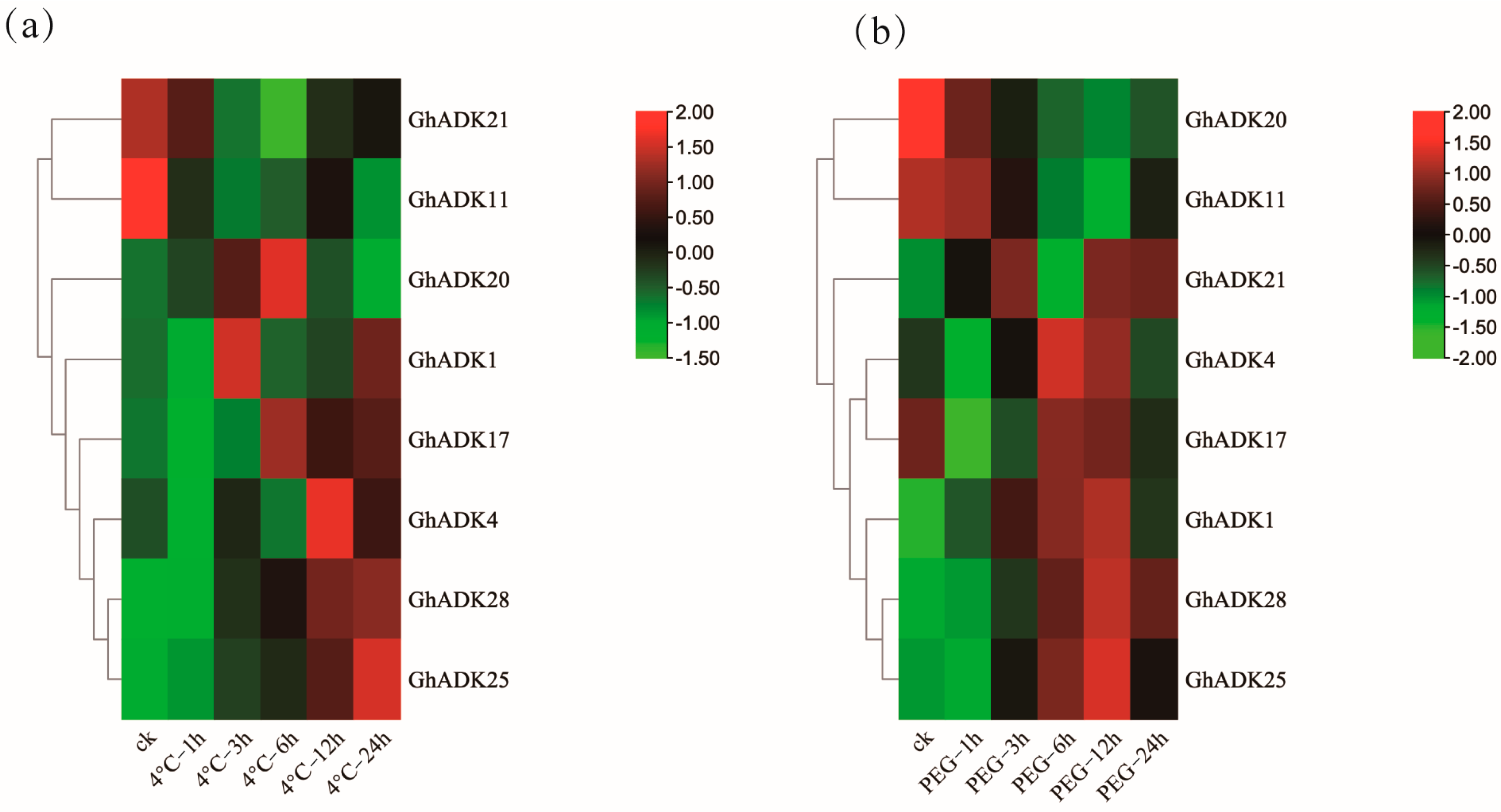
2.6. Expression Profiles of GhADK Genes under Cold Treatment and Drought Stress
2.7. qRT-PCR Verification of Upland Cotton ADK Family Members
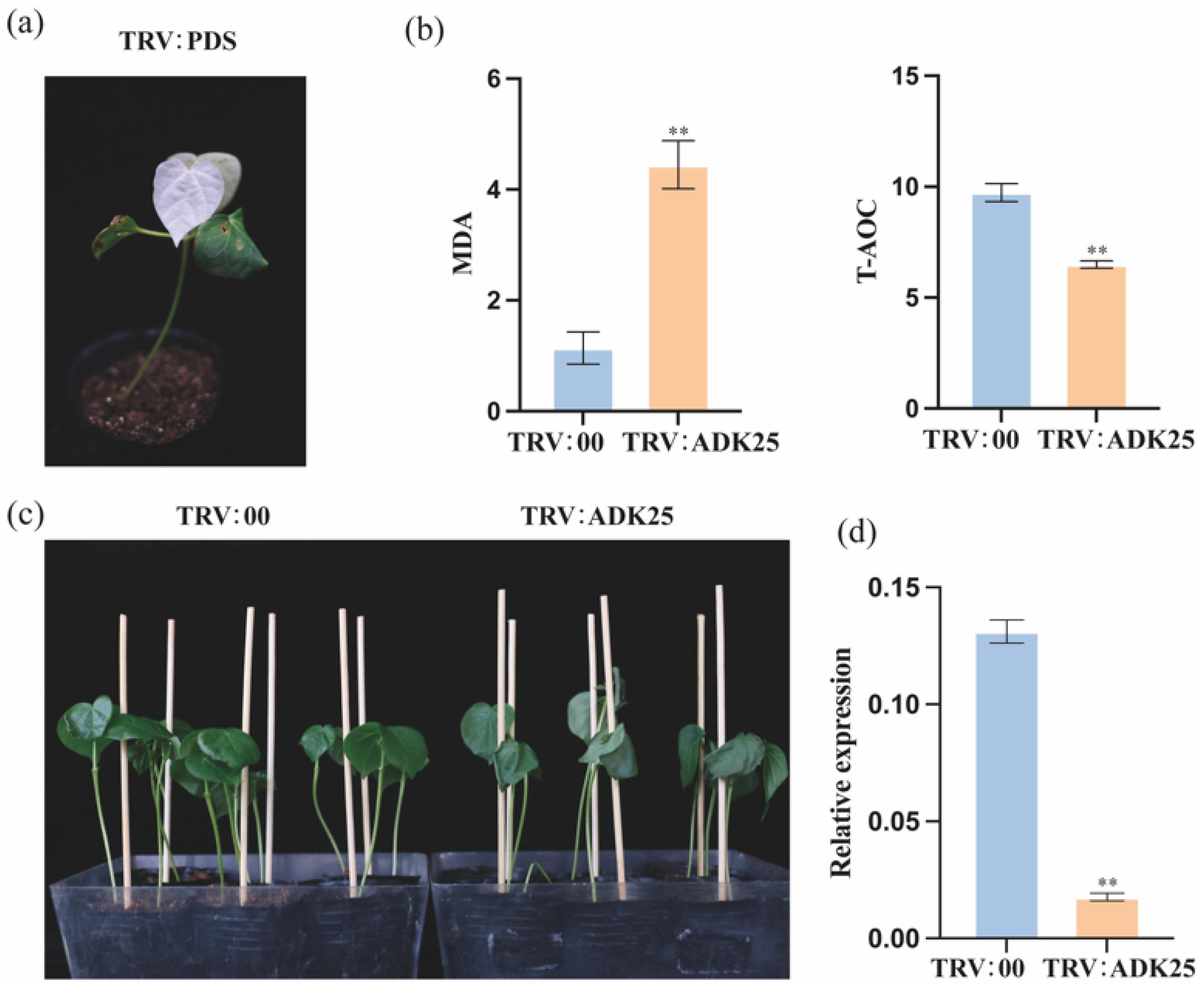
2.8. Functional Analysis of GhADK25 Silencing in Cotton under Drought and Cold Stress
3. Discussion
3.1. Systematical and Comprehensive Analyses of ADK Proteins
3.2. Evolution and Expansion of the ADK Gene Family in Cotton
3.3. Gene Expression Profiles and Functional Divergence of ADKs in G. hirsutum
3.4. The Potential Roles of ADK Genes in Response to Cold and Drought Stress
4. Materials and Methods
4.1. Identification and Bioinformatics Analysis of the ADK Gene Family in Cotton
4.2. Multiple Alignments and Phylogenetic Analysis
4.3. Chromosomal Localization and Gene Duplication Analysis
4.4. Gene Structure Analysis and Conserved Motif Detection of the ADK Gene Family in Gossypium hirsutum
4.5. Promoter Cis-Regulatory Analysis of the ADK Gene Family in Gossypium hirsutum
4.6. Expression Analysis of GhADKs
4.7. Plant Materials and Treatments, RNA Isolation, and Quantitative Real-Time PCR Analysis
4.8. Virus-Induced Gene Silencing (VIGS) Treatment of ADK25 in Cotton Seedlings
4.9. Measurement of Physio-Biochemical Attributes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dzheia, P.; Kal’venas, A.; Toleĭkis, A.; Prashkiavichius, A. The role of adenylate kinase in the regulation of the rate and effectiveness of energy transfer from mitochondria to hexokinase in vitro. Biokhimiia 1986, 51, 974–979. [Google Scholar] [PubMed]
- Lange, P.R.; Geserick, C.; Tischendorf, G.; Zrenner, R. Functions of chloroplastic adenylate kinases in Arabidopsis. Plant Physiol. 2008, 146, 492. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, D.E. Energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry 1968, 7, 4030–4034. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Fu, C. Adenylate kinase. In Encyclopedia of Food Microbiology: Second Edition; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 18–23. [Google Scholar]
- Noda, L. 8 adenylate kinase. In The Enzymes; Elsevier: Amsterdam, The Netherlands, 1973; Volume 8, pp. 279–305. [Google Scholar]
- Pradet, A.; Raymond, P. Adenine nucleotide ratios and adenylate energy charge in energy metabolism. Annu. Rev. Plant Physiol. 1983, 34, 199–224. [Google Scholar] [CrossRef]
- Arora, K.; Brooks, C.L., III. Large-scale allosteric conformational transitions of adenylate kinase appear to involve a population-shift mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 18496–18501. [Google Scholar] [CrossRef] [PubMed]
- Maragakis, P.; Karplus, M. Large amplitude conformational change in proteins explored with a plastic network model: Adenylate kinase. J. Mol. Biol. 2005, 352, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Birkenhead, K.; Walker, D.; Foyer, C. The intracellular distribution of adenylate kinase in the leaves of spinach, wheat and barley. Planta 1982, 156, 171–175. [Google Scholar] [CrossRef]
- Kawai, M.; Kidou, S.I.; Kato, A.; Uchimiya, H. Molecular characterization of cDNA encoding for adenylate kinase of rice (Oryza sativa L.). Plant J. 1992, 2, 845–854. [Google Scholar] [CrossRef]
- Thieulin-Pardo, G.; Schramm, A.; Lignon, S.; Lebrun, R.; Kojadinovic, M.; Gontero, B. The intriguing CP12-like tail of adenylate kinase 3 from Chlamydomonas reinhardtii. FEBS J. 2016, 283, 3389–3407. [Google Scholar] [CrossRef]
- Boonrueng, C.; Tangpranomkorn, S.; Yazhisai, U.; Sirikantaramas, S. Molecular cloning, subcellular localization and characterization of two adenylate kinases from cassava, Manihot esculenta Crantz cv. KU50. J. Plant Physiol. 2016, 204, 66–73. [Google Scholar] [CrossRef]
- Carrari, F.; Coll-Garcia, D.; Schauer, N.; Lytovchenko, A.; Palacios-Rojas, N.; Balbo, I.; Rosso, M.; Fernie, A.R. Deficiency of a plastidial adenylate kinase in Arabidopsis results in elevated photosynthetic amino acid biosynthesis and enhanced growth. Plant Physiol. 2005, 137, 70–82. [Google Scholar] [CrossRef]
- Zhang, Y.; Launay, H.; Liu, F.; Lebrun, R.; Gontero, B. Interaction between adenylate kinase 3 and glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii. FEBS J. 2018, 285, 2495–2503. [Google Scholar] [CrossRef] [PubMed]
- Peterson, T.A.; Nieman, R.H.; Clark, R.A. Nucleotide metabolism in salt-stressed Zea mays L. Root tips: I. Adenine and uridine nucleotides. Plant Physiol. 1987, 85, 984–989. [Google Scholar] [CrossRef]
- Zhou, S.; Wei, S.; Boone, B.; Levy, S. Microarray analysis of genes affected by salt stress in tomato. Afr. J. Environ. Sci. Technol. 2007, 1, 14–26. [Google Scholar]
- Gong, P.; Zhang, J.; Li, H.; Yang, C.; Zhang, C.; Zhang, X.; Khurram, Z.; Zhang, Y.; Wang, T.; Fei, Z. Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J. Exp. Bot. 2010, 61, 3563–3575. [Google Scholar] [CrossRef]
- Raveneau, M.-P.; Benamar, A.; Macherel, D. Water content, adenylate kinase, and mitochondria drive adenylate balance in dehydrating and imbibing seeds. J. Exp. Bot. 2017, 68, 3501–3512. [Google Scholar] [CrossRef]
- Wilkins, T.A.; Rajasekaran, K.; Anderson, D.M. Cotton biotechnology. Crit. Rev. Plant Sci. 2000, 19, 511–550. [Google Scholar] [CrossRef]
- Pettigrew, W. Physiological consequences of moisture deficit stress in cotton. Crop Sci. 2004, 44, 1265–1272. [Google Scholar] [CrossRef]
- Wendel, J.F.; Brubaker, C.L.; Percival, A.E. Genetic diversity in Gossypium hirsutum and the origin of upland cotton. Am. J. Bot. 1992, 79, 1291–1310. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Rosli, H.G.; Martin, G.B.; Mueller, L.A. The SGN VIGS tool: User-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol. Plant 2015, 8, 486–488. [Google Scholar] [CrossRef]
- Yang, L.; Cao, H.; Zhang, X.; Gui, L.; Chen, Q.; Qian, G.; Xiao, J.; Li, Z. Genome-wide identification and expression analysis of tomato ADK gene family during development and stress. Int. J. Mol. Sci. 2021, 22, 7708. [Google Scholar] [CrossRef]
- Cheng, Y.; Huang, C.; Hu, Y.; Jin, S.; Zhang, X.; Si, Z.; Zhao, T.; Chen, J.; Fang, L.; Dai, F. Gossypium purpurascens genome provides insight into the origin and domestication of upland cotton. J. Adv. Res. 2024, 56, 15–29. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; He, Q.; Wang, J.; Wang, B.; Zhao, J.; Huang, S.; Yang, T.; Tang, Y.; Yang, S.; Aisimutuola, P. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat. Genet. 2023, 55, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef]
- Kakimoto, T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001, 42, 677–685. [Google Scholar] [CrossRef]
- Yu, J.; Jung, S.; Cheng, C.-H.; Ficklin, S.P.; Lee, T.; Zheng, P.; Jones, D.; Percy, R.G.; Main, D. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014, 42, D1229–D1236. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K. The Pfam protein families database. Nucleic Acids Res. 2010, 38 (Suppl. 1), D211–D222. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P.W.; Kayastha, A.M.; Singh, V.K. MFPPI–multi FASTA ProtParam interface. Bioinformation 2016, 12, 74. [Google Scholar] [CrossRef] [PubMed]
- Shahmuradov, I.A.; Gammerman, A.J.; Hancock, J.M.; Bramley, P.M.; Solovyev, V.V. PlantProm: A database of plant promoter sequences. Nucleic Acids Res. 2003, 31, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Mach. Learn. 1995, 21, 51–80. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]



| Gene ID | Gene name | Chr | Start | End | PI | MW (kDa) | AA |
|---|---|---|---|---|---|---|---|
| GhADK1 | Gh_A05G2360 | A05 | 28948631 | 28953225 | 7.14 | 32528.07 | 296 |
| GhADK2 | Gh_A05G2963 | A05 | 73057726 | 73059034 | 6.15 | 29723.98 | 265 |
| GhADK3 | Gh_A05G3557 | A05 | 91270041 | 91271305 | 6.37 | 25040.75 | 222 |
| GhADK4 | Gh_A06G0410 | A06 | 6886337 | 6900641 | 8.56 | 65724.32 | 590 |
| GhADK5 | Gh_A07G0462 | A07 | 5972595 | 5974906 | 7.11 | 30968.44 | 273 |
| GhADK6 | Gh_A07G0465 | A07 | 5995641 | 5998894 | 6.97 | 26899.23 | 245 |
| GhADK7 | Gh_A07G2086 | A07 | 77433461 | 77435110 | 6.15 | 30273.43 | 272 |
| GhADK8 | Gh_A08G1415 | A08 | 89538235 | 89540746 | 8.5 | 24830.9 | 230 |
| GhADK9 | Gh_A09G2184 | A09 | 74822698 | 74825672 | 7.64 | 26854.17 | 244 |
| GhADK10 | Gh_A10G1601 | A10 | 86412249 | 86415506 | 6.37 | 32856.6 | 303 |
| GhADK11 | Gh_A12G1706 | A12 | 78779245 | 78781576 | 7.69 | 26803.97 | 244 |
| GhADK12 | Gh_A12G1810 | A12 | 80690562 | 80693656 | 8.49 | 26316.29 | 236 |
| GhADK13 | Gh_A13G0108 | A13 | 1268419 | 1270512 | 7.03 | 26556.53 | 237 |
| GhADK14 | Gh_D01G1585 | D01 | 49745145 | 49746750 | 6.91 | 26725.94 | 245 |
| GhADK15 | Gh_D04G0048 | D04 | 774522 | 775676 | 7.6 | 24074.71 | 214 |
| GhADK16 | Gh_D04G0753 | D04 | 15612652 | 15613961 | 6.15 | 29856.14 | 267 |
| GhADK17 | Gh_D05G2627 | D05 | 27184500 | 27189101 | 8.41 | 32934.66 | 299 |
| GhADK18 | Gh_D06G0443 | D06 | 6370844 | 6380120 | 8.94 | 66355.03 | 594 |
| GhADK19 | Gh_D07G0526 | D07 | 5948360 | 5959238 | 9.08 | 33159.95 | 290 |
| GhADK20 | Gh_D07G0529 | D07 | 5978760 | 5982018 | 6.55 | 26957.26 | 245 |
| GhADK21 | Gh_D07G2304 | D07 | 54335943 | 54337596 | 6.51 | 30197.38 | 272 |
| GhADK22 | Gh_D09G2390 | D09 | 50793345 | 50796339 | 8.22 | 26720.04 | 244 |
| GhADK23 | Gh_D10G1856 | D10 | 51921697 | 51924967 | 6.24 | 32658.46 | 301 |
| GhADK24 | Gh_D10G2125 | D10 | 58219589 | 58223112 | 8.37 | 32771.56 | 301 |
| GhADK25 | Gh_D12G1868 | D12 | 51286089 | 51288425 | 8.26 | 26892.19 | 245 |
| GhADK26 | Gh_D12G1980 | D12 | 52858886 | 52864409 | 8.52 | 83784.67 | 746 |
| GhADK27 | Gh_D13G0125 | D13 | 1246139 | 1248231 | 8.4 | 26423.47 | 237 |
| GhADK28 | Gh_A01G2126 | scaffold147_A01 | 199905 | 201512 | 6.96 | 26852.05 | 245 |
| GhADK29 | Gh_A10G2351 | scaffold2716_A10 | 5914 | 9425 | 8.37 | 32710.48 | 301 |
| GhADK30 | Gh_A12G2555 | scaffold3185_A12 | 1180857 | 1186323 | 9.68 | 34073.49 | 317 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, P.; Lin, Z.; Zhang, Y.; Gao, Y.; Tan, S.; Wang, S.; Cao, X.; Shi, H.; Sun, C.; Bai, J.; et al. Genome-Wide Identification and Expression Analysis of ADK Gene Family Members in Cotton under Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 7821. https://doi.org/10.3390/ijms25147821
Huang P, Lin Z, Zhang Y, Gao Y, Tan S, Wang S, Cao X, Shi H, Sun C, Bai J, et al. Genome-Wide Identification and Expression Analysis of ADK Gene Family Members in Cotton under Abiotic Stress. International Journal of Molecular Sciences. 2024; 25(14):7821. https://doi.org/10.3390/ijms25147821
Chicago/Turabian StyleHuang, Peijun, Ziwei Lin, Yuzhi Zhang, Yu Gao, Songjuan Tan, Shuai Wang, Xiaoyu Cao, Hongyan Shi, Chao Sun, Jiangping Bai, and et al. 2024. "Genome-Wide Identification and Expression Analysis of ADK Gene Family Members in Cotton under Abiotic Stress" International Journal of Molecular Sciences 25, no. 14: 7821. https://doi.org/10.3390/ijms25147821
APA StyleHuang, P., Lin, Z., Zhang, Y., Gao, Y., Tan, S., Wang, S., Cao, X., Shi, H., Sun, C., Bai, J., & Ma, X. (2024). Genome-Wide Identification and Expression Analysis of ADK Gene Family Members in Cotton under Abiotic Stress. International Journal of Molecular Sciences, 25(14), 7821. https://doi.org/10.3390/ijms25147821





