Genetic Insights into Colorectal Cancer: Evaluating PI3K/AKT Signaling Pathway Genes Expression
Abstract
1. Introduction
2. Results
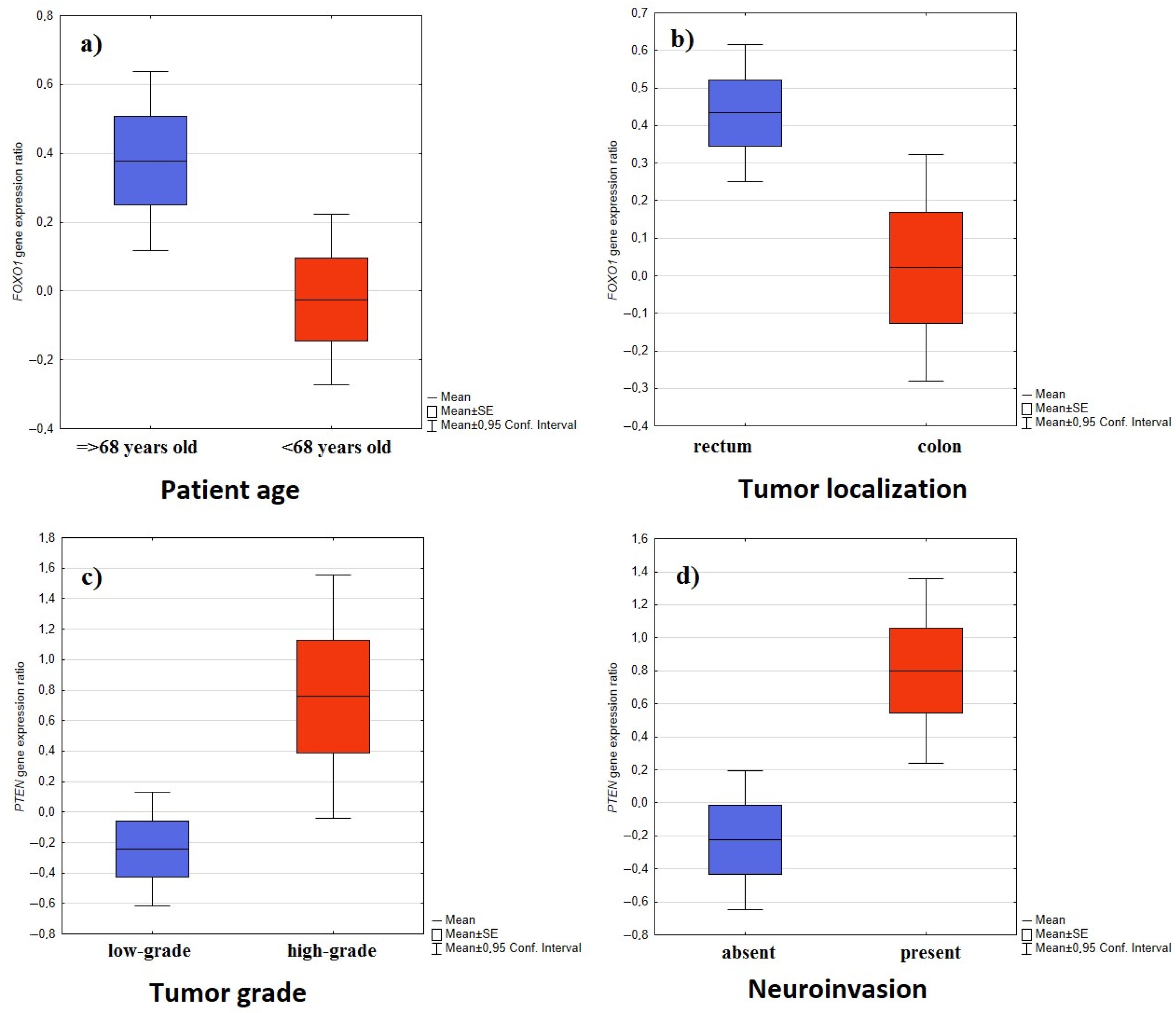
2.1. The Association between Gene Expression and the Sex and Age of Patients
2.2. The Association between Gene Expression and the Cancer Stage According to the TNM Scale (AJCC)
2.3. The Association between Gene Expression and Tumor Localization, Tumor Grading, Presence of Angioinvasion, and Neuroinvasion
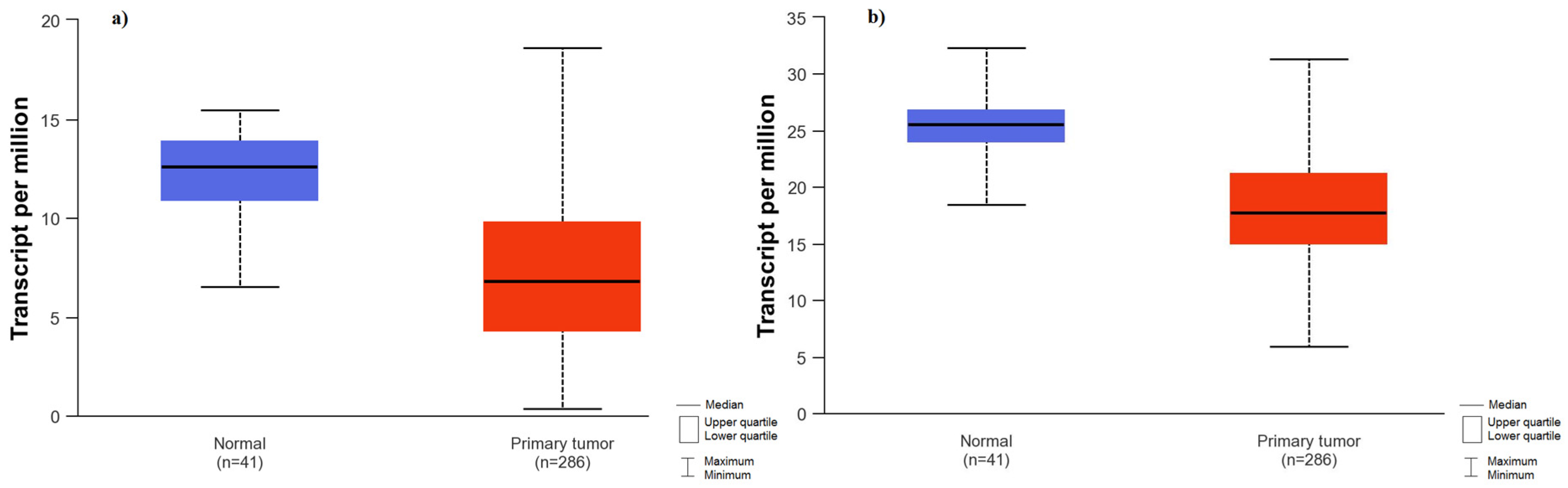
2.4. Comparison of PIK3CA, PTEN, AKT1, FRAP, FOXO1 Gene Expression and Protein Levels between Tumor Tissue from Patients with Colorectal Cancer and Normal Tissue
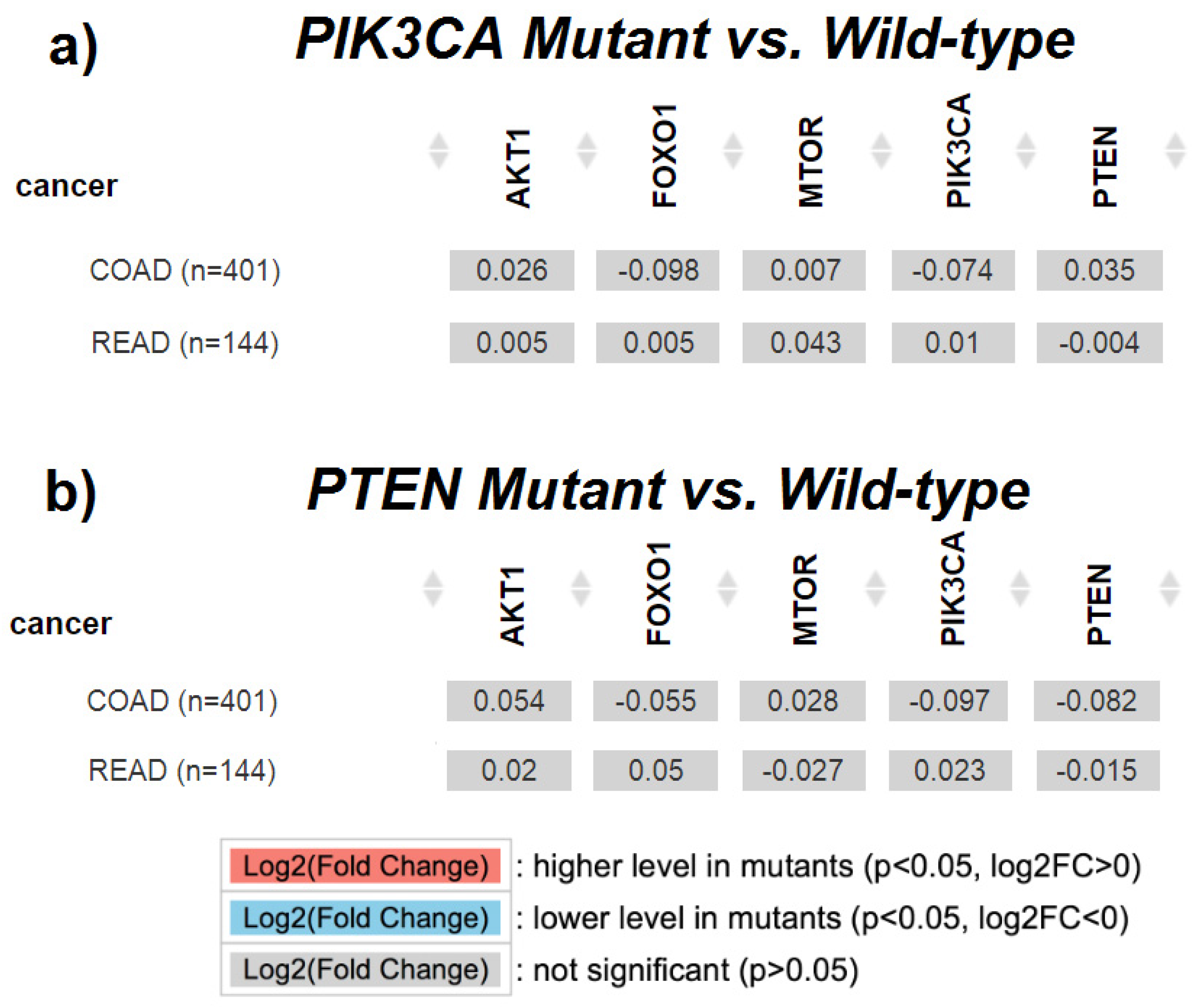
2.5. The Association between PIK3CA, PTEN, AKT1, FRAP, FOXO1 Gene Expressions and Presence of PIK3CA, PTEN Gene Mutation
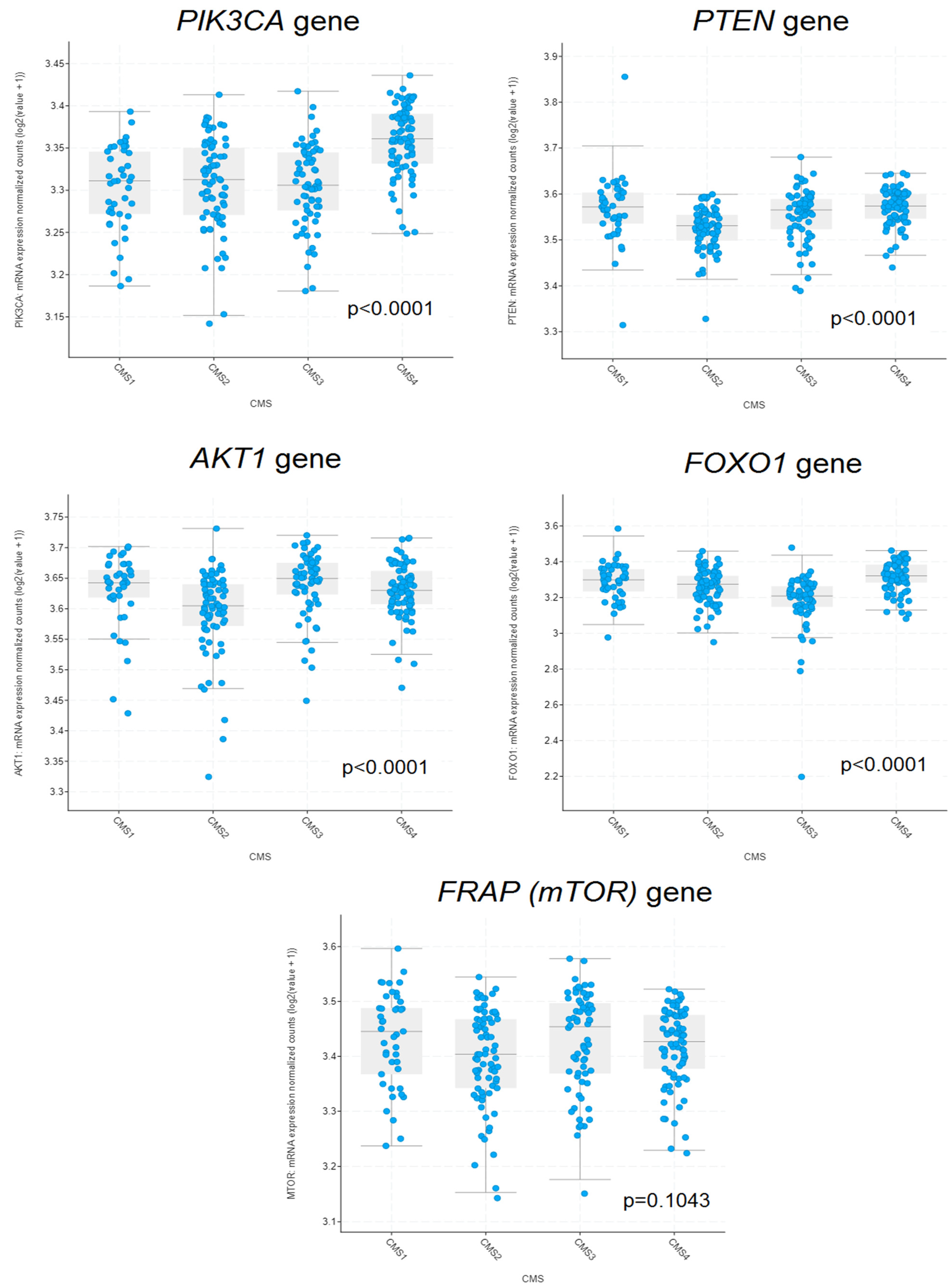
2.6. The Association between PIK3CA, PTEN, AKT1, FRAP, FOXO1 Gene Expressions and Consensus Molecular Subtype (CMS)
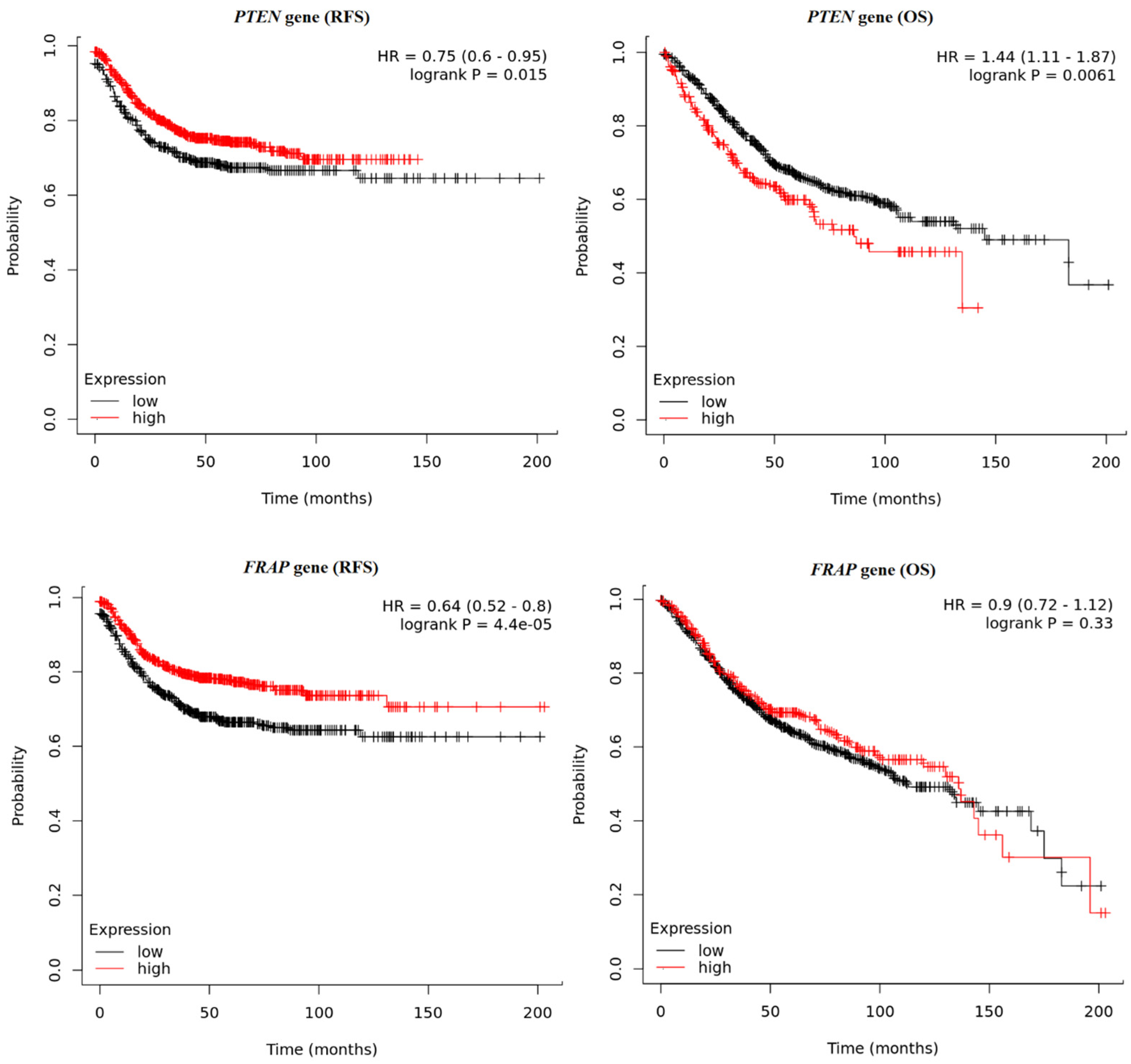
2.7. Survival Analysis Based on the PIKCA, PTEN, AKT1, FRAP, and FOXO1 Gene Expressions in Patients with Colon Cancer
3. Discussion
4. Materials and Methods
4.1. Patients and Material
4.2. RNA Extraction, Quantity, and Quality
4.3. Reverse Transcription Reaction
4.4. qPCR with Taqman Probes
4.5. Bioinformatics Analysis
4.6. Statistical Analysis of Experimental Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dahmani, A.; Delisle, J.-S. TGF-β in T Cell Biology: Implications for Cancer Immunotherapy. Cancers 2018, 10, 194. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Arcaro, A.; Guerreiro, A.S. The phosphoinositide 3-kinase pathway in human cancer: Genetic alterations and therapeutic implications. Curr. Genomics 2007, 8, 271–306. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Papa, A.; Pandolfi, P.P. The PTEN–PI3K Axis in Cancer. Biomolecules 2019, 9, 153. [Google Scholar] [CrossRef] [PubMed]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.K.; Webb, A.E. Regulation of FOXO Factors in Mammalian Cells. Curr. Top. Dev. Biol. 2018, 127, 165–192. [Google Scholar] [CrossRef] [PubMed]
- Papadatos-Pastos, D.; Rabbie, R.; Ross, P.; Sarker, D. The role of the PI3K pathway in colorectal cancer. Crit. Rev. Oncol. Hematol. 2015, 94, 18–30. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cardoso, R.; Guo, F.; Heisser, T.; De Schutter, H.; Van Damme, N.; Nilbert, M.C.; Christensen, J.; Bouvier, A.-M.; Bouvier, V.; Launoy, G.; et al. Overall and stage-specific survival of patients with screen-detected colorectal cancer in European countries: A population-based study in 9 countries. Lancet Reg. Health Eur. 2022, 21, 100458. [Google Scholar] [CrossRef]
- US researchers unveil FR that differentiates facial occlusion. Biom. Technol. Today 2017, 2017, 11. [CrossRef]
- Voutsadakis, I.A. The Landscape of PIK3CA Mutations in Colorectal Cancer. Clin. Color. Cancer 2021, 20, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Fleming-de-Moraes, C.D.; Rocha, M.R.; Tessmann, J.W.; de Araujo, W.M.; Morgado-Diaz, J.A. Crosstalk between PI3K/Akt and Wnt/β-catenin pathways promote colorectal cancer progression regardless of mutational status. Cancer Biol. Ther. 2022, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.T.; Castellano, G.; Stallone, G.; Netti, G.S.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3949. [Google Scholar] [CrossRef]
- Wang, Q.; Shao, X.; Zhang, Y.; Zhu, M.; Wang, F.X.C.; Mu, J.; Li, J.; Yao, H.; Chen, K. Role of tumor microenvironment in cancer progression and therapeutic strategy. Cancer Med. 2023, 12, 11149–11165. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Ding, S.; Zhang, X.; Di, W.; Wang, X.; Zhang, H.; Chen, Y.; Zhang, Y.; Hu, Y. To Investigate the Occurrence and Development of Colorectal Cancer Based on the PI3K/AKT/mTOR Signaling Pathway. Front. Biosci. Landmark 2023, 28, 37. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Tao, T.; Li, H.; Zhu, X. mTOR signaling pathway and mTOR inhibitors in cancer: Progress and challenges. Cell Biosci. 2020, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Jiramongkol, Y.; Lam, E.W.F. FOXO transcription factor family in cancer and metastasis. Cancer Metastasis Rev. 2020, 39, 681–709. [Google Scholar] [CrossRef]
- Khan, M.A.; Massey, S.; Ahmad, I.; Sadaf; Akhter, N.; Habib, M.; Mustafa, S.; Deo, S.V.S.; Husain, S.A. FOXO1 Gene Downregulation and Promoter Methylation Exhibits Significant Correlation With Clinical Parameters in Indian Breast Cancer Patients. Front. Genet. 2022, 13, 842943. [Google Scholar] [CrossRef]
- Ko, Y.S.; Kim, N.Y.; Pyo, J.-S. Clinicopathological significance and angiogenic role of the constitutive phosphorylation of the FOXO1 transcription factor in colorectal cancer. Pathol. Res. Pract. 2020, 216, 153150. [Google Scholar] [CrossRef]
- Beg, S.; Siraj, A.K.; Prabhakaran, S.; Jehan, Z.; Ajarim, D.; Al-Dayel, F.; Tulbah, A.; Al-Kuraya, K.S. Loss of PTEN expression is associated with aggressive behavior and poor prognosis in Middle Eastern triple-negative breast cancer. Breast Cancer Res. Treat. 2015, 151, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Yazdani, Y.; Farazmandfar, T.; Azadeh, H.; Zekavatian, Z. The prognostic effect of PTEN expression status in colorectal cancer development and evaluation of factors affecting it: miR-21 and promoter methylation. J. Biomed. Sci. 2016, 23, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, X.H.; Zheng, H.C.; Takahashi, H.; Masuda, S.; Yang, X.H.; Takano, Y. PTEN expression and mutation in colorectal carcinomas. Oncol. Rep. 2009, 22, 757–764. [Google Scholar] [CrossRef][Green Version]
- Salvatore, L.; Calegari, M.A.; Loupakis, F.; Fassan, M.; Di Stefano, B.; Bensi, M.; Bria, E.; Tortora, G. PTEN in Colorectal Cancer: Shedding Light on Its Role as Predictor and Target. Cancers 2019, 11, 1765. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Li, T.; Yang, Z.; Hu, W.; Yang, Y. Deciphering the roles of FOXO1 in human neoplasms. Int. J. Cancer 2018, 143, 1560–1568. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Rejali, L.; Seifollahi Asl, R.; Sanjabi, F.; Fatemi, N.; Asadzadeh Aghdaei, H.; Saeedi Niasar, M.; Ketabi Moghadam, P.; Nazemalhosseini Mojarad, E.; Mini, E.; Nobili, S. Principles of Molecular Utility for CMS Classification in Colorectal Cancer Management. Cancers 2023, 15, 2746. [Google Scholar] [CrossRef] [PubMed]
- Frąckowiak, M.; Lewandowski, T.; Stelmasiak, P. Molecular subtypes of colorectal cancer as a potential prognostic and predictive factor in the selection of the optimal treatment strategy. Oncol. Clin. Pract. 2019, 15, 320–325. [Google Scholar] [CrossRef]
- da Costa, A.A.B.A.; D’Almeida Costa, F.; Ribeiro, A.R.; Guimarães, A.P.; Chinen, L.T.; Lopes, C.A.P.; de Lima, V.C.C. Low PTEN expression is associated with worse overall survival in head and neck squamous cell carcinoma patients treated with chemotherapy and cetuximab. Int. J. Clin. Oncol. 2014, 20, 282–289. [Google Scholar] [CrossRef]
- Yndestad, S.; Austreid, E.; Knappskog, S.; Chrisanthar, R.; Lilleng, P.K.; Lønning, P.E.; Eikesdal, H.P. High PTEN gene expression is a negative prognostic marker in human primary breast cancers with preserved p53 function. Breast Cancer Res. Treat. 2017, 163, 177–190. [Google Scholar] [CrossRef]
- Johnson, S.M.; Gulhati, P.; Rampy, B.A.; Han, Y.; Rychahou, P.G.; Doan, H.Q.; Weiss, H.L.; Evers, B.M. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J. Am. Coll. Surg. 2010, 210, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Malinowsky, K.; Nitsche, U.; Janssen, K.P.; Bader, F.G.; Späth, C.; Drecoll, E.; Keller, G.; Höfler, H.; Slotta-Huspenina, J.; Becker, K.F. Activation of the PI3K/AKT pathway correlates with prognosis in stage II colon cancer. Br. J. Cancer 2014, 110, 2081–2089. [Google Scholar] [CrossRef] [PubMed]
- Leiphrakpam, P.D.; Are, C. PI3K/Akt/mTOR Signaling Pathway as a Target for Colorectal Cancer Treatment. Int. J. Mol. Sci. 2024, 25, 3178. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Győrffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience 2023, 45, 1889–1898. [Google Scholar] [CrossRef]



| Characteristic | Categories | n (%) |
|---|---|---|
| Sex | Male | 31 (51.6) |
| Female | 29 (48.4) | |
| Tumor localization | Rectum | 29 (48.4) |
| Sigmoid colon | 13 (21.7) | |
| Ascending colon | 5 (8.3) | |
| Rectosigmoid junction | 4 (6.7) | |
| Splenic flexure | 4 (6.7) | |
| Hepatic flexure | 4 (6.7) | |
| Tumor size (TNM) | Tis | 1 (1.7) |
| T2 | 16 (26.7) | |
| T3 | 30 (50.0) | |
| T4 | 9 (15.0) | |
| Missing | 4 (6.7) | |
| Lymph node status (TNM) | N0 | 33 (55.0) |
| N1 | 11 (18.3) | |
| N2 | 12 (20.0) | |
| Missing | 4 (6.7) | |
| Distant Metastasis (TNM) | M0 | 50 (83.3) |
| M1 | 6 (10.0) | |
| Missing | 4 (6.7) | |
| AJCC stage | I | 16 (26.7) |
| II | 17 (28.3) | |
| III | 17 (28.3) | |
| IV | 6 (10.0) | |
| Missing | 4 (6.7) | |
| Tumor grading | Low-Grade | 44 (73.3) |
| High-Grade | 14 (23.4) | |
| Missing | 2 (3.3) | |
| Angioinvasion | − | 32 (53.3) |
| + | 24 (40.0) | |
| Missing | 4 (6.7) | |
| Neuroinvasion | − | 43 (71.7) |
| + | 13 (21.6) | |
| Missing | 4 (6.7) |
| Gene Symbol | Assay ID | Dye |
|---|---|---|
| GAPDH | Hs02758991_g1 | FAM |
| ACTB | Hs01060665_g1 | VIC |
| PIK3CA | Hs00907957_m1 | FAM |
| PTEN | Hs02621230_s1 | FAM |
| FOXO1 | Hs00231106_m1 | VIC |
| FRAP | Hs00234508_m1 | VIC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Świechowski, R.; Pietrzak, J.; Wosiak, A.; Mik, M.; Balcerczak, E. Genetic Insights into Colorectal Cancer: Evaluating PI3K/AKT Signaling Pathway Genes Expression. Int. J. Mol. Sci. 2024, 25, 5806. https://doi.org/10.3390/ijms25115806
Świechowski R, Pietrzak J, Wosiak A, Mik M, Balcerczak E. Genetic Insights into Colorectal Cancer: Evaluating PI3K/AKT Signaling Pathway Genes Expression. International Journal of Molecular Sciences. 2024; 25(11):5806. https://doi.org/10.3390/ijms25115806
Chicago/Turabian StyleŚwiechowski, Rafał, Jacek Pietrzak, Agnieszka Wosiak, Michał Mik, and Ewa Balcerczak. 2024. "Genetic Insights into Colorectal Cancer: Evaluating PI3K/AKT Signaling Pathway Genes Expression" International Journal of Molecular Sciences 25, no. 11: 5806. https://doi.org/10.3390/ijms25115806
APA StyleŚwiechowski, R., Pietrzak, J., Wosiak, A., Mik, M., & Balcerczak, E. (2024). Genetic Insights into Colorectal Cancer: Evaluating PI3K/AKT Signaling Pathway Genes Expression. International Journal of Molecular Sciences, 25(11), 5806. https://doi.org/10.3390/ijms25115806





