Concanavalin A Delivers a Photoactive Protein to the Bacterial Wall
Abstract
1. Introduction
2. Results and Discussion
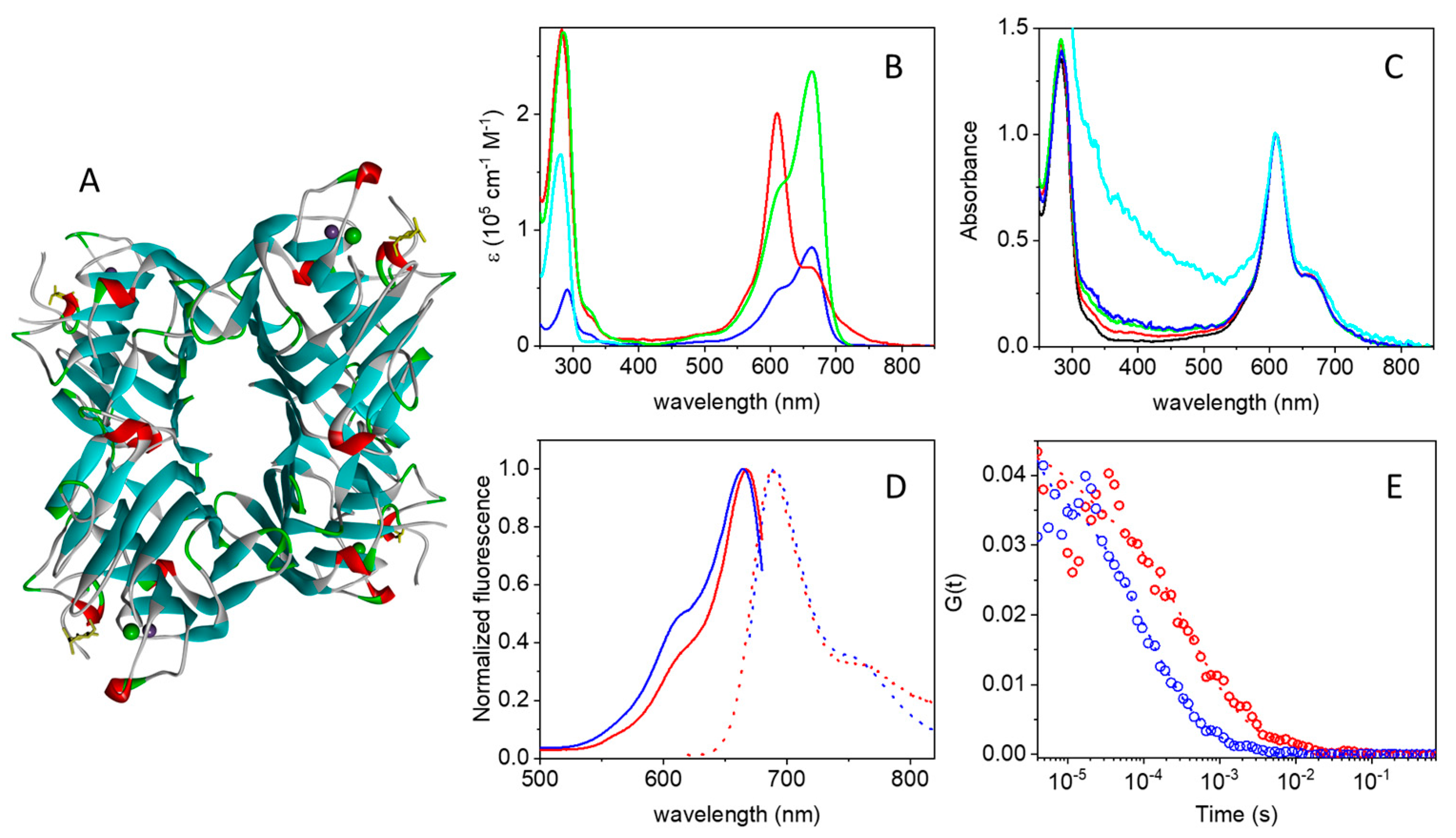
2.1. Photophysics of the MB-Streptavidin Conjugate
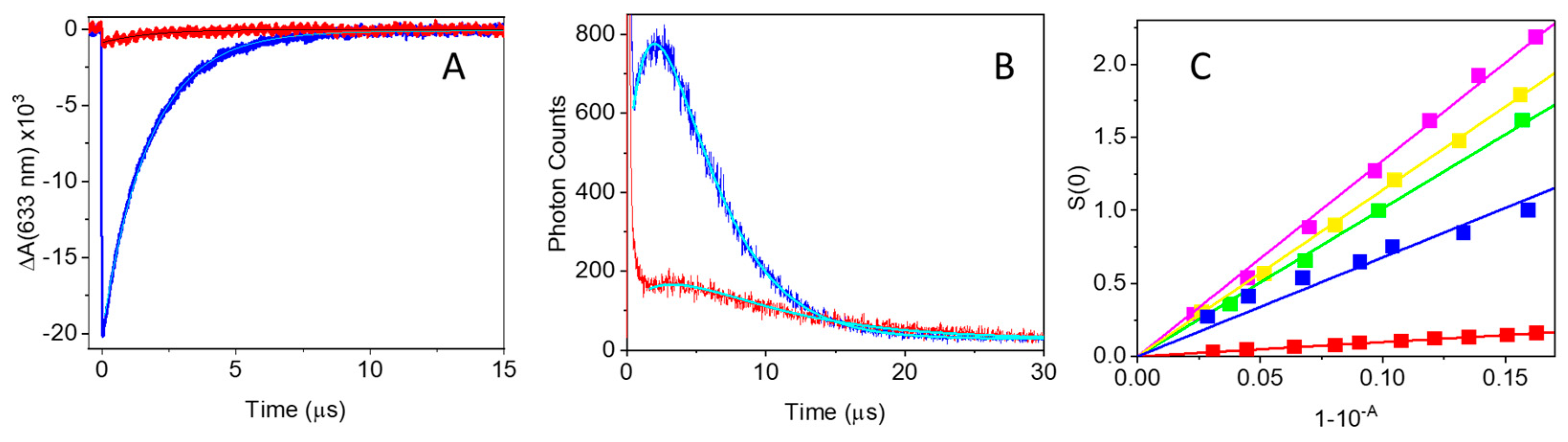
2.2. Photoactivity of MB-Streptavidin
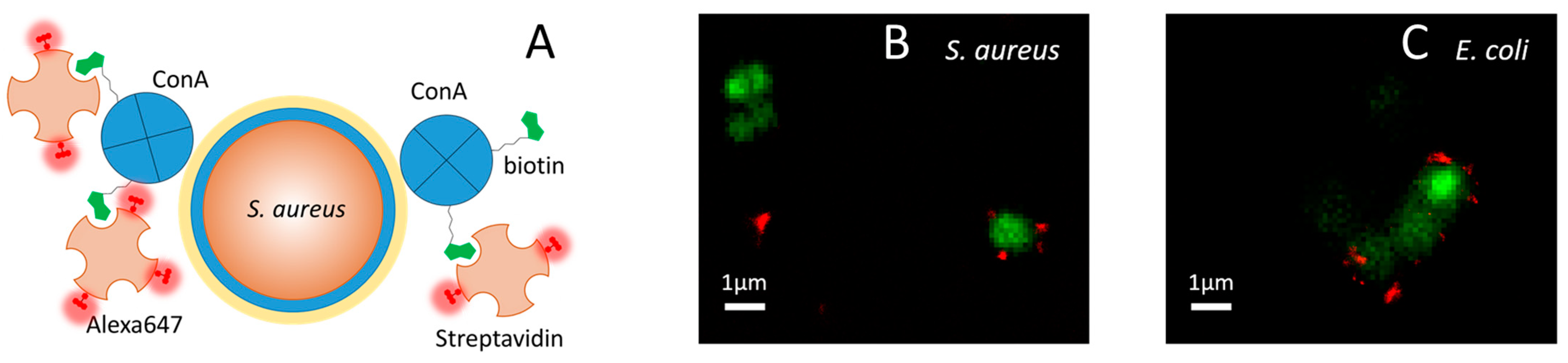
2.3. ConA Effectively Targets Bacteria
2.4. Fluorescence Correlation Spectroscopy Detects Binding of ConA to Bacteria
2.5. ConA Drives the Full Supramolecular Construct to the Bacterial Wall
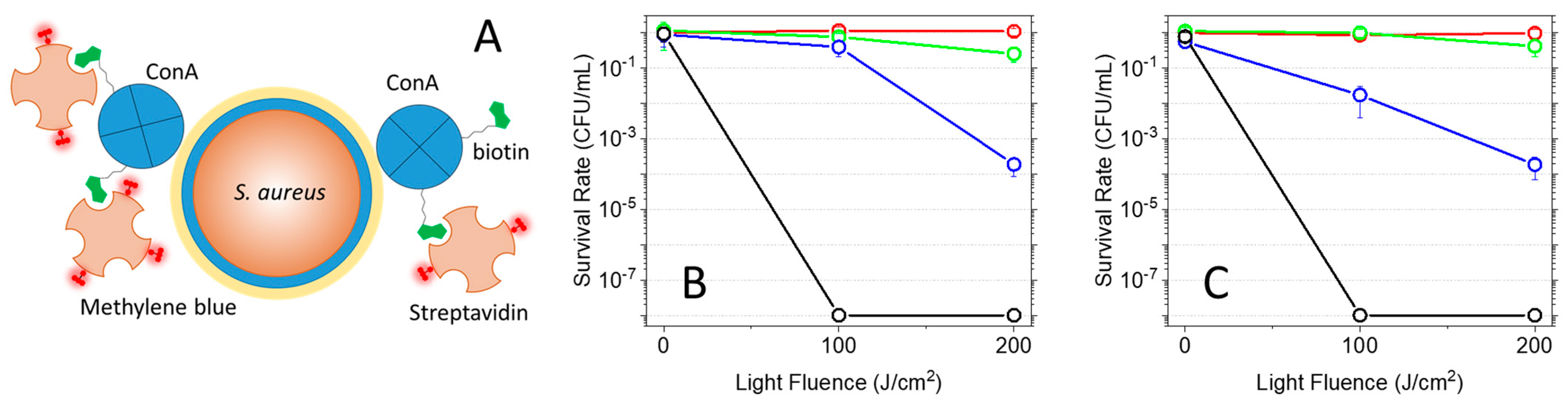
2.6. Photoinactivation of Bacteria
3. Materials and Methods
3.1. Laser Flash Photolysis
3.2. Fluorescence Quantum Yield
3.3. Singlet Oxygen Quantum Yields
3.4. Fluorescence Correlation Spectroscopy
3.5. Analysis of Cross-Correlation Curves Using the Maximum Entropy Method (MEM)
3.6. Widefield Fluorescence Imaging
3.7. dSTORM Fluorescence Imaging
3.8. Photoinactivation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance: The need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; The Review on Antimicrobial Resistance; Government of the United Kingdom: London, UK, 2016.
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R. A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, M.; Maisch, T.; Nonell, S.; Plaetzer, K.; Almeida, A.; Tegos, G.P.; Hamblin, M.R. Photoantimicrobial: Are we afraid of the light? Lancet Infect. Dis. 2017, 17, e49–e55. [Google Scholar] [CrossRef] [PubMed]
- Klausen, M.; Ucuncu, M.; Bradley, M. Design of Photosensitizing Agents for Targeted Antimicrobial Photodynamic Therapy. Molecules 2020, 25, 5239. [Google Scholar] [CrossRef] [PubMed]
- Maisch, T. Photoantimicrobials—An update. Transl. Biophotonics 2020, 2, e201900033. [Google Scholar] [CrossRef]
- Cieplik, F.; Deng, D.; Crielaard, W.; Buchalla, W.; Hellwig, E.; Al-Ahmad, A.; Maisch, T. Antimicrobial photodynamic therapy—What we know and what we don’t. Crit. Rev. Microbiol. 2018, 44, 571–589. [Google Scholar] [CrossRef] [PubMed]
- Delcanale, P.; Abbruzzetti, S.; Viappiani, C. Photodynamic treatment of pathogens. La Riv. Del Nuovo C. 2022, 405, 407–459. [Google Scholar] [CrossRef]
- Jiang, J.; Lv, X.; Cheng, H.; Yang, D.; Xu, W.; Hu, Y.; Song, Y.; Zeng, G. Type I photodynamic antimicrobial therapy: Principles, progress, and future perspectives. Acta Biomater. 2024, 177, 1–19. [Google Scholar] [CrossRef]
- Wilkinson, F.; Helman, W.P.; Ross, A.B. Rate Constants for the Decay and Reactions of the Lowest Electronically Excited Singlet State of Molecular Oxygen in Solution. An Expanded and Revised Compilation. J. Phys. Chem. Ref. Data 1995, 24, 663–1021. [Google Scholar] [CrossRef]
- Schweitzer, C.; Schmidt, R. Physical Mechanisms of Generation and Deactivation of Singlet Oxygen. Chem. Rev. 2003, 103, 1685–1758. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Banzo, A.; Ragàs, X.; Kapusta, P.; Nonell, S. Time-resolved methods in biophysics. 7. Photon counting vs. analog time-resolved singlet oxygen phosphorescence detection. Photochem. Photobiol. Sci. 2008, 7, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, E.F.F.; Pedersen, B.W.; Breitenbach, T.; Toftegaard, R.; Kuimova, M.K.; Arnaut, L.G.; Ogilby, P.R. Irradiation- and Sensitizer-Dependent Changes in the Lifetime of Intracellular Singlet Oxygen Produced in a Photosensitized Process. J. Phys. Chem. B 2012, 116, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Ogilby, P.R. Singlet oxygen: There is indeed something new under the sun. Chem. Soc. Rev. 2010, 39, 3181–3209. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.; Faustino, M.A.; Tomé, J.P. Photodynamic inactivation of bacteria: Finding the effective targets. Future Med. Chem. 2015, 7, 1221–1224. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, M.R.; Hasan, T. Photodynamic therapy: A new antimicrobial approach to infectious disease? Photochem. Photobiol. Sci. 2004, 3, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Alves, E.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Cunha, A.; Tome, J.; Almeida, A. An insight on bacterial cellular targets of photodynamic inactivation. Future Med. Chem. 2014, 6, 141–164. [Google Scholar] [CrossRef] [PubMed]
- Aydın Tekdaş, D.; Viswanathan, G.; Zehra Topal, S.; Looi, C.Y.; Wong, W.F.; Min Yi Tan, G.; Zorlu, Y.; Gürek, A.G.; Lee, H.B.; Dumoulin, F. Antimicrobial activity of a quaternized BODIPY against Staphylococcus strains. Org. Biomol. Chem. 2016, 14, 2665–2670. [Google Scholar] [CrossRef]
- Mamone, L.; Ferreyra, D.D.; Gándara, L.; Di Venosa, G.; Vallecorsa, P.; Sáenz, D.; Calvo, G.; Batlle, A.; Buzzola, F.; Durantini, E.N.; et al. Photodynamic inactivation of planktonic and biofilm growing bacteria mediated by a meso-substituted porphyrin bearing four basic amino groups. J. Photochem. Photobiol. B Biol. 2016, 161, 222–229. [Google Scholar] [CrossRef]
- Li, M.; Mai, B.; Wang, A.; Gao, Y.; Wang, X.; Liu, X.; Song, S.; Liu, Q.; Wei, S.; Wang, P. Photodynamic antimicrobial chemotherapy with cationic phthalocyanines against Escherichia coli planktonic and biofilm cultures. RSC Adv. 2017, 7, 40734–40744. [Google Scholar] [CrossRef]
- Bresolí-Obach, R.; Gispert, I.; Peña, D.G.; Boga, S.; Gulias, Ó.; Agut, M.; Vázquez, M.E.; Nonell, S. Triphenylphosphonium cation: A valuable functional group for antimicrobial photodynamic therapy. J. Biophotonics 2018, 11, e201800054. [Google Scholar] [CrossRef] [PubMed]
- Felgenträger, A.; Maisch, T.; Dobler, D.; Späth, A. Hydrogen Bond Acceptors and Additional Cationic Charges in Methylene Blue Derivatives: Photophysics and Antimicrobial Efficiency. BioMed Res. Int. 2013, 2013, 482167. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, M.; Phoenix, D.A.; Marland, J.; Wareing, D.R.A.; Bolton, F.J. A study of photobactericidal activity in the phenothiazinium series. FEMS Immunol. Med. Microbiol. 1997, 19, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Merchat, M.; Spikes, J.D.; Bertoloni, G.; Jori, G. Studies on the mechanism of bacteria photosensitization by meso-substituted cationic porphyrins. J. Photochem. Photobiol. B Biol. 1996, 35, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Vecchio, D.; Dai, T.; Huang, L.; Fantetti, L.; Roncucci, G.; Hamblin, M.R. Antimicrobial photodynamic therapy with RLP068 kills methicillin-resistant Staphylococcus aureus and improves wound healing in a mouse model of infected skin abrasion PDT with RLP068/Cl in infected mouse skin abrasion. J. Biophotonics 2013, 6, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Planas, O.; Boix-Garriga, E.; Rodríguez-Amigo, B.; Torra, J.; Bresolí-Obach, R.; Flors, C.; Viappiani, C.; Agut, M.; Ruiz-González, R.; Nonell, S. Newest approaches to singlet oxygen photosensitisation in biological media. In Photochemistry; Albini, A., Fasani, E., Eds.; The Royal Society of Chemistry: London, UK, 2014; Volume 42. [Google Scholar]
- Hally, C.; Delcanale, P.; Nonell, S.; Viappiani, C.; Abbruzzetti, S. Photosensitizing proteins for antibacterial photodynamic inactivation. Transl. Biophotonics 2020, 2, e201900031. [Google Scholar] [CrossRef]
- Pezzuoli, D.; Cozzolino, M.; Montali, C.; Brancaleon, L.; Bianchini, P.; Zantedeschi, M.; Bonardi, S.; Viappiani, C.; Abbruzzetti, S. Serum albumins are efficient delivery systems for the photosensitizer hypericin in photosensitization-based treatments against Staphylococcus aureus. Food Control 2018, 94, 254–262. [Google Scholar] [CrossRef]
- Dharmaratne, P.; Sapugahawatte, D.N.; Wang, B.; Chan, C.L.; Lau, K.-M.; Lau, C.B.S.; Fung, K.P.; Ng, D.K.P.; Ip, M. Contemporary approaches and future perspectives of antibacterial photodynamic therapy (aPDT) against methicillin-resistant Staphylococcus aureus (MRSA): A systematic review. Eur. J. Med. Chem. 2020, 200, 112341. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.-J.; Fang, Y.; Yao, M. Antimicrobial Photodynamic Therapy for Methicillin-Resistant Staphylococcus aureus Infection. BioMed Res. Int. 2013, 2013, 159157. [Google Scholar] [CrossRef]
- Embleton, M.L.; Nair, S.P.; Cookson, B.D.; Wilson, M. Antibody-Directed Photodynamic Therapy of Methicillin Resistant Staphylococcus aureus. Microb. Drug Resist. 2004, 10, 92–97. [Google Scholar] [CrossRef]
- Embleton, M.L.; Nair, S.P.; Cookson, B.D.; Wilson, M. Selective lethal photosensitization of methicillin-resistant Staphylococcus aureus using an IgG–tin (IV) chlorin e6 conjugate. J. Antimicrob. Chemother. 2002, 50, 857–864. [Google Scholar] [CrossRef] [PubMed]
- De Morais, F.A.P.; Gonçalves, R.S.; Campanholi, K.S.; de França, B.M.; Capeloto, O.A.; Lazarin-Bidoia, D.; Balbinot, R.B.; Nakamura, C.V.; Malacarne, L.C.; Caetano, W.; et al. Photophysical characterization of Hypericin-loaded in micellar, liposomal and copolymer-lipid nanostructures based F127 and DPPC liposomes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 248, 119173. [Google Scholar]
- Nieves, I.; Hally, C.; Viappiani, C.; Agut, M.; Nonell, S. A porphycene-gentamicin conjugate for enhanced photodynamic inactivation of bacteria. Bioorg. Chem. 2020, 97, 103661. [Google Scholar] [CrossRef] [PubMed]
- Bispo, M.; Anaya-Sanchez, A.; Suhani, S.; Raineri, E.J.M.; López-Álvarez, M.; Heuker, M.; Szymański, W.; Romero Pastrana, F.; Buist, G.; Horswill, A.R.; et al. Fighting Staphylococcus aureus infections with light and photoimmunoconjugates. JCI Insight 2020, 5, e139512. [Google Scholar] [CrossRef]
- Mussini, A.; Uriati, E.; Hally, C.; Nonell, S.; Bianchini, P.; Diaspro, A.; Pongolini, S.; Delcanale, P.; Abbruzzetti, S.; Viappiani, C. Versatile Supramolecular Complex for Targeted Antimicrobial Photodynamic Inactivation. Bioconjug. Chem. 2022, 33, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Cantelli, A.; Piro, F.; Pecchini, P.; Di Giosia, M.; Danielli, A.; Calvaresi, M. Concanavalin A-Rose Bengal bioconjugate for targeted Gram-negative antimicrobial photodynamic therapy. J. Photochem. Photobiol. B Biol. 2020, 206, 111852. [Google Scholar] [CrossRef] [PubMed]
- Huldani, H.; Rashid, A.I.; Turaev, K.N.; Opulencia, M.J.C.; Abdelbasset, W.K.; Bokov, D.O.; Mustafa, Y.F.; Al-Gazally, M.E.; Hammid, A.T.; Kadhim, M.M.; et al. Concanavalin A as a promising lectin-based anti-cancer agent: The molecular mechanisms and therapeutic potential. Cell Commun. Signal. 2022, 20, 167. [Google Scholar] [CrossRef] [PubMed]
- Hardman, K.D.; Ainsworth, C.F. Structure of concanavalin A at 2.4-Ang resolution. Biochemistry 1972, 11, 4910–4919. [Google Scholar] [CrossRef]
- Reeder, W.J.; Ekstedt, R.D. Study of the Interaction of Concanavalin A with Staphylococcal Teichoic Acids1. J. Immunol. 1971, 106, 334–340. [Google Scholar] [CrossRef]
- Siukstaite, L.; Imberty, A.; Römer, W. Structural Diversities of Lectins Binding to the Glycosphingolipid Gb3. Front. Mol. Biosci. 2021, 8, 704685. [Google Scholar] [CrossRef]
- Da Silva Junior, A.G.; Frias, I.A.M.; Lima-Neto, R.G.; Sá, S.R.; Oliveira, M.D.L.; Andrade, C.A.S. Concanavalin A differentiates gram-positive bacteria through hierarchized nanostructured transducer. Microbiol. Res. 2021, 251, 126834. [Google Scholar] [CrossRef] [PubMed]
- Lis, H.; Sharon, N. Lectins: Carbohydrate-Specific Proteins That Mediate Cellular Recognition. Chem. Rev. 1998, 98, 637–674. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly, F.S.; Youan, B.-B.C. Concanavalin A–Polysaccharides binding affinity analysis using a quartz crystal microbalance. Biosens. Bioelectron. 2014, 59, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Archibald, A.R.; Coapes, H.E. Blocking of Bacteriophage Receptor Sites by Concanavalin A. Microbiology 1972, 73, 581–585. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tardivo, J.P.; Del Giglio, A.; de Oliveira, C.S.; Gabrielli, D.S.; Junqueira, H.C.; Tada, D.B.; Severino, D.; de Fátima Turchiello, R.; Baptista, M.S. Methylene blue in photodynamic therapy: From basic mechanisms to clinical applications. Photodiagn. Photodyn. Ther. 2005, 2, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pérez, A.; Marbán, G. Visible Light Spectroscopic Analysis of Methylene Blue in Water; What Comes after Dimer? ACS Omega 2020, 5, 29801–29815. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.C.; von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Florence, N.; Naorem, H. Dimerization of methylene blue in aqueous and mixed aqueous organic solvent: A spectroscopic study. J. Mol. Liq. 2014, 198, 255–258. [Google Scholar] [CrossRef]
- Junqueira, H.C.; Severino, D.; Dias, L.G.; Gugliotti, M.S.; Baptista, M.S. Modulation of methylene blue photochemical properties based on adsorption at aqueous micelle interfaces. Phys. Chem. Chem. Phys. 2002, 4, 2320–2328. [Google Scholar] [CrossRef]
- Hemdan, S.S. The Shift in the Behavior of Methylene Blue Toward the Sensitivity of Medium: Solvatochromism, Solvent Parameters, Regression Analysis and Investigation of Cosolvent on the Acidity Constants. J. Fluor. 2023, 33, 2489–2502. [Google Scholar] [CrossRef]
- Olmsted, J. Calorimetric determinations of absolute fluorescence quantum yields. J. Phys. Chem. 1979, 83, 2581–2584. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006; p. 496. [Google Scholar]
- Redmond, R.W.; Gamlin, J.N. A Compilation of Singlet Oxygen Yields from Biologically Relevant Molecules. Photochem. Photobiol. 1999, 70, 391–475. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, F.; Helman, W.P.; Ross, A.B. Quantum yields for the photosensitized production of the lowest electronically excited singlet state of molecular oxygen in solution. J. Phys. Chem. Ref. Data 1993, 22, 113–263. [Google Scholar] [CrossRef]
- Van de Linde, S.; Löschberger, A.; Klein, T.; Heidbreder, M.; Wolter, S.; Heilemann, M.; Sauer, M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011, 6, 991–1009. [Google Scholar] [CrossRef] [PubMed]
- Komine, C.; Tsujimoto, Y. A Small Amount of Singlet Oxygen Generated via Excited Methylene Blue by Photodynamic Therapy Induces the Sterilization of Enterococcus faecalis. J. Endod. 2013, 39, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Usacheva, M.N.; Teichert, M.C.; Biel, M.A. Comparison of the methylene blue and toluidine blue photobactericidal efficacy against gram-positive and gram-negative microorganisms. Lasers Surg. Med. 2001, 29, 165–173. [Google Scholar] [CrossRef]
- Gollmer, A.; Felgenträger, A.; Bäumler, W.; Maisch, T.; Späth, A. A novel set of symmetric methylene blue derivatives exhibits effective bacteria photokilling—A structure—Response study. Photochem. Photobiol. Sci. 2015, 14, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Menezes, S.; Capella, M.A.M.; Caldas, L.R. Photodynamic action of methylene blue: Repair and mutation in Escherichia coli. J. Photochem. Photobiol. B Biol. 1990, 5, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Blaylock, W.K.; Yue, B.Y.; Robin, J.B. The use of concanavalin A to competitively inhibit Pseudomonas aeruginosa adherence to rabbit corneal epithelium. CLAO J. 1990, 16, 223–227. [Google Scholar]
- Pinnock, A.; Shivshetty, N.; Roy, S.; Rimmer, S.; Douglas, I.; MacNeil, S.; Garg, P. Ex vivo rabbit and human corneas as models for bacterial and fungal keratitis. Graefe’s Arch. Clin. Exp. Ophthalmol. 2017, 255, 333–342. [Google Scholar] [CrossRef]
- Gyimah, J.E.; Panigrahy, B. Adhesin-receptor interactions mediating the attachment of pathogenic Escherichia coli to chicken tracheal epithelium. Avian Dis. 1988, 32, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, A.J.; Amundsen, S.K.; Jones, J.M. Effect of carbohydrates on adherence of Escherichica coli to human urinary tract epithelial cells. Infect. Immun. 1980, 30, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Cinco, M.; Banfi, E.; Ruaro, E.; Crevatin, D.; Crotti, D. Evidence for l-fucose (6-deoxy-l-galactopyranose)-mediated adherence of Campylobacter spp. to epithelial cells. FEMS Microbiol. Lett. 1984, 21, 347–351. [Google Scholar] [CrossRef]
- Abbruzzetti, S.; Bruno, S.; Faggiano, S.; Grandi, E.; Mozzarelli, A.; Viappiani, C. Time-resolved methods in Biophysics. 2. Monitoring haem proteins at work with nanosecond laser flash photolysis. Photochem. Photobiol. Sci. 2006, 5, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Abbruzzetti, S.; Sottini, S.; Viappiani, C.; Corrie, J.E.T. Acid-induced unfolding of myoglobin triggered by a laser pH-jump method. Photochem. Photobiol. Sci. 2006, 5, 621–628. [Google Scholar] [CrossRef]
- Nonell, S.; Braslavsky, S.E. Time-resolved singlet oxygen detection. Methods Enzymol. 2000, 319, 37–49. [Google Scholar] [PubMed]
- Gensch, T.; Braslavsky, S.E. Volume changes related to triplet formation of water-soluble porphyrins. A laser-induced optoacoustic spectroscopy (LIOAS) study. J. Phys. Chem. 1997, 101, 101–108. [Google Scholar] [CrossRef]
- Schwille, P.; Haustein, E. Fluorescence Correlation Spectroscopy: An Introduction to its Concepts and Applications. Biophys. Textb. Online 2004. [Google Scholar]
- Haustein, E.; Schwille, P. Fluorescence Correlation Spectroscopy: Novel Variations of an Established Technique. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 151–169. [Google Scholar] [CrossRef]
- Steinbach, P.J.; Ionescu, R.; Matthews, C.R. Analysis of kinetics using a hybrid maximum-entropy/nonlinear-least-squares method: Application to protein folding. Biophys. J. 2002, 82, 2244–2255. [Google Scholar] [CrossRef]
- Chakraborty, S.; Steinbach, P.J.; Paul, D.; Mu, H.; Broyde, S.; Min, J.-H.; Ansari, A. Enhanced spontaneous DNA twisting/bending fluctuations unveiled by fluorescence lifetime distributions promote mismatch recognition by the Rad4 nucleotide excision repair complex. Nucleic Acids Res. 2017, 46, 1240–1255. [Google Scholar] [CrossRef]
- Connolly, M.; Arra, A.; Zvoda, V.; Steinbach, P.J.; Rice, P.A.; Ansari, A. Static Kinks or Flexible Hinges: Multiple Conformations of Bent DNA Bound to Integration Host Factor Revealed by Fluorescence Lifetime Measurements. J. Phys. Chem. B 2018, 122, 11519–11534. [Google Scholar] [CrossRef] [PubMed]
- Sternisha, S.M.; Whittington, A.C.; Martinez Fiesco, J.A.; Porter, C.; McCray, M.M.; Logan, T.; Olivieri, C.; Veglia, G.; Steinbach, P.J.; Miller, B.G. Nanosecond-Timescale Dynamics and Conformational Heterogeneity in Human GCK Regulation and Disease. Biophys. J. 2020, 118, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Jin, S.; Nagasaka, S.; Higgins, D.A.; Ito, T. Investigation of Molecular Diffusion at Block Copolymer Thin Films Using Maximum Entropy Method-Based Fluorescence Correlation Spectroscopy and Single Molecule Tracking. J. Fluor. 2022, 32, 1779–1787. [Google Scholar] [CrossRef] [PubMed]
- Wachsmuth, M.; Waldeck, W.; Langowski, J. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy11Edited by W. Baumeister. J. Mol. Biol. 2000, 298, 677–689. [Google Scholar] [CrossRef]
- Skilling, J. Classic Maximum Entropy. In Maximum Entropy and Bayesian Methods; Springer: Dordrecht, The Netherlands, 1989; pp. 45–52. [Google Scholar]
- Comas-Barceló, J.; Rodríguez-Amigo, B.; Abbruzzetti, S.; Rey-Puech, P.D.; Agut, M.; Nonell, S.; Viappiani, C. A self-assembled nanostructured material with photosensitising properties. RSC Adv. 2013, 3, 17874–17879. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mussini, A.; Delcanale, P.; Berni, M.; Pongolini, S.; Jordà-Redondo, M.; Agut, M.; Steinbach, P.J.; Nonell, S.; Abbruzzetti, S.; Viappiani, C. Concanavalin A Delivers a Photoactive Protein to the Bacterial Wall. Int. J. Mol. Sci. 2024, 25, 5751. https://doi.org/10.3390/ijms25115751
Mussini A, Delcanale P, Berni M, Pongolini S, Jordà-Redondo M, Agut M, Steinbach PJ, Nonell S, Abbruzzetti S, Viappiani C. Concanavalin A Delivers a Photoactive Protein to the Bacterial Wall. International Journal of Molecular Sciences. 2024; 25(11):5751. https://doi.org/10.3390/ijms25115751
Chicago/Turabian StyleMussini, Andrea, Pietro Delcanale, Melissa Berni, Stefano Pongolini, Mireia Jordà-Redondo, Montserrat Agut, Peter J. Steinbach, Santi Nonell, Stefania Abbruzzetti, and Cristiano Viappiani. 2024. "Concanavalin A Delivers a Photoactive Protein to the Bacterial Wall" International Journal of Molecular Sciences 25, no. 11: 5751. https://doi.org/10.3390/ijms25115751
APA StyleMussini, A., Delcanale, P., Berni, M., Pongolini, S., Jordà-Redondo, M., Agut, M., Steinbach, P. J., Nonell, S., Abbruzzetti, S., & Viappiani, C. (2024). Concanavalin A Delivers a Photoactive Protein to the Bacterial Wall. International Journal of Molecular Sciences, 25(11), 5751. https://doi.org/10.3390/ijms25115751







