B Cell Lymphocytes as a Potential Source of Breast Carcinoma Marker Candidates
Abstract
1. Introduction
B Cells: Physiological Differentiation, Maturation, and Function
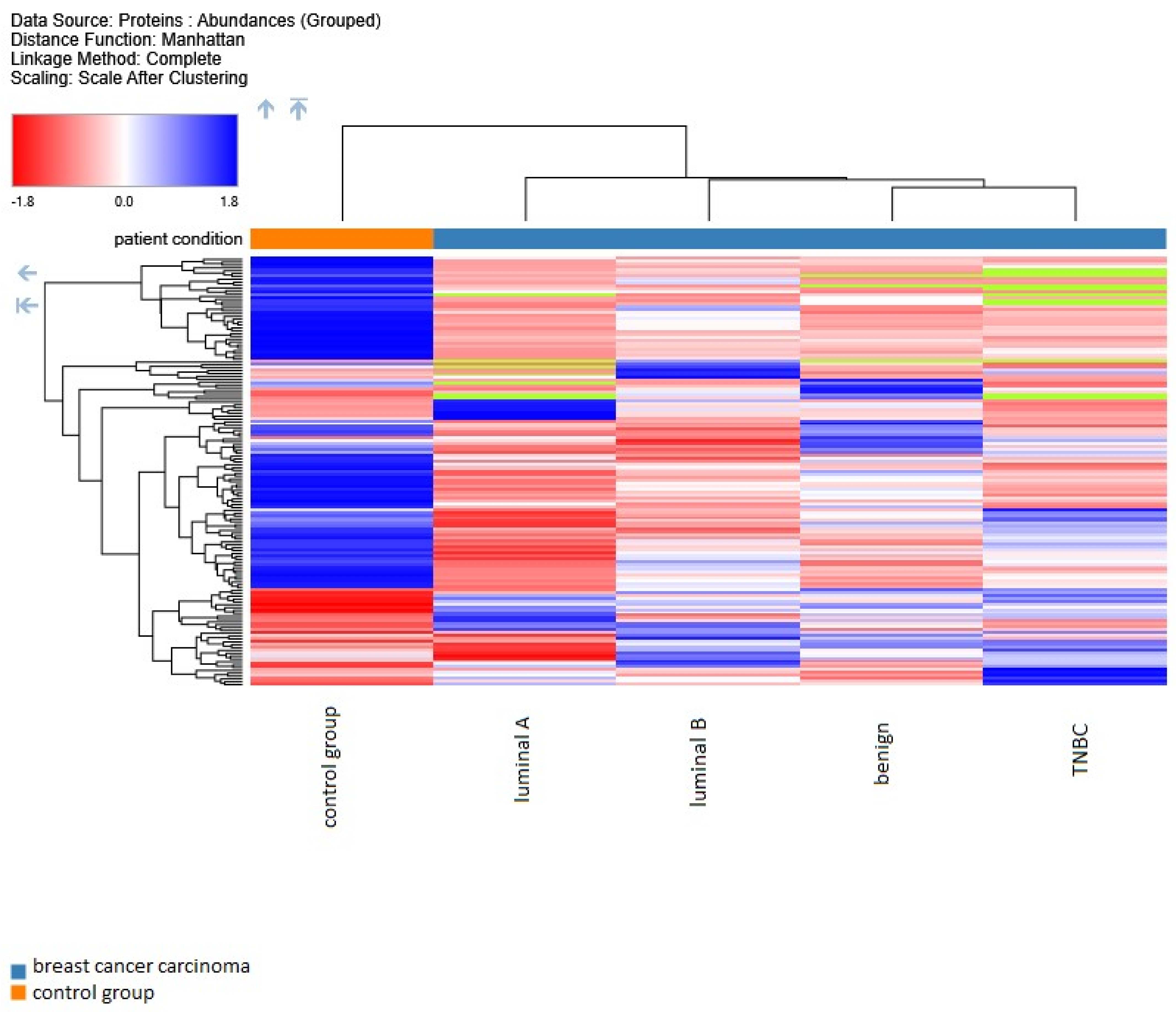
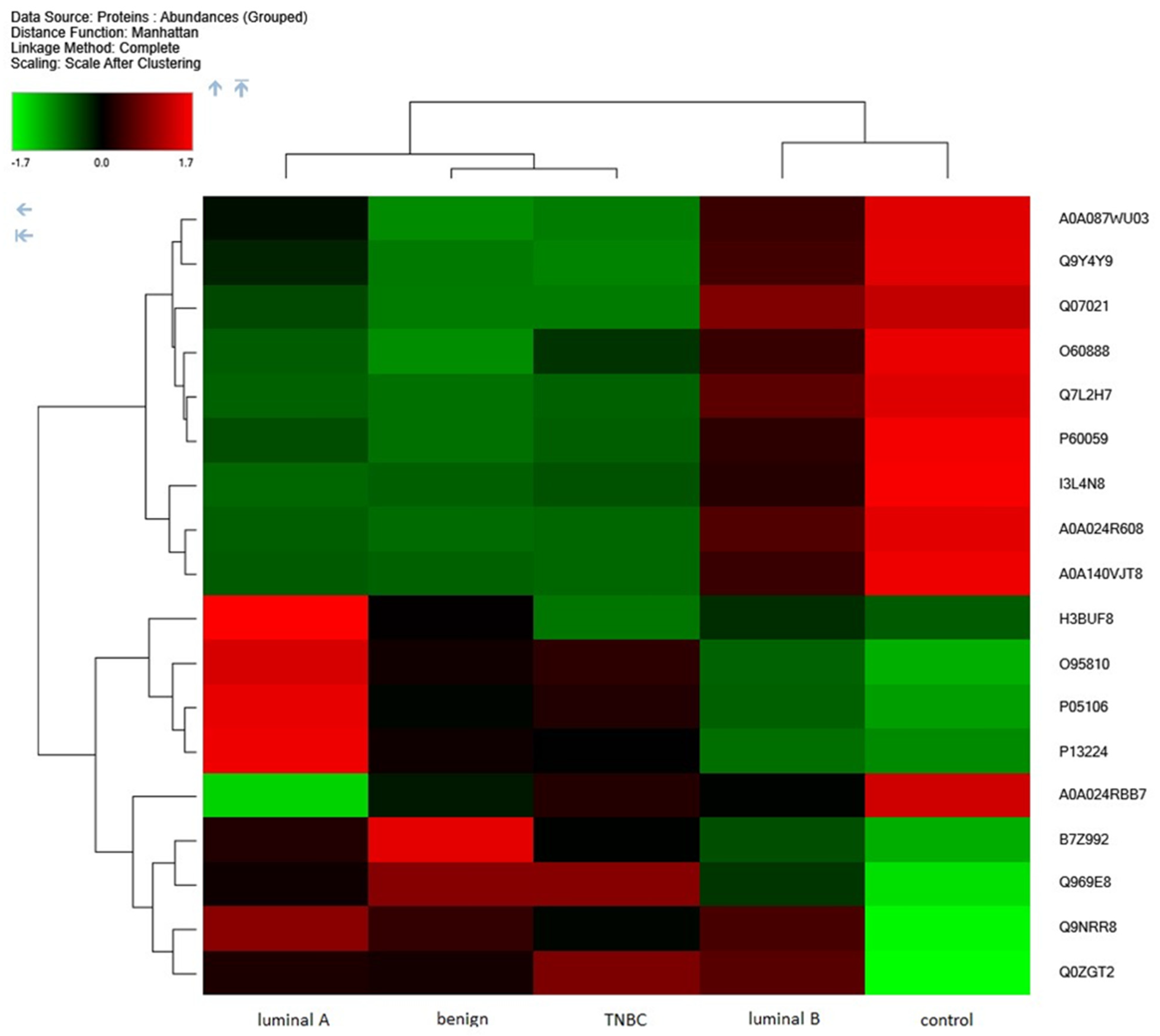
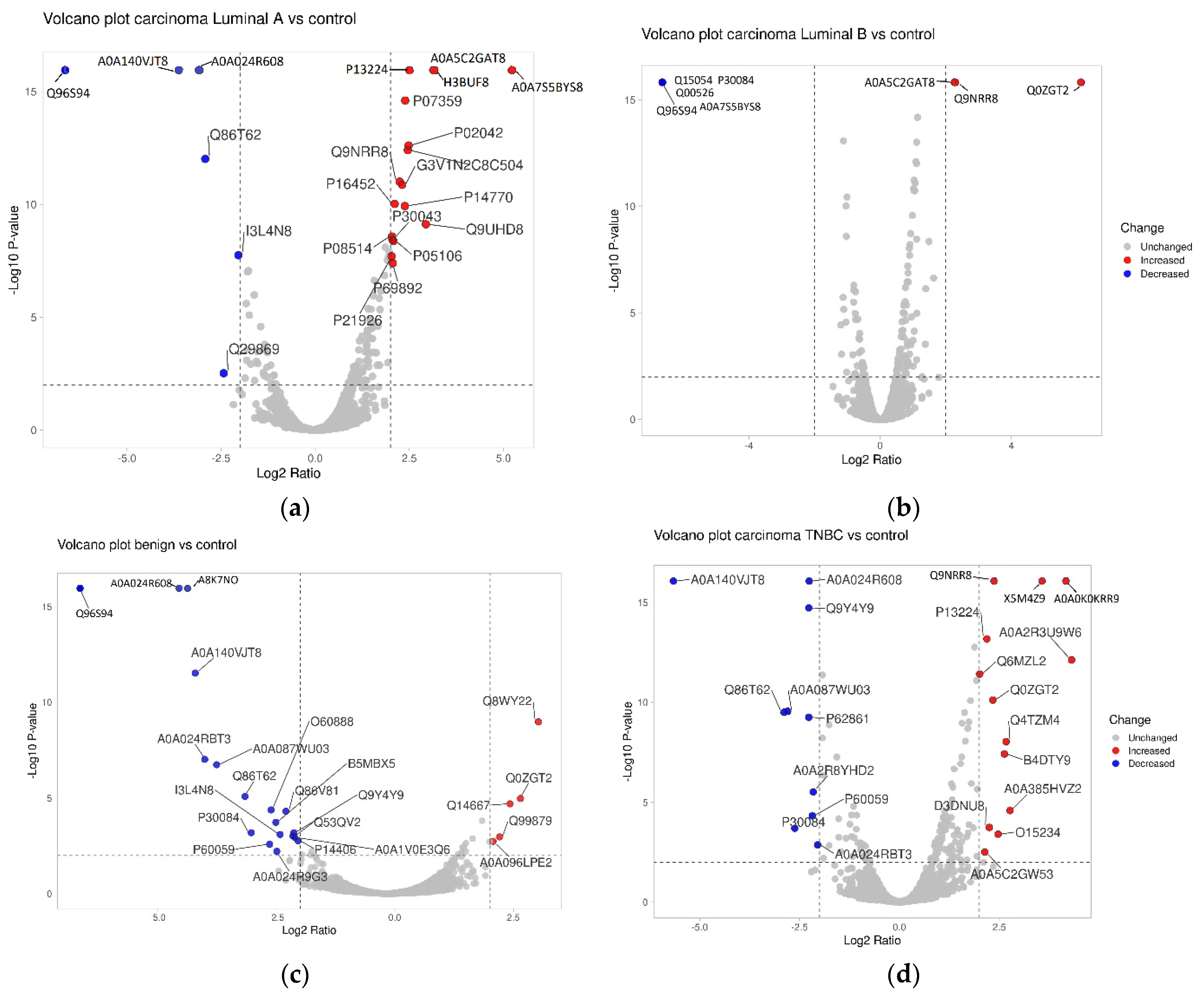
2. Results
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. Blood Sampling and B Cell Isolation
4.3. LC-MS/MS Analysis and Database Search
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN, Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ősz, Á.; Lánczky, A.; Győrffy, B. Survival analysis in breast cancer using proteomic data from four independent datasets. Sci. Rep. 2021, 11, 16787. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kuroda, H.; Jamiyan, T.; Yamaguchi, R.; Kakumoto, A.; Abe, A.; Harada, O.; Enkhbat, B.; Masunaga, A. Prognostic value of tumor-infiltrating B lymphocytes and plasma cells in triple-negative breast cancer. Breast Cancer 2021, 28, 904–914. [Google Scholar] [CrossRef]
- Onitilo, A.A.; Engel, J.M.; Greenlee, R.T.; Mukesh, B.N. Breast cancer subtypes based on ER/PR and Her2 expression: Comparison of clinicopathologic features and survival. Clin. Med. Res. 2009, 7, 4–13. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Wang, W.; Xu, L.; Duan, X.; Cheng, Y.; Xin, L.; Zhang, H.; Zhang, S.; Li, T.; Liu, Y. A retrospective prognostic evaluation analysis using the 8th edition of American Joint Committee on Cancer (AJCC) cancer staging system for luminal A breast cancer. Chin. J. Cancer Res. 2017, 29, 351–360. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Weigelt, B.; Baehner, F.L.; Reis-Filho, J.S. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: A retrospective of the last decade. J. Pathol. 2010, 220, 263–280. [Google Scholar] [CrossRef]
- Doll, S.; Gnad, F.; Mann, M. The Case for Proteomics and Phospho-Proteomics in Personalized Cancer Medicine. Proteom. Clin. Appl. 2019, 13, e1800113. [Google Scholar] [CrossRef]
- Asleh, K.; Negri, G.L.; Spencer Miko, S.E.; Colborne, S.; Hughes, C.S.; Wang, X.Q.; Gao, D.; Gilks, C.B.; Chia, S.K.L.; Nielsen, T.O.; et al. Proteomic analysis of archival breast cancer clinical specimens identifies biological subtypes with distinct survival outcomes. Nat. Commun. 2022, 16, 896. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, M.; Quintana, A.; Alberts, E.; Hung, M.S.; Boulat, V.; Ripoll, M.M.; Grigoriadis, A. B Cells in Breast Cancer Pathology. Cancers 2023, 15, 1517. [Google Scholar] [CrossRef]
- Peng, S.; Hebert, L.L.; Eschbacher, J.M.; Kim, S. Single-Cell RNA Sequencing of a Postmenopausal Normal Breast Tissue Identifies Multiple Cell Types That Contribute to Breast Cancer. Cancers 2020, 12, 3639. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, B.; Miyamoto, A.; Yokoyama, K.; Ogiya, R.; Oshitanai, R.; Terao, M.; Morioka, T.; Niikura, N.; Okamura, T.; Miyako, H.; et al. B-cell populations are expanded in breast cancer patients compared with healthy controls. Breast Cancer 2018, 25, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Michiels, S.; Salgado, R.; Sirtaine, N.; Jose, V.; Fumagalli, D.; Kellokumpu-Lehtinen, P.L.; Bono, P.; Kataja, V.; Desmedt, C.; et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. 2014, 25, 1544–1550. [Google Scholar] [CrossRef]
- Hu, Q.; Hong, Y.; Qi, P. Atlas of breast cancer infiltrated B-lymphocytes revealed by paired single-cell RNA-sequencing and antigen receptor profiling. Nat. Commun. 2021, 12, 2186. [Google Scholar] [CrossRef]
- Lam, B.M.; Verrill, C. Clinical Significance of Tumour-Infiltrating B Lymphocytes (TIL-Bs)in Breast Cancer: A Systematic Literature Review. Cancers 2023, 15, 1164. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, E.; van den Biggelaar, M.; de Kivit, S.; Chen, Y.Y.; Slot, M.; Doubal, I.; Meijer, A.; van Lier, R.A.W.; Borst, J.; Amsen, D. Proteomic Analyses of Human Regulatory T Cells Reveal Adaptations in Signaling Pathways that Protect Cellular Identity. Immunity 2018, 48, 1046–1059.e6. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Osborne, C.K.; Yochmowitz, M.G.; Knight, W.A.; McGuire, W.L. The value of estrogen and progesterone receptors in the treatment of breast cancer. Cancer 1980, 46, 2884–2888. [Google Scholar] [CrossRef]
- He, Z.; Xu, Q.; Wang, X.; Wang, J.; Mu, X.; Cai, Y.; Qian, Y.; Shao, W.; Shao, Z. RPLP1 promotes tumor metastasis and is associated with a poor prognosis in triple-negative breast cancer patients. Cancer Cell Int. 2018, 18, 170. [Google Scholar] [CrossRef] [PubMed]
- Michaut, M.; Chin, S.F.; Majewski, I.; Severson, T.M.; Bismeijer, T.; de Koning, L.; Peeters, J.K.; Schouten, P.C.; Rueda, O.M.; Bosma, A.J.; et al. Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci. Rep. 2016, 6, 18517. [Google Scholar] [CrossRef]
- Tang, Y.; Ren, F.; Cong, X.; Kong, Y.; Tian, Y.; Xu, Y.; Fan, J. Overexpression of ribonuclease inhibitor induces autophagy in human colorectal cancer cells via the Akt/mTOR/ULK1 pathway. Mol. Med. Rep. 2019, 19, 3519–3526. [Google Scholar] [CrossRef] [PubMed]
- Sudhakaran, M.; Doseff, A.I. Role of Heterogeneous Nuclear Ribonucleoproteins in the Cancer-Immune Landscape. Int. J. Mol. Sci. 2023, 24, 5086. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Niu, W.; He, Z.; Gao, C.; Peng, C.; Niu, J. SEC61G plays an oncogenic role in hepatocellular carcinoma cells. Cell Cycle 2020, 19, 3348–3361. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Wang, Z.; Zhang, M.; Yu, Y.; Chen, R.; Lu, T.; Liu, L.; Ma, J.; Liu, T.; Zheng, H.; et al. Prognostic value of SEC61G in lung adenocarcinoma: A comprehensive study based on bioinformatics and in vitro validation. BMC Cancer 2021, 21, 1216. [Google Scholar] [CrossRef]
- Blockhuys, S.; Celauro, E.; Hildesjo, C.; Feizi, A.; Stål, O.; Fierro-Gonzaález, J.C.; Wittung-Stafshede, P. Defining the human copper proteome and analysis of its expression variation in cancers. Metallomics 2017, 9, 112. [Google Scholar] [CrossRef]
- Dugina, V.B.; Shagieva, G.S.; Kopnin, P.B. Cytoplasmic Beta and Gamma Actin Isoforms Reorganization and Regulation in Tumor Cells in Culture and Tissue. Front. Pharmacol. 2022, 13, 895703. [Google Scholar] [CrossRef] [PubMed]
- Zali, M.R.; Azodi, M.Z.; Razzaghi, Z.; Heydari, M.H. Gall bladder Cancer Integrated Bioinformatics Analysis of Protein Profile Data. Gastroenterol. Hepatol. Bed Bench 2019, 12, S66. [Google Scholar] [CrossRef]
- Yang, H.; Beutler, B.; Zhang, D. Emerging roles of spliceosome in cancer and immunity. Protein Cell 2022, 13, 559–579. [Google Scholar] [CrossRef]
- Scully, O.J.; Shyamasundar, S.; Matsumoto, K.; Dheen, S.T.; Yip, G.W.; Bay, B.H. C1QBP Mediates Breast Cancer Cell Proliferation and Growth via Multiple Potential Signalling Pathways. Int. J. Mol. Sci. 2023, 24, 1343. [Google Scholar] [CrossRef]
- Gul, G.; Aydin, M.A.; Algul, S.; Kiziltan, R.; Kemik, O. Nucleosome assembly protein 1-like 1 (NAP1L1) in gastric cancer patients: A potential biomarker with diagnostic and prognostic utility. Biomarkers 2024, 29, 30–35. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Zhang, Y.; Tang, Y.; Fang, W.; Liu, X.; Liu, Z. NAP1L1 targeting suppresses the proliferation of nasopharyngeal carcinoma. Biomed. Pharmacother. 2021, 143, 112096. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Liu, J.; Zhang, M.; Gao, X.; Sun, W.; Liu, P.; Wang, Y.; Li, J. Eukaryotic translation initiation factor 3 subunit B could serve as a potential prognostic predictor for breast cancer. Bioengineered 2022, 13, 2762–2776. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kovacheva, M.; Zepp, M.; Berger, S.; Berger, M.R. Conditional knockdown of integrin beta-3 reveals its involvement in osteolytic and soft tissue lesions of breast cancer skeletal metastasis. J. Cancer Res. Clin. Oncol. 2021, 147, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Low, J.Y.; Laiho, M. Caveolae-Associated Molecules; Tumor Stroma; and Cancer Drug Resistance: Current Findings and Future Perspectives. Cancers 2022, 14, 589. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.M.; Karbstein, K. The chaperone Tsr2 regulates Rps26 release and reincorporation from mature ribosomes to enable a reversible; ribosome-mediated response to stress. Sci. Adv. 2022, 8, eabl4386. [Google Scholar] [CrossRef]
- Yap, S.Q.; Mathavarajah, S.; Huber, R.J. The converging roles of Batten disease proteins in neurodegeneration and cancer. iScience 2021, 24, 102337. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Murphy, N.P.; Binti Ahmad Mokhtar, A.M.; Mott, H.R.; Owen, D. Molecular subversion of Cdc42 signalling in cancer. Biochem. Soc. Trans. 2021, 49, 1425–1442. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hsieh, C.H.; Wang, Y.C. Emerging roles of plasma gelsolin in tumorigenesis and modulating the tumor microenvironment. Kaohsiung J. Med. Sci. 2022, 38, 819–825. [Google Scholar] [CrossRef]
- Dang, E.; Yang, S.; Song, C.; Jiang, D.; Li, Z.; Fan, W.; Sun, Y.; Tao, L.; Wang, J.; Liu, T.; et al. BAP31, a newly defined cancer/testis antigen, regulates proliferation, migration, and invasion to promote cervical cancer progression. Cell Death Dis. 2018, 9, 791. [Google Scholar] [CrossRef]
- Taban, Q.; Mumtaz, P.T.; Masoodi, K.Z.; Haq, E.; Ahmad, S.M. Scavenger receptors in host defense: From functional aspects to mode of action. Cell Commun. Signal. 2022, 20, 2. [Google Scholar] [CrossRef]
- Crusz, S.M.; Balkwill, F.R. Inflammation and cancer: Advances and new agents. Nat. Rev. Clin. Oncol. 2015, 12, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Semple, J.; Italiano, J.; Freedman, J. Platelets and the immune continuum. Nat. Rev. Immunol. 2011, 11, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Zamora, C.; Toniolo, E.; Diaz-Torné, C.; Cantó, E.; Magallares, B.; Ortiz, M.A.; Perea, L.; Corominas, H.; Vidal, S. Association of Platelet Binding to Lymphocytes with B Cell Abnormalities and Clinical Manifestations in Systemic Lupus Erythematosus. Mediat. Inflamm. 2019, 2019, 2473164. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Oleksowicz, L.; Dutcher, J.P.; Deleon-Fernandez, M.; Paietta, E.; Etkind, P. Human breast carcinoma cells synthesize a immunorelated to platelet glycoprotein-lbα different functional properties. J. Lab. Clin. Med. 1997, 129, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, M.; Janus-Bell, E.; Schaff, M.; Gachet, C.; Mangin, P.H. Platelet Integrins in Tumor Metastasis: Do They Represent a Therapeutic Target? Cancers 2017, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Tao, D.L.; Tassi Yunga, S.; Williams, C.D.; McCarty, O.J.T. Aspirin and antiplatelet treatments in cancer. Blood 2021, 137, 3201–3211. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Plantureux, L.; Mège, D.; Crescence, L.; Dignat-George, F.; Dubois, C.; Panicot-Dubois, L. Impacts of Cancer on Platelet Production; Activation and Education and Mechanisms of Cancer-Associated Thrombosis. Cancers 2018, 10, 441. [Google Scholar] [CrossRef] [PubMed]
- Bijak, M.; Saluk, J.; Ponczek, M.; Nowak, P.; Wachowicz, B. The synthesis of proteins in unnucleated blood platelets. Adv. Hyg. Exp. Med. 2013, 67, 672–679. [Google Scholar] [CrossRef]
- Yari, F.; Motafakker, M.; Nikougoftar, M.; Khayati, Z. Interaction of Platelet-Derived Microparticles with a Human B-Lymphoblast Cell Line: A Clue for the Immunologic Function of the Microparticles. Transfus. Med. Hemotherapy 2018, 45, 55–61. [Google Scholar] [CrossRef]
- Guo, T.; Luna, A.; Rajapakse, V.N.; Koh, C.C.; Wu, Z.; Liu, W.; Sun, Y.; Gao, H.; Menden, M.P.; Xu, C.; et al. Quantitative Proteome Landscape of the NCI-60 Cancer Cell Lines. iScience 2019, 21, 664–680. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

| B Cell Subtype | Immunohistochemical Markers |
|---|---|
| Naïve B cell | CD19+, CD20+, IgM+IgD−, CD38−/− |
| Naïve activated B cell | CD19+, CD20+, IgM+, IgD+, CD38+ |
| Germinal center B cell | CD19+, CD20+, IgM+/−, IgD+, CD38++ |
| Plasmablast | CD19+, CD20−, CD38++, CD27++, IgD−, IgM/G/A/E+ |
| Plasma cell | CD19+/−, CD20−, CD38++, CD138+, CD27+ IgD−, IgM/G/A/E+ |
| Memory B cell | CD19+ CD20+ CD38−, CD27+, IgD−, IgM/G/A/E+ |
| Accession | Description | Coverage [%] | Peptides | Unique Peptides | MW [kDa] |
|---|---|---|---|---|---|
| B7Z992 | Gelsolin | 61 | 37 | 2 | 78.8 |
| P13224 | Platelet glycoprotein Ib beta chain | 25 | 6 | 6 | 21.7 |
| P05106 | Integrin beta-3 | 58 | 40 | 4 | 87 |
| Q9NRR8 | CDC42 small effector protein | 24 | 2 | 2 | 8.9 |
| O95810 | Caveolae-associated protein 2 | 54 | 21 | 21 | 47.1 |
| Q969E8 | Pre-rRNA-processing protein TSR2 homolog | 19 | 5 | 5 | 20.9 |
| Q0ZGT2 | Nexilin | 12 | 7 | 7 | 80.6 |
| Accession | Description | Coverage [%] | Peptides | Unique Peptides | MW [kDa] |
|---|---|---|---|---|---|
| A0A024R608 | Ribosomal protein, large, P1, isoform CRA_a | 85 | 7 | 6 | 11.5 |
| A0A024RBB7 | Nucleosome assembly protein 1-like 1, isoform CRA_a | 56 | 18 | 16 | 45.3 |
| A0A087WU03 | Heterogeneous nuclear ribonucleoprotein D-like | 40 | 2 | 2 | 6.7 |
| A0A140VJT8 | Ribonuclease inhibitor | 88 | 34 | 2 | 49.9 |
| I3L4N8 | Actin, cytoplasmic 2 | 99 | 49 | 4 | 39 |
| O60888 | Protein CutA | 49 | 5 | 5 | 19.1 |
| P60059 | Protein transport protein Sec61 subunit gamma | 29 | 2 | 2 | 7.7 |
| Q07021 | Complement component 1 Q subcomponent-binding protein, mitochondrial | 54 | 10 | 10 | 31.3 |
| Q7L2H7 | Eukaryotic translation initiation factor 3 subunit M | 51 | 15 | 15 | 42.5 |
| Q9Y4Y9 | U6 snRNA-associated Sm-like protein LSm5 | 56 | 6 | 6 | 9.9 |
| Description | Gene Symbol | Accession | AR Benign/ Control | CV [%] Benign | AR Lumi-nal A Control | CV [%] Lumi-nal A | AR Lumi-nal B/Control | CV [%] Lumi-nal B | AR TNBC/Control | CV [%] TNBC |
|---|---|---|---|---|---|---|---|---|---|---|
| DOWN REGULATED | ||||||||||
| Ribosomal protein, large, P1, isoform CRA | RPLP1 | A0A024R608 | 0.043 | 233.65 | 0.118 | 162.09 | 0.461 | 86.08 | 0.208 | 153.7 |
| Ribonuclease inhibitor | A0A140VJT8 | 0.054 | 196.44 | 0.082 | 128.71 | 0.456 | 60.23 | 0.02 | n/a | |
| Heterogeneous nuclear ribonucleoprotein D-like | HNRNPDL | A0A087WU03 | 0.074 | 223.7 | 0.347 | 43.3 | 0.581 | 94.49 | 0.143 | 108.33 |
| Protein transport protein Sec61 subunit gamma | SEC61G | P60059 | 0.161 | 147.94 | 0.287 | 48.63 | 0.567 | 60.72 | 0.221 | 50.32 |
| Protein CutA | CUTA | O60888 | 0.164 | 229.51 | 0.282 | 80.01 | 0.614 | 76.12 | 0.511 | 87.57 |
| Actin, cytoplasmic 2 | ACTG1 | I3L4N8 | 0.187 | 157.77 | 0.243 | 77.41 | 0.571 | 83.21 | 0.262 | 88.8 |
| U6 snRNA-associated Sm-like protein LSm5 | LSM5 | Q9Y4Y9 | 0.228 | 207.85 | 0.395 | 71.81 | 0.498 | 78.14 | 0.208 | 117.05 |
| Complement component 1 Q subcomponent-binding protein, mitochondrial | C1QBP | Q07021 | 0.296 | 168.91 | 0.412 | 84.96 | 0.699 | 41.6 | 0.51 | 89.27 |
| Nucleosome assembly protein 1-like 1, isoform CRA | NAP1L1 | A0A024RBB7 | 0.32 | 69.93 | 0.399 | 80.34 | 0.708 | 59.95 | 0.511 | 93.97 |
| Eukaryotic translation initiation factor 3 subunit M | EIF3M | Q7L2H7 | 0.351 | 171.11 | 0.367 | 61.06 | 0.734 | 50.91 | 0.456 | 84.82 |
| UP REGULATED | ||||||||||
| Integrin beta-3 | ITGB3 | P05106 | 2.224 | 94.2 | 4.228 | 97.92 | 1.583 | 91.35 | 3.679 | 117.97 |
| Caveolae-associated protein 2 | CAVIN2 | O95810 | 2.262 | 80.52 | 2.785 | 122.56 | 1.535 | 48.04 | 2.583 | 97.02 |
| Pre-rRNA-processing protein TSR2 homolog | TSR2 | Q969E8 | 2.385 | 30.62 | 2.1 | 19.4 | 1.649 | 16.28 | 2.323 | 29.61 |
| Platelet glycoprotein Ib beta chain | GP1BB | P13224 | 2.428 | 101.72 | 5.664 | 97.69 | 2.053 | 115.73 | 4.556 | 127.94 |
| CDC42 small effector protein 1 | CDC42SE1 | Q9NRR8 | 2.519 | 44.69 | 4.718 | 24.56 | 4.898 | 34.17 | 5.186 | 21.57 |
| Gelsolin | B7Z992 | 3.568 | 83.17 | 2.911 | 77.88 | 1.779 | 42.36 | 2.488 | 65.93 | |
| Nexilin | NEXN | Q0ZGT2 | 6.257 | 46.96 | 9.158 | 49.51 | 69.689 | 24.14 | 5.048 | 19.48 |
| Pathway Identifier | Pathway Name | #Entities Found | #Entities Total |
|---|---|---|---|
| R-HSA-76002 | Platelet activation, signaling, and aggregation | 7 | 265 |
| R-HSA-114608 | Platelet degranulation | 5 | 128 |
| R-HSA-76005 | Response to elevated platelet cytosolic Ca2+ | 5 | 133 |
| R-HSA-9673221 | Defective F9 activation | 2 | 6 |
| R-HSA-9846298 | Defective binding of VWF variant to GPIb:IX:V | 2 | 7 |
| R-HSA-9845620 | Enhanced binding of GP1BA variant to VWF multimer:collagen | 2 | 7 |
| R-HSA-381426 | Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) | 4 | 124 |
| R-HSA-9823587 | Defects of platelet adhesion to exposed collagen | 2 | 8 |
| R-HSA-2173782 | Binding and uptake of ligands by scavenger receptors | 4 | 129 |
| R-HSA-9668250 | Defective factor IX causes hemophilia B | 2 | 9 |
| R-HSA-430116 | GP1b-IX-V activation signaling | 2 | 12 |
| R-HSA-109582 | Hemostasis | 7 | 727 |
| R-HSA-75892 | Platelet adhesion to exposed collagen | 2 | 16 |
| R-HSA-9651496 | Defects of Contact Activation System (CAS) and Kallikrein/Kinin System (KKS) | 2 | 16 |
| R-HSA-9671793 | Diseases of hemostasis | 2 | 19 |
| R-HSA-140837 | Intrinsic pathway of fibrin clot formation | 2 | 23 |
| R-HSA-2168880 | Scavenging of heme from plasma | 3 | 99 |
| R-HSA-8957275 | Post-translational protein phosphorylation | 3 | 107 |
| R-HSA-140877 | Formation of fibrin clot (clotting cascade) | 2 | 39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tkáčiková, S.; Marcin, M.; Bober, P.; Kacírová, M.; Šuliková, M.; Parnica, J.; Tóth, D.; Lenárt, M.; Radoňak, J.; Urdzík, P.; et al. B Cell Lymphocytes as a Potential Source of Breast Carcinoma Marker Candidates. Int. J. Mol. Sci. 2024, 25, 7351. https://doi.org/10.3390/ijms25137351
Tkáčiková S, Marcin M, Bober P, Kacírová M, Šuliková M, Parnica J, Tóth D, Lenárt M, Radoňak J, Urdzík P, et al. B Cell Lymphocytes as a Potential Source of Breast Carcinoma Marker Candidates. International Journal of Molecular Sciences. 2024; 25(13):7351. https://doi.org/10.3390/ijms25137351
Chicago/Turabian StyleTkáčiková, Soňa, Miroslav Marcin, Peter Bober, Mária Kacírová, Michaela Šuliková, Jozef Parnica, Dávid Tóth, Marek Lenárt, Jozef Radoňak, Peter Urdzík, and et al. 2024. "B Cell Lymphocytes as a Potential Source of Breast Carcinoma Marker Candidates" International Journal of Molecular Sciences 25, no. 13: 7351. https://doi.org/10.3390/ijms25137351
APA StyleTkáčiková, S., Marcin, M., Bober, P., Kacírová, M., Šuliková, M., Parnica, J., Tóth, D., Lenárt, M., Radoňak, J., Urdzík, P., Fedačko, J., & Sabo, J. (2024). B Cell Lymphocytes as a Potential Source of Breast Carcinoma Marker Candidates. International Journal of Molecular Sciences, 25(13), 7351. https://doi.org/10.3390/ijms25137351







