Toxicity of the New Psychoactive Substance (NPS) Clephedrone (4-Chloromethcathinone, 4-CMC): Prediction of Toxicity Using In Silico Methods for Clinical and Forensic Purposes
Abstract
1. Introduction
- (1)
- A basic β-keto phenethylamine structure: 4-CMC possesses a basic β-keto phenethylamine structure; this structure is similar to that of amphetamines and plays a significant role in toxicology due to its impact on the interactions of the substance with various neurotransmitter systems in the body, including dopaminergic, serotonergic and adrenergic systems.
- (2)
- The position of the chlorine atom: the presence of a chlorine atom in the para position (fourth position) of the phenyl ring (hence, ‘4-chloro’) is a key feature; this modification can affect the lipophilicity of the compound, how it crosses the blood–brain barrier, and its overall metabolic stability, which are important for determining its toxicological profile.
- (3)
- A methyl group linked to the nitrogen atom (amino group): the term ‘meth’ in 4-CMC indicates a methyl group attached to the nitrogen atom in the amine group; this modification is common in many stimulants and can influence the pharmacokinetics of the compound, including absorption, distribution, metabolism, and excretion (ADME), often increasing the lipid solubility of the compound and potentially leading to a faster onset of action.
- (4)
- A ketone group: The presence of a ketone group, as indicated by the suffix ‘one’ in the name, is characteristic of cathinones; in 4-CMC, this ketone group is linked to the first carbon of the ethyl chain. The ketone functionality is crucial for the substance’s reactivity and interaction with enzymatic systems in the body, affecting its metabolic pathways and potential toxicity.
- (5)
- An ethyl chain: the ethyl chain that links the amine group and the aromatic ring in 4-CMC affects the spatial configuration; this can influence how the drug binds to neural receptors, affecting its potency and the nature of its toxic effects.

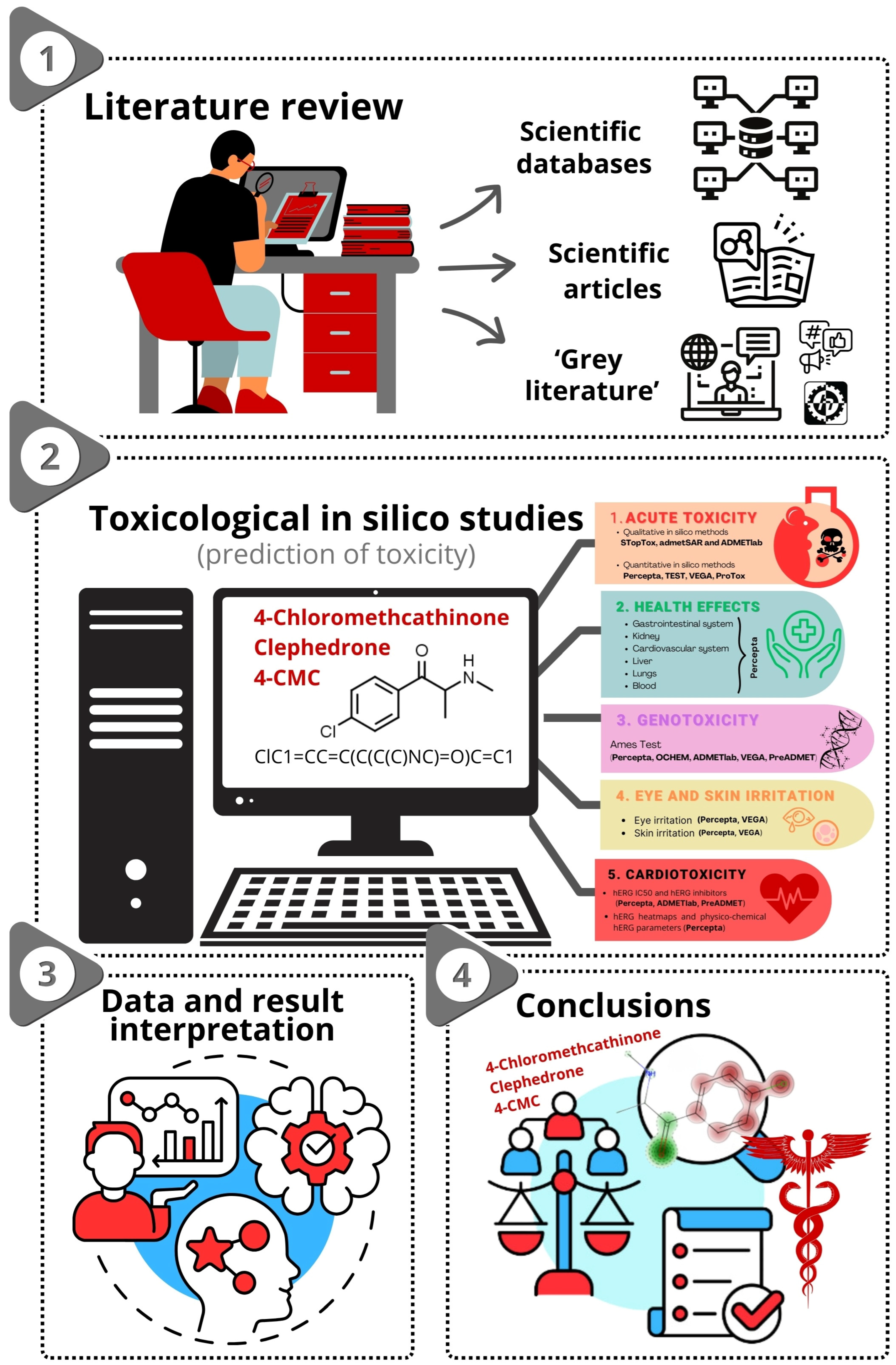
2. Results
2.1. Qualitative In Silico Methods
Acute Toxicity/Eye and Skin Irritation
2.2. Quantitative In Silico Methods
2.2.1. Acute Toxicity
2.2.2. Health Effects
2.2.3. Genotoxicity
2.2.4. Eye and Skin Irritation
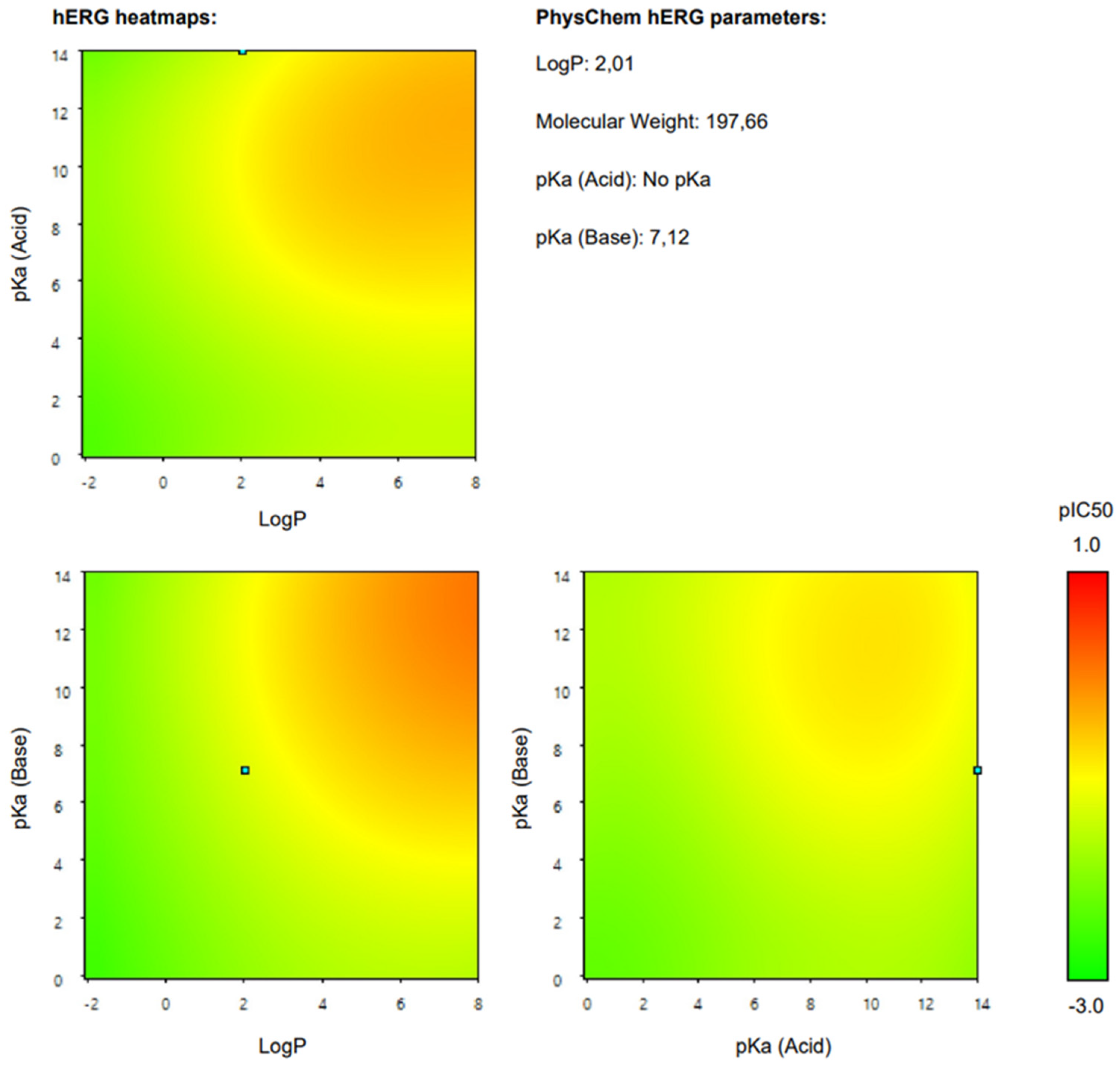
2.2.5. Cardiotoxicity
2.2.6. Endocrine System Disruption
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Toxicity Endpoints
- Acute toxicity and health effects: the prediction of acute toxicity and health effects is vital in assessing the immediate and long-term risks associated with exposure to 4-CMC; in forensic toxicology, understanding acute toxicity helps determine the cause of acute poisoning incidents, while in clinical toxicology, it guides treatment strategies for overdose cases;
- Genotoxicity: assessing the genotoxic potential of 4-CMC is critical, as genetic mutations or DNA damage can have profound implications for human health; this is particularly relevant in forensic toxicology, to evaluate long-term exposure risks and potential carcinogenicity;
- Eye and skin irritation: the prediction of eye and skin irritation is crucial for understanding the risks of accidental exposure; this information is essential in both forensic and clinical toxicology, to assess cases of dermal or ocular exposure and to provide appropriate medical interventions;
- Cardiotoxicity: given the increasing concern about the cardiovascular effects of NPSs, evaluating the cardiotoxic potential of 4-CMC is essential; this parameter assists in assessing the risk of heart-related complications, which is significant both for emergency medical responses and for forensic investigations;
- Endocrine system disruption: investigating the potential of 4-CMC to disrupt the endocrine system is important due to the critical role that hormones play in bodily functions; endocrine disruption can lead to a range of health issues, making this parameter highly relevant in clinical assessments and forensic analysis.
4.3. Utilized In Silico Software
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 4-CMC | 4-Chloromethcathinone |
| ADME | Absorption, distribution, metabolism, and excretion |
| hERG | Human ether-a-go-go-related gene |
| IC50 | Half-maximal inhibitory concentration |
| LD50 | Median lethal dose rate 50 |
| LogP | Logarithm of the partition coefficient |
| LogRBA | Logarithm of relative binding affinity |
| NPSs | New psychoactive substances |
| pKa | Acid dissociation constant |
| RI | Reliability index |
| QSAR | Quantitative structure–activity relationships |
References
- Smith, J.P.; Metters, J.P.; Khreit, O.I.G.; Sutcliffe, O.B.; Banks, C.E. Forensic Electrochemistry Applied to the Sensing of New Psychoactive Substances: Electroanalytical Sensing of Synthetic Cathinones and Analytical Validation in the Quantification of Seized Street Samples. Anal. Chem. 2014, 86, 9985–9992. [Google Scholar] [CrossRef] [PubMed]
- Grifell, M.; Ventura, M.; Carbón, X.; Quintana, P.; Galindo, L.; Palma, Á.; Fornis, I.; Gil, C.; Farre, M.; Torrens, M. Patterns of Use and Toxicity of New Para-halogenated Substituted Cathinones: 4-CMC (Clephedrone), 4-CEC (4-chloroethcatinone) and 4-BMC (Brephedrone). Hum. Psychopharmacol. 2017, 32, e2621. [Google Scholar] [CrossRef] [PubMed]
- Taschwer, M.; Weiß, J.A.; Kunert, O.; Schmid, M.G. Analysis and Characterization of the Novel Psychoactive Drug 4-Chloromethcathinone (Clephedrone). Forensic Sci. Int. 2014, 244, e56–e59. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, E.; Woźniak, M.K.; Kata, M.; Wiergowski, M.; Szpiech, B.; Biziuk, M. Blood Concentrations of a New Psychoactive Substance 4-Chloromethcathinone (4-CMC) Determined in 15 Forensic Cases. Forensic Toxicol 2018, 36, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.P.; Ferro, R.A.; Milhazes, M.; Figueira, M.; Caldeira, M.J.; Antunes, A.M.M.; Gaspar, H. Metabolic Stability and Metabolite Profiling of Emerging Synthetic Cathinones. Front. Pharmacol. 2023, 14, 1145140. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://pubchem.ncbi.nlm.nih.gov/compound/82100418 (accessed on 30 April 2023).
- Available online: https://comptox.epa.gov/dashboard/chemical/details/DTXSID201014163 (accessed on 30 April 2023).
- Wojcieszak, J.; Kuczyńska, K.; Zawilska, J.B. Four Synthetic Cathinones: 3-Chloromethcathinone, 4-Chloromethcathinone, 4-Fluoro-α-Pyrrolidinopentiophenone, and 4-Methoxy-α-Pyrrolidinopentiophenone Produce Changes in the Spontaneous Locomotor Activity and Motor Performance in Mice with Varied Profiles. Neurotox. Res. 2020, 38, 536–551. [Google Scholar] [CrossRef] [PubMed]
- Frączak, O.; Kądzioła-Długołęcka, K.; Kijewska, I.; Wilczek, R.; Tkacz-Szczęsna, B.; Makowski, K.; Komorowski, P.; Bachliński, R.; Trynda, A.; Walkowiak, B. Raman Spectroscopy and Gas Chromatography with Flame Ionization Detection as Analysis Tools in Stability Tests of Selected Synthetic Psychoactive Substances: 5-IT, MT-45 and 4-CMC. Vib. Spectrosc. 2020, 111, 103176. [Google Scholar] [CrossRef]
- Borba, J.V.B.; Alves, V.M.; Braga, R.C.; Korn, D.R.; Overdahl, K.; Silva, A.C.; Hall, S.U.S.; Overdahl, E.; Kleinstreuer, N.; Strickland, J.; et al. STopTox: An In Silico Alternative to Animal Testing for Acute Systemic and Topical Toxicity. Environ. Health Perspect. 2022, 130, 027012. [Google Scholar] [CrossRef] [PubMed]
- Botham, P.A. Acute Systemic Toxicity—Prospects for Tiered Testing Strategies. Toxicol. In Vitro 2004, 18, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Bureau, R. Nontest Methods to Predict Acute Toxicity: State of the Art for Applications of In Silico Methods. In Computational Toxicology; Nicolotti, O., Ed.; Methods in Molecular Biology; Springer New York: New York, NY, USA, 2018; Volume 1800, pp. 519–534. ISBN 978-1-4939-7898-4. [Google Scholar]
- Martin, T. User’s Guide for T. E. S. T. (Toxicity Estimation Software Tool) Version 5.1 A Java Application to Estimate Toxicities and Physical Properties from Molecular Structure; U.S. Environmental Protection Agency: Washington, DC, USA, 2020. [Google Scholar]
- Melnikov, F.; Kostal, J.; Voutchkova-Kostal, A.; Zimmerman, J.B.; Anastas, P.T. Assessment of Predictive Models for Estimating the Acute Aquatic Toxicity of Organic Chemicals. Green Chem. 2016, 18, 4432–4445. [Google Scholar] [CrossRef]
- Niu, X.; Chen, G.; Chen, Y.; Luo, N.; Wang, M.; Hu, X.; Gao, Y.; Ji, Y.; An, T. Estrogenic Effect Mechanism and Influencing Factors for Transformation Product Dimer Formed in Preservative Parabens Photolysis. Toxics 2023, 11, 186. [Google Scholar] [CrossRef] [PubMed]
- Zeiger, E. The Test That Changed the World: The Ames Test and the Regulation of Chemicals. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 2019, 841, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Fournier, M.; Vroland, C.; Megy, S.; Aguero, S.; Chemelle, J.; Defoort, B.; Jacob, G.; Terreux, R. In Silico Genotoxicity Prediction by Similarity Search and Machine Learning Algorithm: Optimization and Validation of the Method for High Energetic Materials. Propellants Explos. Pyrotech. 2023, 48, e202200259. [Google Scholar] [CrossRef]
- Vinardell, M.P.; Mitjans, M. Alternative Methods for Eye and Skin Irritation Tests: An Overview. J. Pharm. Sci. 2008, 97, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Babcock, J.; Liu, L.; Li, M.; Gao, Z. Activation of Human Ether-a-Go-Go Related Gene (hERG) Potassium Channels by Small Molecules. Acta Pharmacol. Sin. 2011, 32, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Recanatini, M.; Poluzzi, E.; Masetti, M.; Cavalli, A.; De Ponti, F. QT Prolongation through hERG K+ Channel Blockade: Current Knowledge and Strategies for the Early Prediction during Drug Development. Med. Res. Rev. 2005, 25, 133–166. [Google Scholar] [CrossRef]
- Martin, T.M.; Harten, P.; Venkatapathy, R.; Das, S.; Young, D.M. A Hierarchical Clustering Methodology for the Estimation of Toxicity. Toxicol. Mech. Methods 2008, 18, 251–266. [Google Scholar] [CrossRef]
- Tusiewicz, K.; Chłopaś-Konowałek, A.; Wachełko, O.; Zawadzki, M.; Szpot, P. A Fatal Case Involving the Highest Ever Reported 4-CMC Concentration. J. Forensic Sci. 2023, 68, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.C.; Borba, J.V.V.B.; Alves, V.M.; Hall, S.U.S.; Furnham, N.; Kleinstreuer, N.; Muratov, E.; Tropsha, A.; Andrade, C.H. Novel Computational Models Offer Alternatives to Animal Testing for Assessing Eye Irritation and Corrosion Potential of Chemicals. Artif. Intell. Life Sci. 2021, 1, 100028. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. admetSAR: A Comprehensive Source and Free Tool for Assessment of Chemical ADMET Properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Lou, C.; Tang, Y. admetSAR—A Valuable Tool for Assisting Safety Evaluation. In QSAR in Safety Evaluation and Risk Assessment; Elsevier: Amsterdam, The Netherlands, 2023; pp. 187–201. ISBN 978-0-443-15339-6. [Google Scholar]
- Moon, A.; Khan, D.; Gajbhiye, P.; Jariya, M. Insilico Prediction of Toxicity of Ligands Utilizing Admetsar. Int. J. Pharm. Bio Sci. 2017, 8, 674–677. [Google Scholar] [CrossRef]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-Service for Prediction and Optimization of Chemical ADMET Properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Martin, T.M.; Ye, L.; Sedykh, A.; Young, D.M.; Tropsha, A. Quantitative Structure−Activity Relationship Modeling of Rat Acute Toxicity by Oral Exposure. Chem. Res. Toxicol. 2009, 22, 1913–1921. [Google Scholar] [CrossRef] [PubMed]
- Drwal, M.N.; Banerjee, P.; Dunkel, M.; Wettig, M.R.; Preissner, R. ProTox: A Web Server for the in Silico Prediction of Rodent Oral Toxicity. Nucleic Acids Res. 2014, 42, W53–W58. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A Webserver for the Prediction of Toxicity of Chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wang, N.-N.; Yao, Z.-J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.-S. ADMETlab: A Platform for Systematic ADMET Evaluation Based on a Comprehensively Collected ADMET Database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An Integrated Online Platform for Accurate and Comprehensive Predictions of ADMET Properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
- Sushko, I.; Novotarskyi, S.; Körner, R.; Pandey, A.K.; Rupp, M.; Teetz, W.; Brandmaier, S.; Abdelaziz, A.; Prokopenko, V.V.; Tanchuk, V.Y.; et al. Online Chemical Modeling Environment (OCHEM): Web Platform for Data Storage, Model Development and Publishing of Chemical Information. J. Comput. Aided. Mol. Des. 2011, 25, 533–554. [Google Scholar] [CrossRef] [PubMed]
- Sushko, I.; Salmina, E.; Potemkin, V.A.; Poda, G.; Tetko, I.V. ToxAlerts: A Web Server of Structural Alerts for Toxic Chemicals and Compounds with Potential Adverse Reactions. J. Chem. Inf. Model. 2012, 52, 2310–2316. [Google Scholar] [CrossRef]
- Tetko, I.V.; Novotarskyi, S.; Sushko, I.; Ivanov, V.; Petrenko, A.E.; Dieden, R.; Lebon, F.; Mathieu, B. Development of Dimethyl Sulfoxide Solubility Models Using 163 000 Molecules: Using a Domain Applicability Metric to Select More Reliable Predictions. J. Chem. Inf. Model. 2013, 53, 1990–2000. [Google Scholar] [CrossRef]
- Oprisiu, I.; Novotarskyi, S.; Tetko, I.V. Modeling of Non-Additive Mixture Properties Using the Online CHEmical Database and Modeling Environment (OCHEM). J. Cheminform. 2013, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Martin, T. WebTEST (Web-Services Toxicity Estimation Software Tool). In Proceedings of the American Chemical Society, New Orleans, LA, USA, 18–22 March 2018. [Google Scholar] [CrossRef]
- European Commission; Joint Research Centre; Institute for Health and Consumer Protection. Review of Software Tools for Toxicity Prediction; Publications Office of the EU: Luxembourg, 2010. [Google Scholar]
- European Commission; Joint Research Centre; Institute for Health and Consumer Protection. Review of QSAR Models and Software Tools for Predicting Acute and Chronic; Publications Office of the EU: Luxembourg, 2010. [Google Scholar]
- Chavan, S.; Friedman, R.; Nicholls, I. Acute Toxicity-Supported Chronic Toxicity Prediction: A k-Nearest Neighbor Coupled Read-Across Strategy. Int. J. Mol. Sci. 2015, 16, 11659–11677. [Google Scholar] [CrossRef] [PubMed]
- Lunghini, F.; Marcou, G.; Azam, P.; Horvath, D.; Patoux, R.; Van Miert, E.; Varnek, A. Consensus Models to Predict Oral Rat Acute Toxicity and Validation on a Dataset Coming from the Industrial Context. SAR QSAR Environ. Res. 2019, 30, 879–897. [Google Scholar] [CrossRef] [PubMed]
- Benfenati, E.; Roncaglioni, A.; Lombardo, A.; Manganaro, A. Integrating QSAR, Read-Across, and Screening Tools: The VEGAHUB Platform as an Example. In Advances in Computational Toxicology; Hong, H., Ed.; Challenges and Advances in Computational Chemistry and Physics; Springer International Publishing: Cham, Switzerland, 2019; Volume 30, pp. 365–381. ISBN 978-3-030-16442-3. [Google Scholar]
| Name(s) | 4-Chloromethcathinone, Clephedrone |
|---|---|
| IUPAC name | 1-(4-chlorophenyl)-2-(methylamino)-1-propanone |
| Acronym | 4-CMC |
| Molecular formula | C10H12ClNO |
| Structural formula |  |
| CAS | 1225843-86-6 |
| SMILES | ClC1=CC=C(C(C(C)NC)=O)C=C1 |
| Molecular weight, g/mol | 197.66 |
| Density, g/mL | 1.1 |
| Melting point, °C | 198 |
| Country/Region | Status |
|---|---|
| Germany | List I controlled drug |
| Sweden | Suggested as illegal narcotic (June 2015) |
| China | Controlled substance (as of October 2015) |
| Virginia, USA | Schedule 1 substance |
| USA | Schedule II (December 2019) |
| Poland | Listed among new psychoactive substances (since August 2018) |
| Type of Acute Toxicity | STopTox (https://stoptox.mml.unc.edu/, accessed on 30 April 2023) | AdmetSAR 3.0 | ADMETlab 2.0 | ||||
|---|---|---|---|---|---|---|---|
| Prediction | Confidence (%) | Applicability Domain | Predicted Toxicophore(s) * | Classification | Probability (%) | Probability of Being Toxic | |
| Oral acute toxicity | Toxic (+) | 82.0 |  |  | III | 85.47 | 0.651 |
| Dermal acute toxicity | Toxic (+) | 67.0 |  |  | NA | NA | NA |
| Inhalation acute toxicity | Non-Toxic (−) | 50.0 |  |  | NA | NA | NA |
| Eye irritation | Negative (−) | 62.0 | 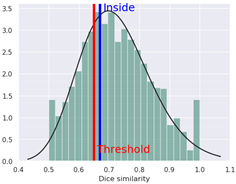 |  | − | 58.38 | NA |
| Skin irritation | Negative (−) | 80.0 |  | 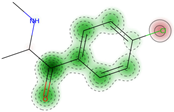 | + | 53.28 | NA |
| Software | Species | Administration Route | LD50, mg/kg bw. | Reliability/Similarity Coefficient |
|---|---|---|---|---|
| Percepta 2023.1.2 | Rat | Oral | 470.00 | Moderate, RI = 0.65 |
| Intraperitoneal | 330.00 | Moderate, RI = 0.58 | ||
| Mouse | Oral | 620.00 | Moderate, RI = 0.61 | |
| Intraperitoneal | 260.00 | High, RI = 0.83 | ||
| Intravenous | 120.00 | Moderate, RI = 0.70 | ||
| Subcutaneous | 310.00 | Moderate, RI = 0.71 | ||
| TEST 5.1.2. (Consensus) | Rat | Oral | 242.07 | SC ≥ 0.9 |
| TEST 5.1.2. (Hierarchical clustering) | Rat | Oral | 169.67 | SC ≥ 0.9 |
| TEST 5.1.2. (Nearest neighbor) | Rat | Oral | 345.38 | SC ≥ 0.9 |
| VEGA QSAR 1.2.3 | Rat | Oral | 201.85 | SC ≥ 0.9 |
| ProTox 3.0 | Rat | Oral | 340.00 | SC = 70.97% |
| Health Effect | Probability of Health Effects, % | Atomic/Functional Group Contributions to the Calculated Parameter Values | Details |
|---|---|---|---|
| Gastrointestinal system | 90 | 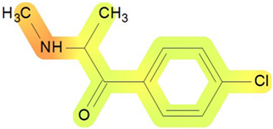 | Toxic effects: nausea or vomiting Changes in structure or function of salivary glands Route: oral Species: human, female; rat; dog Studies: acute; chronic |
| Lungs | 80 | 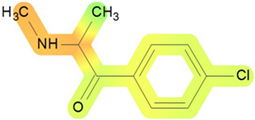 | Toxic effects: dyspnea Route: oral Species: rat; mouse Studies: acute |
| Cardiovascular system | 39 | 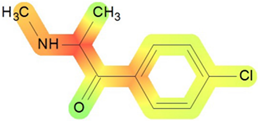 | Toxic effects: pulse rate increase without fall in blood pressure and other changes Route: oral Species: human, male and female Studies: acute |
| Blood | 33 | 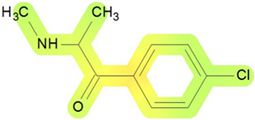 | No specific values given |
| Liver | 17 |  | Toxic effects: changes in liver weight Route: oral Species: rat Studies: chronic |
| Kidney | 15 | 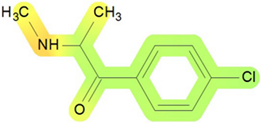 | Toxic effects: changes in bladder weight and other changes Route: oral Species: rat Studies: chronic |
| Predicted Ames Test Results | Probability (%) | Structure with Toxicophores | Software |
|---|---|---|---|
| Mutagen | 45 (Moderate reliability, RI = 0.51) | 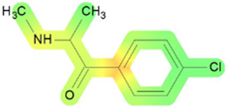 | Percepta 2023.1.2 |
| Inactive | 80 | N/A | OCHEM (https://ochem.eu/predictor/show.do, accessed on 30 April 2023) |
| Nontoxic | 88 | ADMETlab 3.0 | |
| Non-mutagen | N/A | VEGA QSAR 1.2.3 |
| Eye Irritation | Skin Irritation | Software |
|---|---|---|
| Probability: 57% | Probability: 74% | Percepta 2023.1.2 |
| Irritant (out of AD) | Not irritant (possibly out of AD) | VEGA QSAR 1.2.3 |
| Cardiotoxicity Predictions | Probability | Reliability | Software |
|---|---|---|---|
| hERG half-maximal inhibitory concentration | IC50 = 240.2 µM | N/A | Percepta 2023.1.2 |
| hERG inhibition (Ki < 10 µM, patch clamp) | 3% | Moderate (RI = 0.55) | Percepta 2023.1.2 |
| hERG inhibition (Ki < 40 µM, patch clamp) | 40% | N/A | ADMETlab 3.0 |
| Probability of Estrogen Receptor Binding | |||
|---|---|---|---|
| LogRBA > −3 | Reliability | LogRBA > 0 | Reliability |
| 0.03 | Moderate (RI = 0.62) | 0.00 | High (RI = 0.79) |
| Software | Description | Toxicity Endpoints |
|---|---|---|
| StopTox (https://stoptox.mml.unc.edu/, accessed on 30 April 2023) | STopTox is a sophisticated software designed to predict human acute toxicity tests known as the ‘6-pack’ [10]. These tests cover various aspects such as oral, dermal, and inhalation toxicity, as well as irritation and sensitization [23]. The predictions are based on quantitative structure–activity relationship (QSAR) models validated using animal experimental data. The machine learning algorithm, random forest, along with MACCS descriptors, is employed for prediction. It offers a swift screening method for assessing chemical toxicity, identifying molecular components that increase or decrease toxicity, and providing fragment contribution predictions. | Qualitative: acute toxicity, skin and eye irritation. |
| AdmetSar 3.0 | AdmetSAR has emerged as a comprehensive software solution for predicting the ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties of chemicals [24,25]. Its latest version, admetSAR 3.0, integrates over 40 predictive models using in silico filtering techniques to evaluate new chemical properties related to ADMET [26,27]. The model uses a dataset of 10,207 molecules with LD50 values determined in rat [28]. | Qualitative: oral toxicity, skin and eye irritation |
| ACD/Labs Percepta 2023.1.2 | The ACD/Labs Percepta platform is commercially available scientific software for predicting different toxicological properties using computational methods. It provides insight into how chemical structures correlate with a wide array of ADME, toxicological, and physicochemical characteristics. The platform includes a structure optimization module with comprehensive ADMET filters and takes advantage of various data inputs to generate predictions. The reliability and accuracy of Percepta models are continually evaluated and refined based on new scientific data. Research was carried out in Percepta version 2023.1.2 with a license purchased by the Institute of Medical Expertises in Łódź. | Quantitative: acute toxicity (LD50), health effects (blood, cardiovascular system, gastrointestinal system, kidneys, liver, lungs), genotoxicity (mutagenicity as Ames test), eye and skin irritation, cardiotoxicity (hERG inhibition), endocrine system disruption. |
| ProTox 3.0 | ProTox 3.0, updated scientific software released in March 2024, utilizes diverse machine learning models and databases to predict toxicological properties [29,30]. Mainly applied in drug development and chemical safety evaluation, ProTox 3.0 provides valuable insights into the potential toxic effects of novel compounds. Predictions include similarity- and fragment-based approaches, generating alerts for potential toxicity targets, with results provided with confidence scores, a comprehensive toxicity radar chart, and details on analogous known toxic compounds. | Quantitative: acute toxicity |
| ADMETlab 2.0 | ADMETlab 2.0 is an expansive scientific platform dedicated to predicting the ADMET properties of compounds [31,32]. Designed for user-friendliness and efficiency, ADMETlab 2.0 facilitates batch computation and features an intuitive interface. The prediction models utilize graph-based neural networks with an attention mechanism, producing precise and nuanced predictions across various ADMET-related endpoints. | Qualitative: acute toxicityQuantitative: genotoxicity (mutagenicity by Ames test), cardiotoxicity (hERG inhibition) |
| OCHEM (https://ochem.eu/predictor/show.do, accessed on 30 April 2023) | The online chemical modeling environment (OCHEM) is a web-based platform strategically crafted to simplify and automate traditional processes in quantitative structure–activity relationship (QSAR) modeling [33,34]. This database is intricately linked with the modeling framework, which assists users through all necessary steps in developing a predictive model: data retrieval, calculation and selection of various molecular descriptors, application of machine learning methods, validation, model analysis, and assessment of the applicability domain [35,36]. | Quantitative: genotoxicity (mutagenicity as Ames Test) |
| TEST 5.1.2 | The toxicity estimation software tool (TEST) was created as an open-source application by the U.S. EPA [37,38,39]. The software utilizes various molecular descriptors sourced from the EPA ECOTOX databases. TEST employs three QSAR approaches: the hierarchical method groups compounds based on structural similarities [21], the nearest-neighbor method averages values from structurally similar chemicals [40], and the consensus method combines predictions from all preceding methods [41]. | Quantitative: acute toxicity |
| VEGA QSAR 1.2.3 | VEGA QSAR, a collaborative effort from EU projects, integrates QSAR models and rule-based expert systems to predict various human toxicity endpoints [42]. The platform includes an applicability domain index for reliability assessment and conducts automatic checks to identify potential issues that could compromise prediction accuracy or reliability. These checks include verifying similarity between the target compound and those in the dataset, assessing concordance between experimental values and predictions, evaluating precision, and more. VEGA’s comprehensive approach ensures robust and reliable toxicity predictions, aiding in risk assessment. | Quantitative: acute toxicity, genotoxicity (mutagenicity by Ames test), eye and skin irritation. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jurowski, K.; Niżnik, Ł. Toxicity of the New Psychoactive Substance (NPS) Clephedrone (4-Chloromethcathinone, 4-CMC): Prediction of Toxicity Using In Silico Methods for Clinical and Forensic Purposes. Int. J. Mol. Sci. 2024, 25, 5867. https://doi.org/10.3390/ijms25115867
Jurowski K, Niżnik Ł. Toxicity of the New Psychoactive Substance (NPS) Clephedrone (4-Chloromethcathinone, 4-CMC): Prediction of Toxicity Using In Silico Methods for Clinical and Forensic Purposes. International Journal of Molecular Sciences. 2024; 25(11):5867. https://doi.org/10.3390/ijms25115867
Chicago/Turabian StyleJurowski, Kamil, and Łukasz Niżnik. 2024. "Toxicity of the New Psychoactive Substance (NPS) Clephedrone (4-Chloromethcathinone, 4-CMC): Prediction of Toxicity Using In Silico Methods for Clinical and Forensic Purposes" International Journal of Molecular Sciences 25, no. 11: 5867. https://doi.org/10.3390/ijms25115867
APA StyleJurowski, K., & Niżnik, Ł. (2024). Toxicity of the New Psychoactive Substance (NPS) Clephedrone (4-Chloromethcathinone, 4-CMC): Prediction of Toxicity Using In Silico Methods for Clinical and Forensic Purposes. International Journal of Molecular Sciences, 25(11), 5867. https://doi.org/10.3390/ijms25115867






