Integrated Methylome and Transcriptome Analysis between Wizened and Normal Flower Buds in Pyrus pyrifolia Cultivar ‘Sucui 1’
Abstract
1. Introduction
2. Results
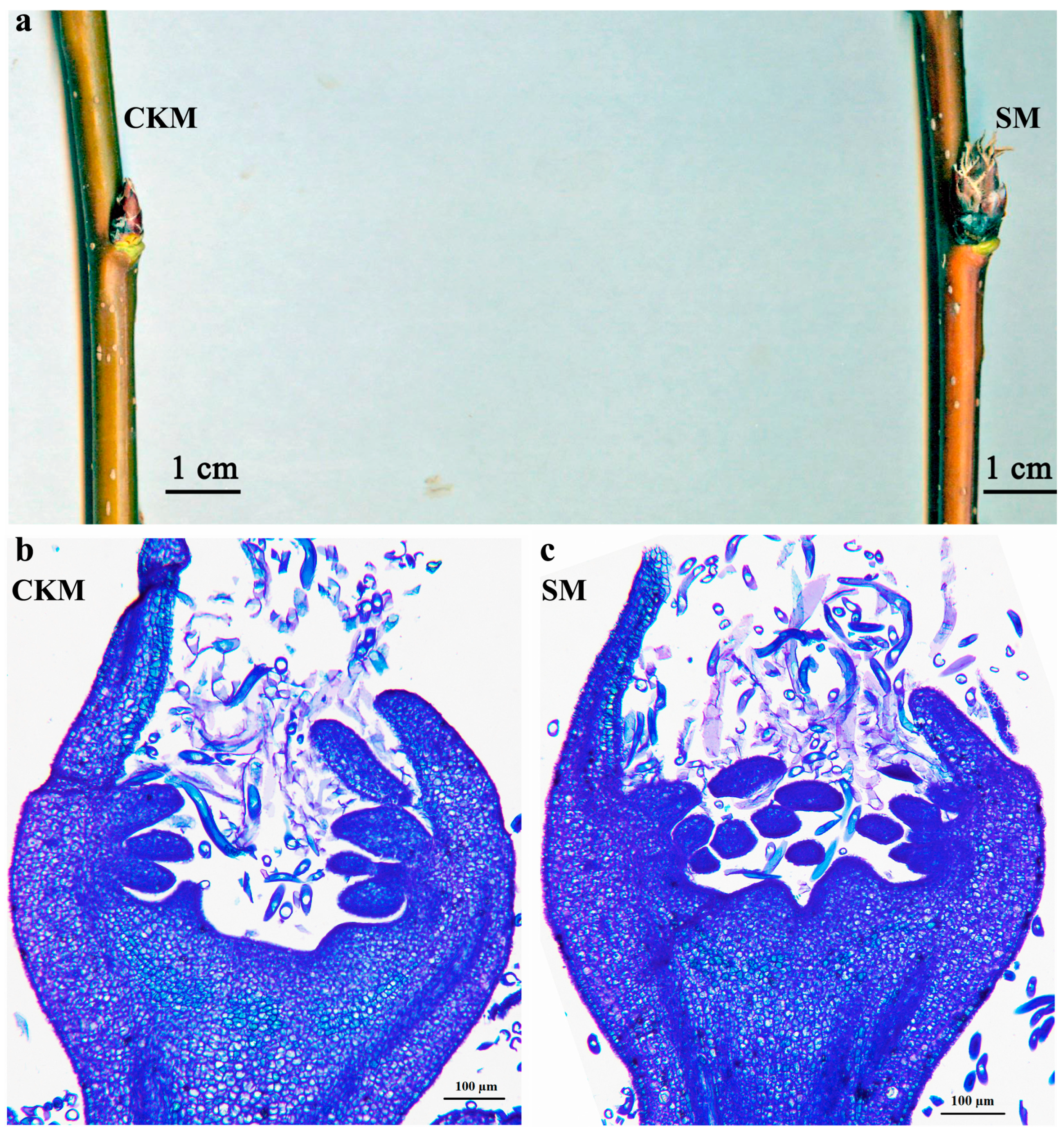
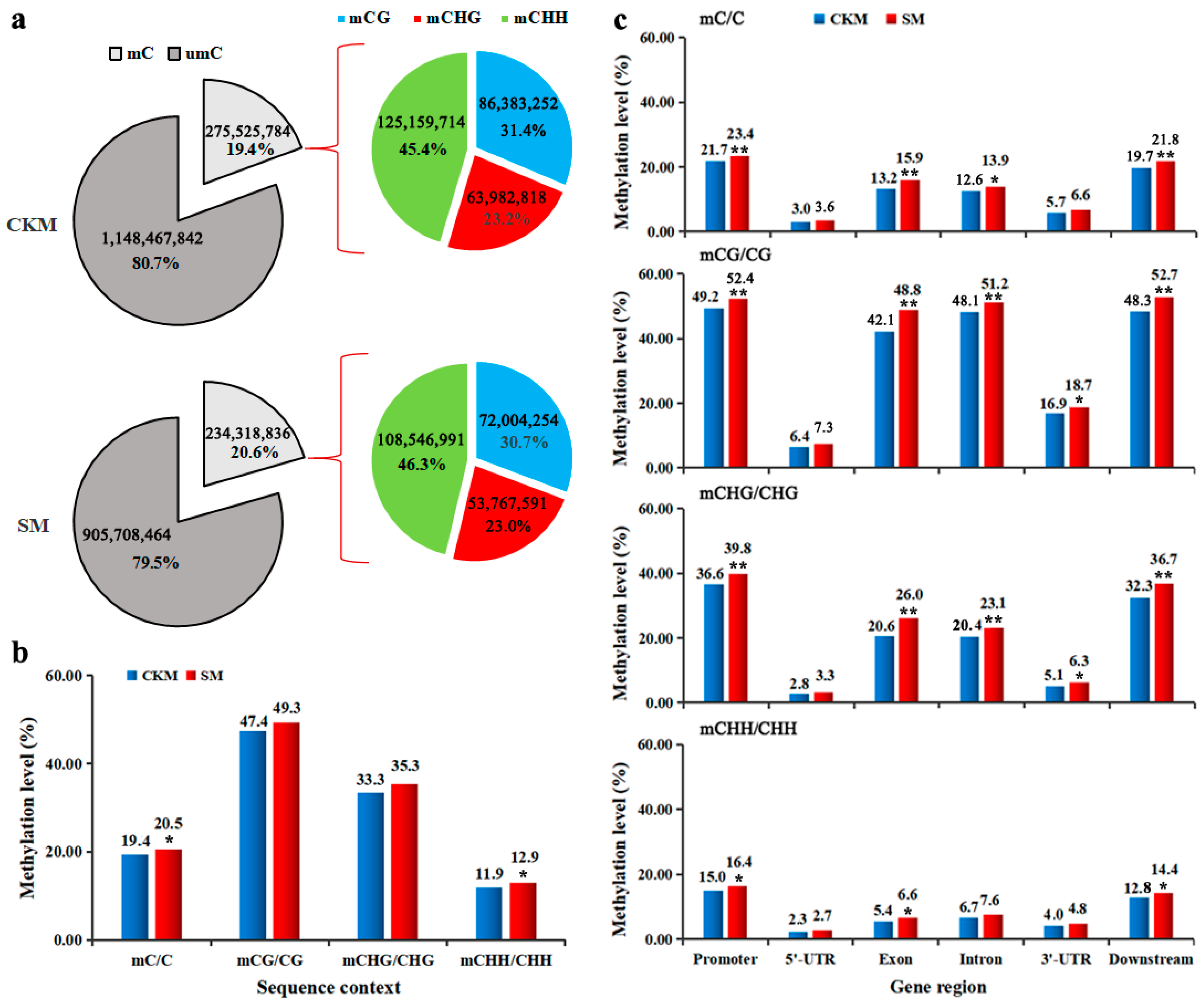
2.1. Methylation Patterns of Pear Flower Buds
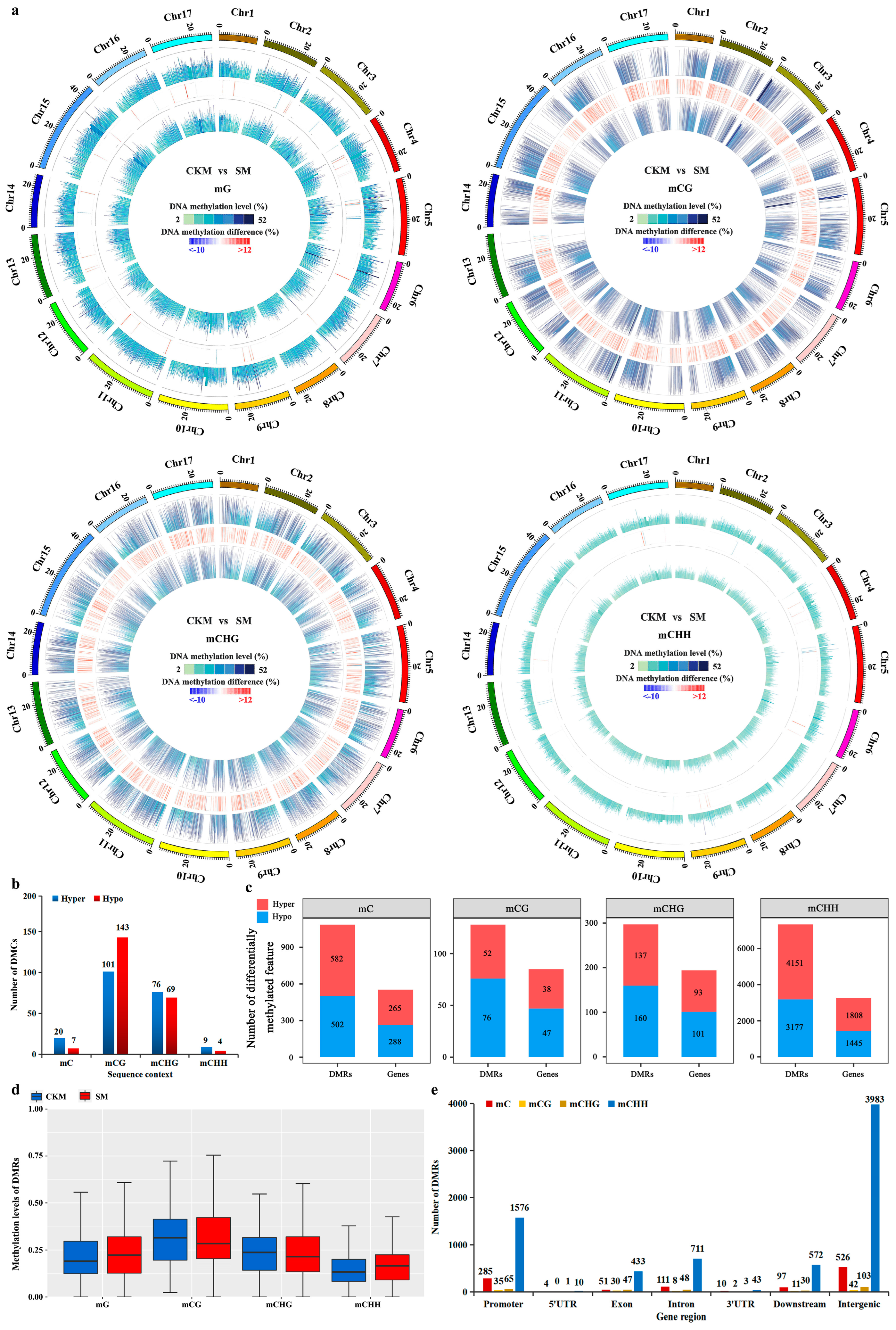
2.2. Analysis of Differential Methylation between CKM and SM
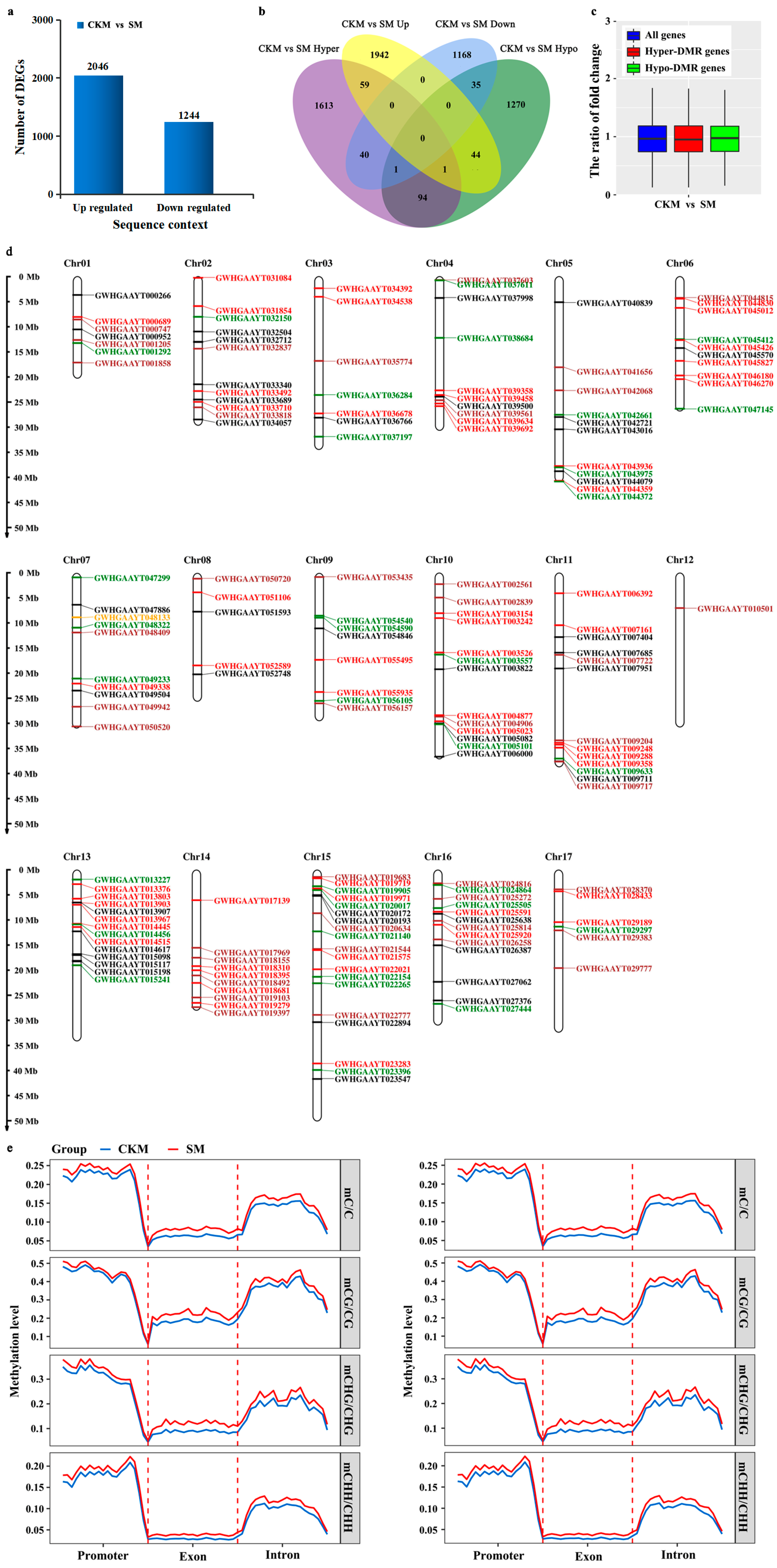
2.3. Differences in Changes in DNA Methylation Patterns and Gene Expression Levels during Flower Bud Wizening
2.4. KEGG and GO Enrichment Analysis of DMEGs
2.5. Association between DNA Methylation, Gene Expression and Hormone Homeostasis to Avoid Bud Wizening
3. Discussion
3.1. Mapping of the DNA Methylome in Pear Flower Buds by WGBS
3.2. Changes in DNA Methylation and Their Association with Various Gene Expression Changes as Buds Wizened
3.3. Possible Regulatory Role of DNA Methylation in Pear Flower Buds
4. Materials and Methods
4.1. Plant Materials
4.2. Branch Scanning and Paraffin Section
4.3. DNA Methylation Sequencing and Data Analysis
4.4. RNA-Seq
4.5. Combined Transcriptome and Methylome Analysis
4.6. McrBC-PCR and Quantitative Real-Time PCR (qPCR)
4.7. Hormone Assays
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, M.Y.; Xue, C.; Hu, H.J.; Li, J.M.; Xue, Y.S.; Wang, R.Z.; Fan, J.; Zou, C.; Tao, S.T.; Qin, M.F.; et al. Genome-wide association studies provide insights into the genetic determination of fruit traits of pear. Nat. Commun. 2021, 12, 1144. [Google Scholar] [CrossRef] [PubMed]
- Li, J.M.; Zhang, M.Y.; Li, X.L.; Khan, A.; Kumar, S.; Allan, A.C.; Kui, L.W.; Espley, R.V.; Wang, C.H.; Wang, R.Z.; et al. Pear genetics: Recent advances, new prospects, and a roadmap for the future. Hortic. Res. 2022, 9, uhab040. [Google Scholar] [CrossRef]
- Du, W.; Shi, C.; Hussain, S.B.; Li, M.; Fan, J.; Chen, Q.; Zhang, J.; Liu, Y.; Yang, X.; Hu, H. Morpho-physiological and transcriptome analyses provide insights into the wizened bud formation in pear trees. Agronomy 2022, 12, 484. [Google Scholar] [CrossRef]
- Yang, S.; Bai, M.D.; Hao, G.W.; Zhang, X.W.; Guo, H.P.; Fu, B.C.; Li, L.L. Effect of endogenous hormone concentrations on wizened bud in pear. J. Am. Pomol. Soc. 2020, 74, 255–263. [Google Scholar]
- Liu, Y.; Zhang, H.P.; Gu, C.; Tao, S.T.; Wang, D.S.; Guo, X.P.; Qi, K.J.; Zhang, S.L. Transcriptome profiling reveals differentially expressed genes associated with wizened flower bud formation in Chinese pear (Pyrus × bretschneideri Rehd.). J. Hortic. Sci. Biotech. 2016, 91, 227–235. [Google Scholar] [CrossRef]
- Landsberg, J.J. Apple fruit bud development and growth; analysis and an empirical model. Ann. Bot. 1974, 38, 1013–1023. [Google Scholar] [CrossRef]
- Okie, W.R.; Werner, D.J. Genetic influence on flower bud density in peach and nectarine exceeds that of environment. HortScience 1996, 31, 1010–1012. [Google Scholar] [CrossRef]
- Chandler, J.W. The hormonal regulation of flower development. J. Plant Growth Regul. 2011, 30, 242–254. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Zhao, Y.D. A role for auxin in flower development. J. Integr. Plant Biol. 2007, 49, 99–104. [Google Scholar] [CrossRef]
- Bai, S.L.; Saito, T.; Sakamoto, D.; Ito, A.; Fujii, H.; Moriguchi, T. Transcriptome analysis of Japanese pear (Pyrus pyrifolia Nakai) flower buds transitioning through endodormancy. Plant Cell Physiol. 2013, 54, 1132–1151. [Google Scholar] [CrossRef]
- Wei, J.; Yang, Q.S.; Ni, J.B.; Gao, Y.H.; Tang, Y.X.; Bai, S.L.; Teng, Y.W. Early defoliation induces auxin redistribution, promoting paradormancy release in pear buds. Plant Physiol. 2022, 190, 2739–2756. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, Y.F.; Zhou, X.Y.; Lin, J.; Liu, C.X.; Li, X.G.; Chang, Y.H. Single-base resolution methylome of different ecotype from Pyrus betulaefolia reveals epigenomic changes in response to salt stress. Sci. Hortic. 2022, 306, 111437. [Google Scholar] [CrossRef]
- Shi, M.M.; Wang, C.L.; Wang, P.; Zhang, M.L.; Liao, W.B. Methylation in DNA, histone, and RNA during flowering under stress condition: A review. Plant Sci. 2022, 324, 111431. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Nery, J.R.; Castanon, R.; Ecker, J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017, 18, 171. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.G.; Wang, Z.; Tian, L.M.; Zhang, Y.; Qi, D.; Huo, H.L.; Xu, J.Y.; Li, Z.; Liao, R.; Shi, M.; et al. De novo assembly of a wild pear (Pyrus betuleafolia) genome. Plant Biotechnol. J. 2020, 18, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Ausin, I.; Greenberg, M.V.C.; Simanshu, D.K.; Hale, C.J.; Vashisht, A.A.; Simon, S.A.; Lee, T.F.; Feng, S.; Espanola, S.D.; Meyers, B.C.; et al. Involved in de novo 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 8374–8381. [Google Scholar] [CrossRef]
- Wybouw, B.; Rybel, B.D. Cytokinin: A developing story. Trends Plant Sci. 2019, 24, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Opio, P.; Omiyama, H.; Saito, T.; Ohkawa, K.; Ohara, H.; Kondo, S. Paclobutrazol elevates auxin and abscisic acid, reduces gibberellins and zeatin and modulates their transporter genes in Marubakaido apple (Malus prunifolia Borkh. var. ringo Asami) rootstocks. Plant Physiol. Biochem. 2020, 155, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.C.; Zhang, T.Y.; Qi, Y.C.; Dong, Z.Y.; Wan, X.Y. Epigenetic dynamics and regulation of plant male reproduction. Int. J. Mol. Sci. 2022, 23, 10420. [Google Scholar] [CrossRef]
- Wang, G.F.; Köhler, C. Epigenetic processes in flowering plant reproduction. J. Exp. Bot. 2017, 68, 797–807. [Google Scholar] [CrossRef]
- Zhang, M.; Kimatu, J.N.; Xu, K.; Liu, B. DNA cytosine methylation in plant development. J. Genet. Genom. 2010, 37, 1–12. [Google Scholar] [CrossRef]
- Xing, L.B.; Li, Y.M.; Qi, S.Y.; Zhang, C.G.; Ma, W.C.; Zuo, X.Y.; Liang, J.Y.; Gao, C.; Jia, P.; Shah, K.; et al. Comparative RNA-Sequencing and DNA methylation analyses of apple (Malus domestica Borkh.) buds with diverse flowering capabilities reveal novel insights into the regulatory mechanisms of flower bud formation. Plant Cell Physiol. 2019, 60, 1702–1721. [Google Scholar] [CrossRef]
- Yang, H.; Chang, F.; You, C.; Cui, J.; Zhu, G.; Wang, L.; Zheng, Y.; Qi, J.; Ma, H. Whole-genome DNA methylation patterns and complex associations with gene structure and expression during flower development in Arabidopsis. Plant J. 2015, 81, 268–281. [Google Scholar] [CrossRef]
- Meijón, M.; Feito, I.; Valledor, L.; Rodríguez, R.; Cañal, M.J. Dynamics of DNA methylation and histone H4 acetylation during floral bud differentiation in azalea. BMC Plant Biol. 2010, 10, 10. [Google Scholar] [CrossRef]
- Cheng, Y.H.; Peng, X.Y.; Yu, Y.C.; Sun, Z.Y.; Han, L. The effects of DNA methylation inhibition on flower development in the dioecious plant Salix viminalis. Forests 2019, 10, 173. [Google Scholar] [CrossRef]
- Li, Z.G.; Li, J.; Liu, Y.H.; Wang, Z.C. DNA demethylation during Chrysanthemum floral transition following short-day treatment. Electron. J. Biotechnol. 2016, 21, 77–81. [Google Scholar] [CrossRef]
- Harris, R.A.; Wang, T.; Coarfa, C.; Nagarajan, R.P.; Hong, C.B.; Downey, S.L.; Johnson, B.E.; Fouse, S.D.; Delaney, A.; Zhao, Y.J.; et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010, 28, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Cokus, S.J.; Feng, S.H.; Zhang, X.Y.; Chen, Z.G.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Wu, Y.; Cao, A.Q.; Mao, B.G.; Zhao, B.R.; Wang, J.B. Genome-wide analysis of DNA methylation during ovule development of female-sterile rice fsv1. G3-Genes Genom. Genet. 2017, 7, 3621–3635. [Google Scholar] [CrossRef]
- Xiang, X.D.; Gao, Y.K.; Cui, J.H.; Ren, G.Z.; Yin, C.P.; Chang, J.H. Methylome and transcriptome analysis of altered leaf phenotype with autotetraploid in grape. Sci. Hortic. 2023, 307, 111534. [Google Scholar] [CrossRef]
- Zhou, P.; Zhang, X.D.; Fatima, M.; Ma, X.Y.; Fang, H.K.; Yan, H.S.; Ming, R. DNA methylome and transcriptome landscapes revealed differential characteristics of dioecious flowers in papaya. Hortic. Res. 2020, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Sehra, B.; Franks, R.G. Auxin and cytokinin act during gynoecial patterning and the development of ovules from the meristematic medial domain. Wires Dev. Biol. 2015, 4, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Campos-Rivero, G.; Osorio-Montalvo, P.; Sánchez-Borges, R.; Us-Camas, R.; Duarte-Aké, F.; De-la-Peña, C. Plant hormone signaling in flowering: An epigenetic point of view. J. Plant Physiol. 2017, 214, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Forghani, A.H.; Almodares, A.; Ehsanpour, A.A. Potential objectives for gibberellic acid and paclobutrazol under salt stress in sweet sorghum (Sorghum bicolor [L.] Moench cv. Sofra). Appl. Biol. Chem. 2018, 61, 113–124. [Google Scholar] [CrossRef]
- Zhang, D.; Ren, L.; Yue, J.H.; Shi, Y.B.; Zhuo, L.H.; Wang, L.; Shen, X.H. RNA-Seq-based transcriptome analysis of stem development and dwarfing regulation in Agapanthus praecox ssp. Orientalis (Leighton) Leighton. Gene 2015, 65, 252–267. [Google Scholar]
- Wang, G.L.; Que, F.; Xu, Z.S.; Wang, F.; Xiong, A.S. Exogenous gibberellin altered morphology, anatomic and transcriptional regulatory networks of hormones in carrot root and shoot. BMC Plant Biol. 2015, 15, 290. [Google Scholar] [CrossRef]
- Xu, Y.; Kong, W.Y.; Wang, F.Q.; Wang, J.; Tao, Y.J.; Li, W.Q.; Chen, Z.H.; Fan, F.J.; Jiang, Y.J.; Zhu, Q.H.; et al. Heterodimer formed by ROC8 and ROC5 modulates leaf rolling in rice. Plant Biotechnol. J. 2021, 19, 2662–2672. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Stroud, H.; Greenberg, M.V.; Feng, S.; Bernatavichute, Y.V.; Jacobsen, S.E. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 2013, 152, 352–364. [Google Scholar] [CrossRef]
- Xu, L.; Yuan, K.; Yuan, M.; Meng, X.B.; Chen, M.; Wu, J.G.; Li, J.Y.; Qi, Y.J. Regulation of rice tillering by RNA-directed DNA methylation at miniature inverted-repeat transposable elements. Mol. Plant 2020, 13, 8518–8563. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.D.; Wang, H.W.; Liu, S.X.; Li, Z.G.; Yang, X.H.; Yan, J.B.; Li, J.S.; Tran, L.S.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef]
- Pan, X.Q.; Welti, R.; Wang, X.M. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Liu, C.; Kan, J.; Lin, J.; Li, X. Integrated Methylome and Transcriptome Analysis between Wizened and Normal Flower Buds in Pyrus pyrifolia Cultivar ‘Sucui 1’. Int. J. Mol. Sci. 2024, 25, 7180. https://doi.org/10.3390/ijms25137180
Li H, Liu C, Kan J, Lin J, Li X. Integrated Methylome and Transcriptome Analysis between Wizened and Normal Flower Buds in Pyrus pyrifolia Cultivar ‘Sucui 1’. International Journal of Molecular Sciences. 2024; 25(13):7180. https://doi.org/10.3390/ijms25137180
Chicago/Turabian StyleLi, Hui, Chunxiao Liu, Jialiang Kan, Jin Lin, and Xiaogang Li. 2024. "Integrated Methylome and Transcriptome Analysis between Wizened and Normal Flower Buds in Pyrus pyrifolia Cultivar ‘Sucui 1’" International Journal of Molecular Sciences 25, no. 13: 7180. https://doi.org/10.3390/ijms25137180
APA StyleLi, H., Liu, C., Kan, J., Lin, J., & Li, X. (2024). Integrated Methylome and Transcriptome Analysis between Wizened and Normal Flower Buds in Pyrus pyrifolia Cultivar ‘Sucui 1’. International Journal of Molecular Sciences, 25(13), 7180. https://doi.org/10.3390/ijms25137180





