Abstract
Recent studies have revealed the impact of human papillomavirus (HPV) infections on the cervicovaginal microbiome; however, few have explored the utility of self-collected specimens (SCS) for microbiome detection, obtained using standardised methods for HPV testing. Here, we present a proof-of-concept analysis utilising Oxford Nanopore sequencing of the 16S rRNA gene in paired samples collected either by the patient using an Evalyn Brush or collected by a physician using liquid-based cytology (LBC). We found no significant differences in the α-diversity estimates between the SCS and LBC samples. Similarly, when analysing β-diversity, we observed a close grouping of paired samples, indicating that both collection methods detected the same microbiome features. The identification of genera and Lactobacillus species in each sample allowed for their classification into community state types (CSTs). Notably, paired samples had the same CST, while HPV-positive and -negative samples belonged to distinct CSTs. As previously described in other studies, HPV-positive samples exhibited heightened bacterial diversity, reduced Lactobacillus abundance, and an increase in genera like Sneathia or Dialister. Altogether, this study showed comparable results between the SCS and LBC samples, underscoring the potential of self-sampling for analysing the microbiome composition in cervicovaginal samples initially collected for HPV testing in the context of cervical cancer screening.
1. Introduction
High-risk human papillomavirus (HR-HPV) infection, the most common sexually transmitted disease, is the primary cause of the development of cervical cancer [1]. It is estimated that over 80% of sexually active women during their lifetime will contract this infection, and while most infections will resolve spontaneously, a minority will progress to cervical dysplasia or cancer. In recent years, the incidence of cervical cancer in high-income countries has declined [2], largely due to the implementation of organised screening programs and human papillomavirus (HPV) vaccination. Strong evidence indicates that clinically validated HPV testing for oncogenic types is more accurate and cost-effective as a primary cervical cancer screening method than traditional cervical cytology (Pap smears) [3,4,5]. In addition, the introduction of self-sampling collection for HPV testing has emerged as a safe, accessible approach, increasing participation among women who might otherwise avoid sample collection by clinicians, therefore enhancing access to the screening test [6,7,8].
The community of microorganisms in a specific environment, also called microbiota, exerts an influence on the metabolic and immune environment of host tissues. Numerous factors such as ethnicity, genetics, lifestyle, diet, hygiene, infections, antibiotic use, sexual activity, and oestrogen have an impact on the microbiota composition. Recent research has highlighted a significant correlation between vaginal microbiota, immune status, and HPV infection [9]. In women of reproductive age, the normal vaginal microbiota is typically Lactobacillus-dominated, playing a protective role by maintaining low pH, producing antimicrobial compounds, and competitive exclusion [10]. Conversely, an increase in non-Lactobacillus-dominated microbiota such as Sneathia, Atopobium, or Megaesphera is associated with vaginal dysbiosis [11]. This dominance is reflected in the cervicovaginal environment, where microbial communities have been grouped into five main community state types (CSTs). Originally identified by Ravel et al. [10], CSTs I, II, III, and V are predominantly dominated by various Lactobacillus species. For instance, CST-I is characterised by the dominance of L. crispatus, CST-II by L. gasseri, CST-III by L. iners, and CST-V by L. jensenii. Alternatively, CST-IV is distinguished by a diverse microbiome composition, characterised by a higher abundance of non-Lactobacillus genera [10].
Abnormal vaginal environments are linked to the development and progression of cervical intraepithelial neoplasia (CIN) [12]. Although all CST types are observed across the different cervical stages including healthy, HPV-infected, LSIL (low-grade squamous intraepithelial lesion), or HSIL (high-grade squamous intraepithelial lesion), the distribution of these groups varies depending on the disease state. Significantly, as the disease stage progresses, the prevalence of CST-I and-V decreases, while CST-IV becomes more prevalent [13].
16S rRNA gene sequencing has become widely utilised to explore microbial diversity across various environments. These methods classify organisms based on phylogenetic differences reflected in their gene sequences, bypassing the need for cultivation [14]. The analysis of the 16S rRNA gene offers detailed insights into microbial populations such as species composition and abundance, and quantifies similarities and differences among microbial communities. The data obtained can be statistically analysed to assess changes over time or between groups. HPV testing using self-collected specimens (SCSs) offers comparable accuracy in detecting HR-HPV genotypes and women with a higher risk of CIN2 or worse [15,16]. However, little is known about the potential utility of self-collection on the determination of the microbiome composition using 16S rRNA gene sequencing methods, and on the identification of previously identified microbiome patterns associated with HPV infection.
Microbiome studies in cervicovaginal samples associated with HPV have mainly used liquid-based cytology (LBC) samples collected by healthcare professionals or alternatively, self-collection methods non-standardised for HPV detection [17].
In this study, we conducted a proof-of-concept analysis to evaluate the cervicovaginal microbiome and ascertain whether HPV-associated features exhibited comparable patterns between the samples collected by physicians with LBC and SCSs in a dry state.
2. Results
Fourteen out of the twenty-two paired SCS and LBC samples collected achieved sufficient sequencing depth for the study and met the sequencing quality criteria. Out of fourteen samples, eight were positive in the HPV test, while the remaining six were negative. The eight positive samples were positive for other HR-HPV types (neither 16 nor 18) (HPV-HR). Additionally, one of those samples was also positive for HPV16 (Supplementary Table S1). From the sequencing of the 16S rRNA gene, we obtained a total of 1,063,730 reads, with a median length per read of 1498 (IQR = 39) base pairs. The taxonomic assignment showed a total of 1036 genera, with a median of 131 (IQR = 88) genera per sample.
The α-diversity analysis showed no differences between the SCS and LBC samples (observed p = 0.41 and Shannon p = 0.73) (Figure 1a). HPV-positive samples showed a higher bacterial diversity than the HPV-negative samples (Figure 1b), although the differences did not reach significance (observed p = 0.10 and Shannon p = 0.39). We measured the similarity of the cervical microbiome composition (β-diversity) between both groups (LBC and SCSs). The PCA plot showed that paired samples (LBC and SCSs) exhibited close grouping (Figure 1c).
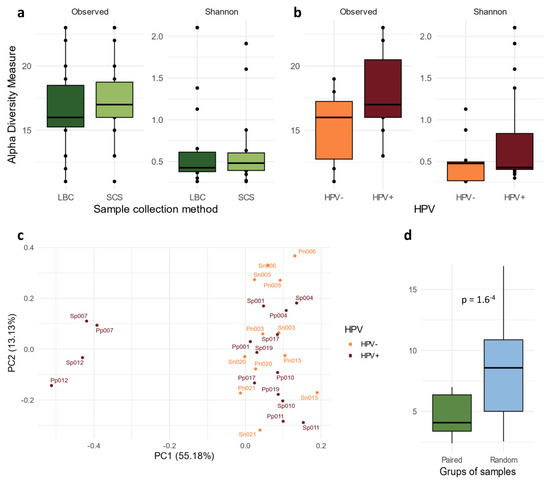
Figure 1.
The α-diversity of the cervicovaginal microbiome comparing the species diversity detected in (a) LBC and SC samples and (b) in HPV-positive and -negative samples, as measured by both the observed and Shannon indices. (c) β-diversity analysis visualised in a PCA plot, showing the genera composition of each sample (paired samples are identified with the same ID). In orange, HPV− samples, and in red, HPV+. (d) Euclidian distance between samples obtained from the same patient was measured and compared with the distance between random pairs of samples. HPV: human papillomavirus; LBC: liquid-based cytology; SCS: self-collected samples; PC: principal component.
Euclidean distance was measured between both paired and random samples to quantify the degree of similarity in microbiome composition. The results revealed that the distances between paired samples were shorter in nearly 90% of the iterations (Figure 1d) than the distance obtained after 1000 random iterations (p < 0.001).
Analysis of the cervicovaginal microbiota revealed Lactobacillus as the predominant bacterial genus across all samples. As seen in the PCA plot, two distinct clusters of samples were identified (Figure 1c). The larger cluster comprised Lactobacillus-dominated samples, whereas the smaller cluster contained two samples where Lactobacillus was not dominant, and the other genus appeared with a higher abundance. However, the abundance of Lactobacillus was lower in the HPV-positive samples, which corresponded with an increase in the abundance of other genera (Figure 2a). Furthermore, a deeper analysis focusing on the Lactobacillus species composition identified distinct bacterial profiles between the two groups. A higher abundance of L. crispatus and L. gasseri were observed in the HPV-negative samples, whereas L. inners displayed the highest abundance in the HPV-positive samples (Figure 2a).
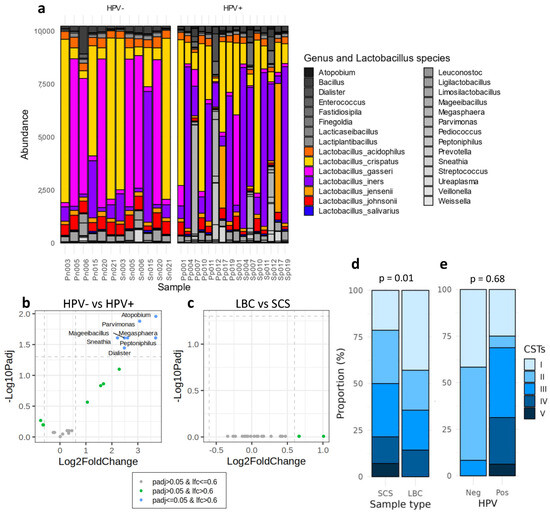
Figure 2.
(a) Microbiome composition and abundance in HPV-negative and -positive samples. (b) Differential analysis at the genus level of the microbiome composition comparing HPV-positive vs. HPV-negative samples and (c) comparing the LBC vs. SC samples. Genera in blue show statistically significant differences in the HPV+ group (c) or in SCSs (b). In the (b,c) plots, dotted lines indicate the Log2(FC) limit (vertical) and the −Log10(p.adj) threshold (horizontal). In green, genera not statistically significant but with a log2(FC) higher than 0.6. (d,e) CST classification between (d) sample collection methods (SCS or LBC) and (e) HPV-positive or -negative samples. HPV: human papillomavirus; LBC: liquid-based cytology; SCS: self-collected samples; CSTs: community state types.
Beyond Lactobacillus abundance, differential microbiome analysis at the genus level between the HPV-positive and-negative samples (Figure 2b) revealed a statistically significant increase in the relative abundance of several genera in the HPV-positive samples. Specifically, Atopobium (p.adj = 0.011), Parvimonas (p.adj = 0.013), Mageeibacillus (p.adj = 0.025), Sneathia (p.adj = 0.025), Megasphaera (p.adj = 0.025), Peptoniphilus (p.adj = 0.025), and Dialister (p.adj = 0.036) exhibited notable increases (Supplementary Table S2). On the other hand, there were no discernible differences in bacterial abundance between the sample collection methods, implying that no genera were inflated or eliminated due to differences in the sampling process (Figure 2c).
No significant differences were found when comparing the CST classification between both sample collection methods (p = 0.68) (Figure 2d). In both the SC and LBC sample types, the community state types (CSTs) were equally assigned in 11 out of 14 samples. Discordant results were observed in three women, whose professional samples were classified as CST-I, and the SCSs as CST-II, CST-III, and CST-V. This discrepancy was caused by a shift in the dominant Lactobacillus species within the sample.
A difference in the CST groups were observed comparing the HPV-positive and -negative samples (p = 0.017) (Figure 2e). Most of the HPV-negative samples were categorized into CST-I or -II types. In contrast, all CST states were found in the HPV-positive samples, with CST-III the most prevalent. Notably, CST-IV and CST-V, commonly associated with vaginal alterations, appeared only in the positive samples (Figure 2e). From all of the samples analysed, only three HPV-positive women showed histological alterations. Two of them were diagnosed with LSIL and classified as CST-IV and CST-III, respectively, using both collection methods, while the sample with an HSIL was classified as CST-I by LBC and CST-V by SCS (Supplementary Table S1).
3. Discussion
This was a proof-of-concept study to determine whether there were differences in the cervicovaginal microbiome analysis when samples were collected by a healthcare professional or self-collected by the patient using the validated methods and devices for HPV-based cervical cancer screening [18]. Alpha- and beta-diversity analysis showed that the microbiome composition and abundance in the liquid-based cytology (LBC) samples collected by a professional were similar to that of the self-collected Evalyn Brush specimens (SCS).
A study by Forney et al. found that the composition of the vaginal microbiome remained similar regardless of the specific sampling device or location within the cervicovaginal subsite [17]. Despite the sampling methods possibly having an impact on the number of cells and DNA collected, the microbiome features were equivalent within the same patient. Other studies compared the microbial diversity between the self-collected swabs or tampons with the physician-collected samples [19,20]. Our results are aligned with previous research, indicating that no differences were found between the SCS and LBC samples, neither in the alpha nor beta diversity analyses (Figure 1b,c).
Moreover, the distinctive microbiome patterns associated with HPV infection or showing an HPV lesion remained discernible in both the SCSs and professionally collected LBC. In agreement with previous published studies [21,22], the HPV-positive samples were enriched in non-Lactobacillus genus such as Atopobium, Parvimonas, Sneathia Megasphaera, Mageeibacillus, Peptoniphilus, and Dialister (Figure 2b). Additionally, clear patterns were distinguished using the CST system in both the SCSs and professionally collected LBC. CST-I and CST-II were more prevalent in the HPV-negative women, while CST-III and CST-IV were dominant in the HPV-positive women (Figure 2d,e), as previously described [13]. Notably, one sample categorised as CST-IV, indicating vaginal dysbiosis, was identified as LSIL on cytology. To conclude, the microbiome composition is mainly influenced by individual or environmental factors, resulting in personalised variations within the cervicovaginal environment [20] instead of how the sample was collected.
Therefore, this study demonstrates that both validated HPV-testing methods commonly employed in cervical cancer screening programs can be equally utilised and implemented to assess the microbiome composition in women. Knowing this, we could consider testing the microbiome using leftover DNA from the collected samples for HPV testing. This opens the door for more research on how the microbiome might affect cancer progression. Moreover, this testing could be easily added to screening procedures, so there that there no need for extra samples or follow-up appointments after receiving an HPV-positive or abnormal cytology result.
We not only demonstrate the equivalence of both collection methods for microbiome detection, but we also identified microbiome HPV-associated features using leftover samples from screening, even after some time had passed since collection. Furthermore, the entire process, from DNA extraction to sequence including the bioinformatic analysis could be conducted in a period of 24-h in the same setting designed for HPV testing. Implementing a molecular test for the accurate triage of HPV-positive women using self-collected specimens could eliminate the need for referring them to primary care centres for a second sample collection for cytological analysis.
This study was a proof-of-concept that clearly shows that the SCS method could be good to analyse the microbiome after HPV detection in cervical cancer screening. The main limitation of the study was the low number of samples analysed; therefore, it would be of considerable interest to repeat it with an expanded sample size of patients to define in more detail the differences observed between SCSs and LBC as well as other aspects such as individual HPV genotyping. Moreover, we can optimise the protocol by increasing the running times and the number of reads per sample. Further research could validate the HPV-associated patterns observed in the microbiome and determine their sensitivity and specificity for identifying women with a HR-HPV infection who are at higher risk of developing CIN2 or more.
We assigned genus and an approximation of Lactobacillus species per sample, facilitating the classification of all samples into CSTs. This enabled the detection of more differences between the HPV-positive and -negative samples that could not be seen with only genera classification. However, 16 RNA seq methods for microbiome analysis are limited by errors in the assignation of species labels, and therefore, the CST classification [23]. We could explore the use of alternative sequencing tools to reduce the error rate, however, such methods could complicate the clinical implementation due to the need for specialised facilities. Conversely, self-sampling methods offer an ease of use and convenience, making them a promising option for further research. However, to gain a more detailed understanding of how a specific microbiome composition relates to cancer progression, larger studies with species-level resolution are needed to identify more specific markers.
The impact of the sample collection method on microbiome composition does not obscure the main features of the individual microbiome profile as our results suggest that SC and LBC samples are comparable in detecting distinctive microbiome profiles.
4. Methods and Materials
A total of 22 pairs of cervicovaginal samples (LBC and SCSs) were collected from women who participated in a study to evaluate self-sampling acceptability [18]. The LBC samples were collected by a healthcare professional using the Rovers Cervex-Brush and eluted in ThinPrep vials containing PreservCyt Solution (Hologic Inc., Marlborough, MA, USA), a medium designed for collecting and preserving cells, and DNA from the patient samples. SCSs were obtained by women using Evalyn Brush (Rovers Medical Devices B·V, Oss, The Netherlands), kept dry, and then eluted in 5 mL of PreservCyt Solution at the reference laboratory.
For the microbiome analysis, we used the leftover PreservCyt solution from the ThinPrep vials and the SC samples after HPV testing with the COBAS system (Roche Molecular Diagnostics, Indianapolis, IN, USA), which detects HPV16, HPV18, and other high-risk HPV (HR-HPV) genotypes. DNA from sample remnants of both sampling methods was isolated with the Maxwell® 16 LEV Blood DNA Kit (Promega, Madison, WI, USA). The 16S rRNA gene sequencing libraries were prepared with the 16S Barcoding Kit (SQK-RAB204, Oxford Nanopore Technologies, Oxford, UK). The full-length 16S rRNA gene was amplified and barcoded from 30 ng input DNA previously extracted from the SCS and LBC samples, using LongAmp® Taq 2× master mix (New England Biolabs, Ipswich, MA, USA) and pooled in equimolar amounts of amplicons per sample. Next, the library was incubated with Library Loading Beads, the mixture was added to Flow Cell version R.9.4, and sequenced using a MinIon sequencer for approximately 4–6 h (Oxford Nanopore Technologies—ONT, Oxford, UK).
The nanopore electrical signal was converted into nucleotides with MinKNOW GUI software (version 4.1.22) (Oxford Nanopore Technologies—ONT, Oxford, UK). Porechop (v0.2.4) [24] was applied to remove the adapters from the raw reads and to eliminate reads with adapters and barcodes in the middle of the sequence, while NanoFilt (v2.7.1) [25] was employed to delete reads falling outside the specified length range of 1300 to 1600 base pairs. Quality control was executed with FASTQC (v0.11.9) [26], and samples containing reads with an average quality score lower than nine were automatically excluded. Samples with less than 10,000 reads after the quality control were excluded. Taxonomy was assigned to the DNA sequences using Kraken2 (v2.1.2) [27] with a 16S rRNA gene reference database (SILVA 138 SSU Parc database) [28].
A customised filtering algorithm was applied to reduce the sequencing noise and taxonomical error. Genera with less than 100 counts in at least two samples out of the total number of samples were excluded. To adjust differences in the library size to aid diversity comparisons, a rarefaction was applied. Read counts were normalised by rarefying the dataset for all samples to a depth of 10,241 reads/sample with phyloseq (v1.42.0).
The α-diversity assessments were conducted to compare the genera richness between groups. To quantify the dissimilarities in the microbial community composition across samples and groups, we first normalised the dataset with the centred log-ratio transformation and then we applied β-diversity analysis with the Euclidian dissimilarity metric. The results were visualised using principal component analysis (PCA), which allowed for the exploration of patterns and relationships within the data. Differential abundance analysis of the microbiome compositional data was performed with the LINDA package, which fits linear regression models on the centred log-ratio transformed data.
For the CST classification, first, we selected all of the sequences previously classified as the Lactobacillus genus with kraken2. Lactobacillus species were classified using BLAST [29]. A local database was constructed with reference 16S rRNA sequences from the following Lactobacillus species: L. acidophilus, L. crispatus, L. gasseri, L. iners, L. jensenii, L. johnsonii, and L. salivarius. Reference sequences from Lactiplantibacillus Plantarum, Limosilactobacillus reuteri, Limosilactobacillus fermentum, and Limosilactobacillus vaginallis were used as controls. Subsequently, our set of samples underwent species classification using “blastn”, where homologous sequences were identified, and each read was assigned to a specific specie. Furthermore, utilising the 13,231 16S rRNA sequences compiled in the VALENCIA software (https://github.com/ravel-lab/VALENCIA/blob/master/CST_centroids_012920.csv, accessed on 22 April 2024) [30], we trained a random forest model for CST sample classification. Once CST prediction was accomplished, we proceeded to compare the CSTs between the HPV-positive and -negative samples as well as between the SC and LBC samples.
The significance level for all p-values was established at 0.05. Data analyses were carried out with R software version 4.2.1.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25115736/s1.
Author Contributions
L.A.-P., Á.d.A.-P. and M.À.P. conceived and designed the protocol for the presented study. Á.d.A.-P. performed the experimental part, and L.A.-P. performed the computations. O.K.-L. and T.G. provided bioinformatical support, R.I. and A.A. kindly provided the sample, and L.B., S.d.S., and L.A. contributed to the conceptualisation of the work. All authors discussed the results and contributed to the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study has been funded by Instituto de Salud Carlos III through the projects PI17/00123 and PI20/01819 (co-funded by European Regional Development Fund. ERDF, a way to build Europe), CIBERESP CB06/02/0073, and the Secretariat for Universities and Research of the Department of Business and Knowledge of the Government of Catalonia grants to support the activities of research groups 2021SGR01029 and 2021SGR1354. We thank the CERCA Programme/Generalitat de Catalunya for institutional support. Á.d.A.-P. is supported by the Contratos Predoctorales de Formación en Investigación en salud (PFIS) program from the Instituto de Salud Carlos III (FI18/00244), and O.K.-L. is supported by the Formación de profesorado universitario (FPU) program from the Spanish Ministerio de Universidades (FPU2020-02907). All authors had full access to all data in the study and had final responsibility for the decision to submit for publication.
Institutional Review Board Statement
The study was approved by the Ethics Committee of Hospital Universitari de Bellvitge (PR382/23), and all subjects provided written informed consent to participate in the study.
Informed Consent Statement
All specimens were fully anonymized and retrospectively selected from a previous study in which participants gave their written informed consent for samples use in future related research. For this reason, it was not necessary to obtain a new written informed consent specifically for the present study.
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The Cancer Epidemiology Research Program (PREC) has received sponsorship for grants from Merck & Co., Roche, GSK, Reig-jofre, IDT, Hologic, and Seegene. None of these entities played a role in the data collection, data analysis, data interpretation, or report writing.
References
- Walboomers, J.M.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.F.; Peto, J.; Meijer, C.J.L.M.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Van Dyne, E.A.; Henley, S.J.; Saraiya, M.; Thomas, C.C.; Markowitz, L.E.; Benard, V.B. Trends in human papillomavirus—Associated cancers? United States, 1999–2015. Morb. Mortal. Wkly. Rep. 2018, 67, 918. [Google Scholar] [CrossRef] [PubMed]
- Meijer, C.J.L.M.; Berkhof, J.; Castle, P.E.; Hesselink, A.T.; Franco, E.L.; Ronco, G.; Arbyn, M.; Bosch, F.X.; Cuzick, J.; Dillner, J.; et al. Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int. J. Cancer 2009, 124, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Koliopoulos, G.; Nyaga, V.N.; Santesso, N.; Bryant, A.; Martin-Hirsch, P.P.L.; Mustafa, R.A.; Schünemann, H.; Paraskevaidis, E.; Arbyn, M. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst. Rev. 2017, 8, CD008587. [Google Scholar] [CrossRef] [PubMed]
- Giorgi Rossi, P.; Baldacchini, F.; Ronco, G. The possible effects on socio-economic inequalities of introducing HPV testing as primary test in cervical cancer screening programs. Front. Oncol. 2014, 4, 20. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Serrano, B.; Ibáñez, R.; Robles, C.; Peremiquel-Trillas, P.; De Sanjosé, S.; Bruni, L. Worldwide use of HPV self-sampling for cervical cancer screening. Prev. Med. 2022, 154, 106900. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Guideline on Self-Care Interventions for Health and Well-Being; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Nishimura, H.; Yeh, P.T.; Oguntade, H.; Kennedy, C.E.; Narasimhan, M. HPV self-sampling for cervical cancer screening: A systematic review of values and preferences. BMJ Glob. Health 2021, 6, e003743. [Google Scholar] [CrossRef]
- Läsche, M.; Urban, H.; Gallwas, J.; Gründker, C. HPV and other microbiota; who’s good and who’s bad: Effects of the microbial environment on the development of cervical cancer—A non-systematic review. Cells 2021, 10, 714. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef]
- Gardella, B.; Pasquali, M.F.; La Verde, M.; Cianci, S.; Torella, M.; Dominoni, M. The complex interplay between vaginal microbiota, HPV infection, and immunological microenvironment in cervical intraepithelial neoplasia: A literature review. Int. J. Mol. Sci. 2022, 23, 7174. [Google Scholar] [CrossRef]
- Audirac-Chalifour, A.; Torres-Poveda, K.; Bahena-Román, M.; Téllez-Sosa, J.; Martínez-Barnetche, J.; Cortina-Ceballos, B.; López-Estrada, G.; Delgado-Romero, K.; Burguete-García, A.I.; Cantú, D.; et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study. PLoS ONE 2016, 11, e0153274. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.A.; Andralojc, K.M.; Huynen, M.A.; Leenders, W.P.; Melchers, W.J. In-depth insights into cervicovaginal microbial communities and hrHPV infections using high-resolution microbiome profiling. NPJ Biofilms Microbiomes 2022, 8, 75. [Google Scholar] [CrossRef]
- Nocker, A.; Burr, M.; Camper, A.K. Genotypic microbial community profiling: A critical technical review. Microb. Ecol. 2007, 54, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Arbyn, M.; Smith, S.B.; Temin, S.; Sultana, F.; Castle, P. Detecting cervical precancer and reaching underscreened women by using HPV testing on self samples: Updated meta-analyses. BMJ 2018, 363, k4823. [Google Scholar] [CrossRef]
- Kamath Mulki, A.; Withers, M. Human Papilloma Virus self-sampling performance in low-and middle-income countries. BMC Womens Health 2021, 21, 12. [Google Scholar] [CrossRef] [PubMed]
- Forney, L.J.; Gajer, P.; Williams, C.J.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Brotman, R.M.; Davis, C.C.; Ault, K.; et al. Comparison of self-collected and physician-collected vaginal swabs for microbiome analysis. J. Clin. Microbiol. 2010, 48, 1741–1748. [Google Scholar] [CrossRef]
- Ibáñez, R.; Roura, E.; Acera, A.; Andújar, M.; Pavón, M.À.; Bruni, L.; De Sanjosé, S. HPV self-sampling among cervical cancer screening users in Spain: A randomized clinical trial of on-site training to increase the acceptability. Prev. Med. 2023, 173, 107571. [Google Scholar] [CrossRef] [PubMed]
- Turner, F.; Drury, J.; Hapangama, D.K.; Tempest, N. Menstrual Tampons Are Reliable and Acceptable Tools to Self-Collect Vaginal Microbiome Samples. Int. J. Mol. Sci. 2023, 24, 14121. [Google Scholar] [CrossRef] [PubMed]
- Wylie, K.M.; Blankenship, S.A.; Tuuli, M.G.; Macones, G.A.; Stout, M.J. Evaluation of patient-versus provider-collected vaginal swabs for microbiome analysis during pregnancy. BMC Res. Notes 2018, 11, 706. [Google Scholar] [CrossRef]
- Fang, B.; Li, Q.; Wan, Z.; OuYang, Z.; Zhang, Q. Exploring the association between cervical microbiota and HR-HPV infection based on 16S rRNA gene and metagenomic sequencing. Front. Cell. Infect. Microbiol. 2022, 12, 922554. [Google Scholar] [CrossRef]
- Usyk, M.; Zolnik, C.P.; Castle, P.E.; Porras, C.; Herrero, R.; Gradissimo, A.; Gonzalez, P.; Safaeian, M.; Schiffman, M.; Burk, R.D.; et al. Cervicovaginal microbiome and natural history of HPV in a longitudinal study. PLoS Pathog. 2020, 16, e1008376. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; MacIntyre, D.A.; Ntritsos, G.; Smith, A.; Tsilidis, K.K.; Marchesi, J.R.; Bennett, P.R.; Moscicki, A.-B.; Kyrgiou, M. The vaginal microbiota associates with the regression of untreated cervical intraepithelial neoplasia 2 lesions. Nat. Commun. 2020, 11, 1999. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- France, M.T.; Ma, B.; Gajer, P.; Brown, S.; Humphrys, M.S.; Holm, J.B.; Waetjen, L.E.; Brotman, R.M.; Ravel, J. VALENCIA: A nearest centroid classification method for vaginal microbial communities based on composition. Microbiome 2020, 8, 166. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).