Effect of Curcumin on Hepatic mRNA and lncRNA Co-Expression in Heat-Stressed Laying Hens
Abstract
1. Introduction
2. Results
2.1. Quality Assessment of Sequencing Data
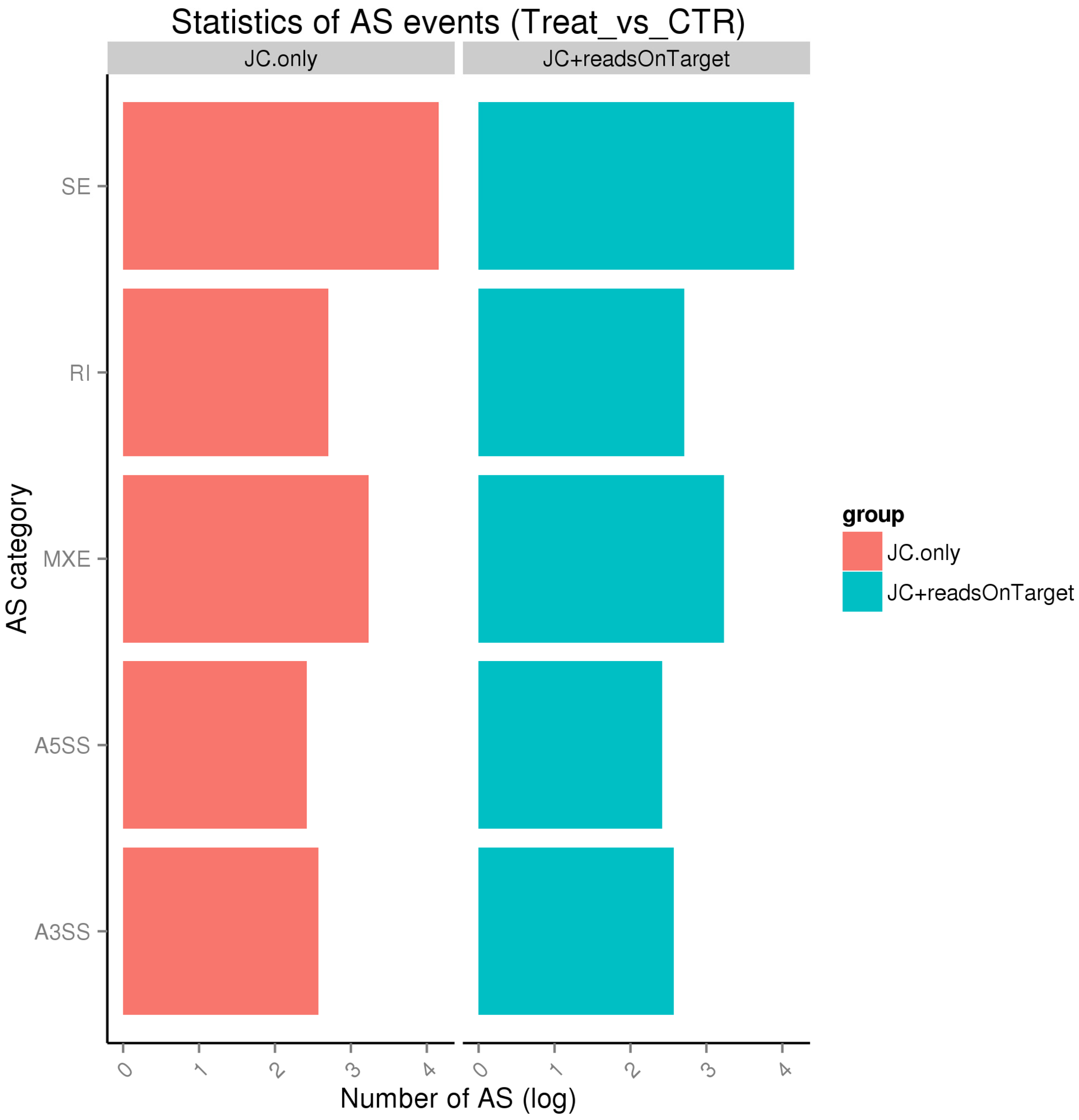
2.2. Reference Sequence Alignment and Splicing of Transcripts
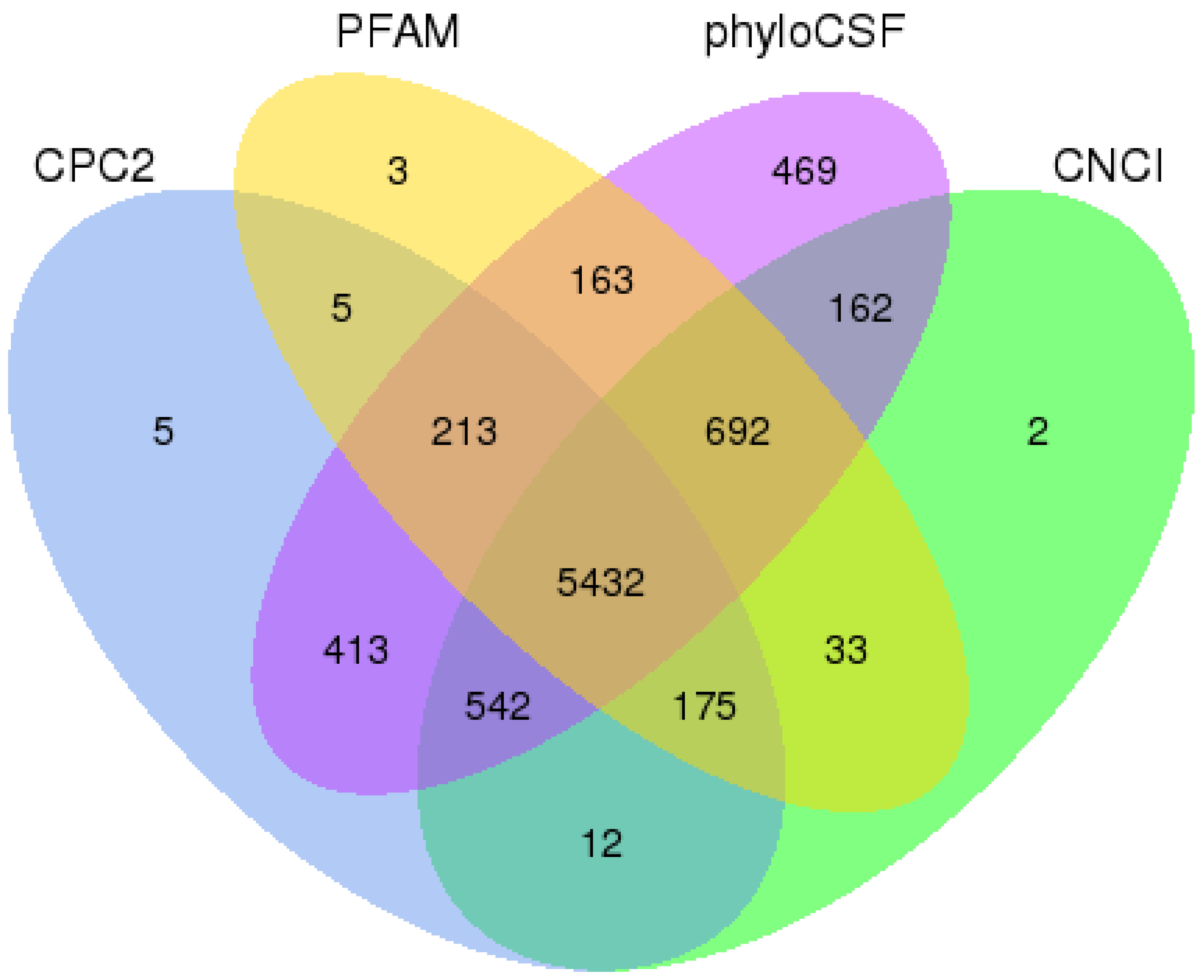
2.3. The Result of lncRNA Filtration
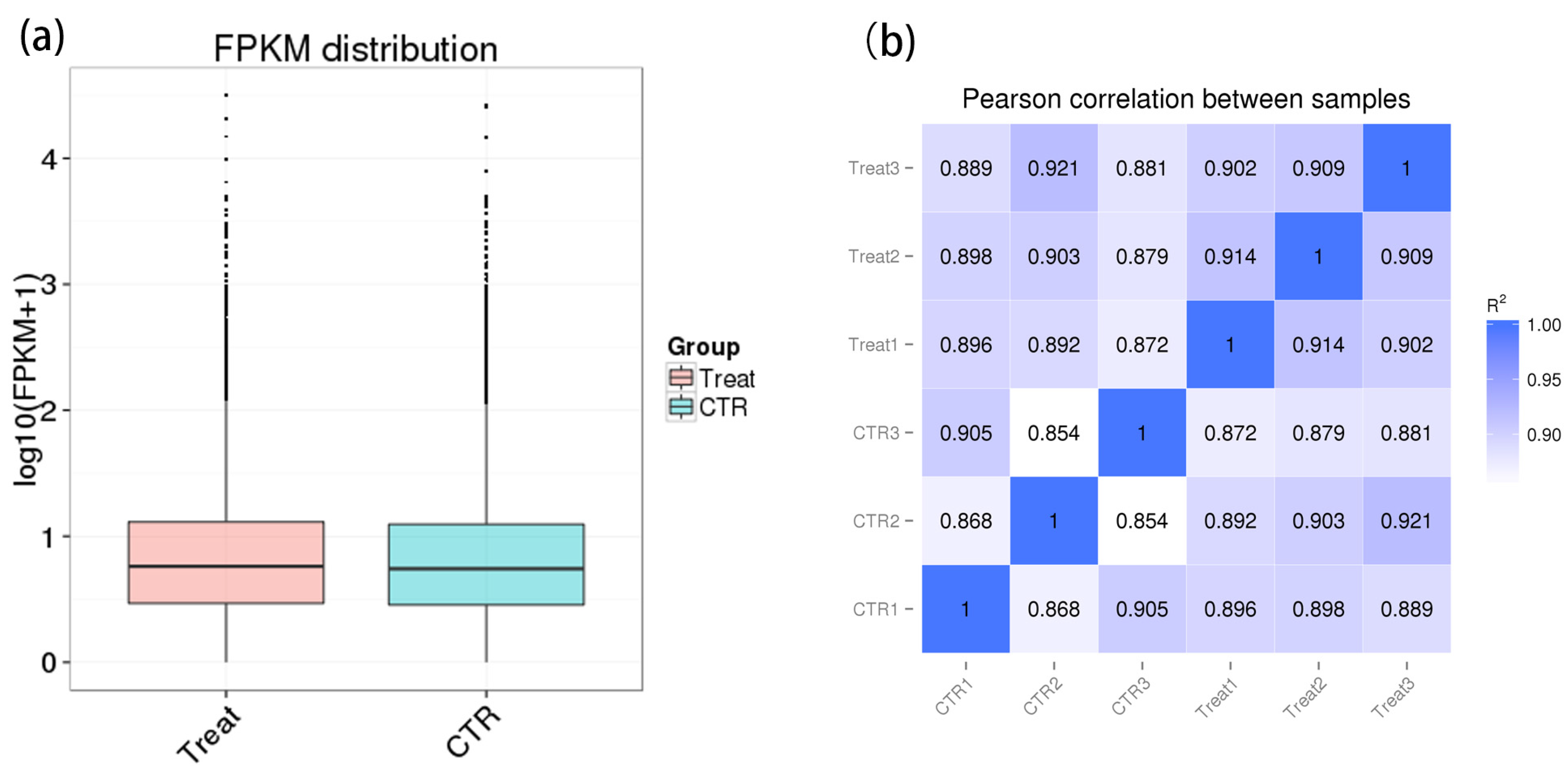
2.4. Correlation Test between Samples
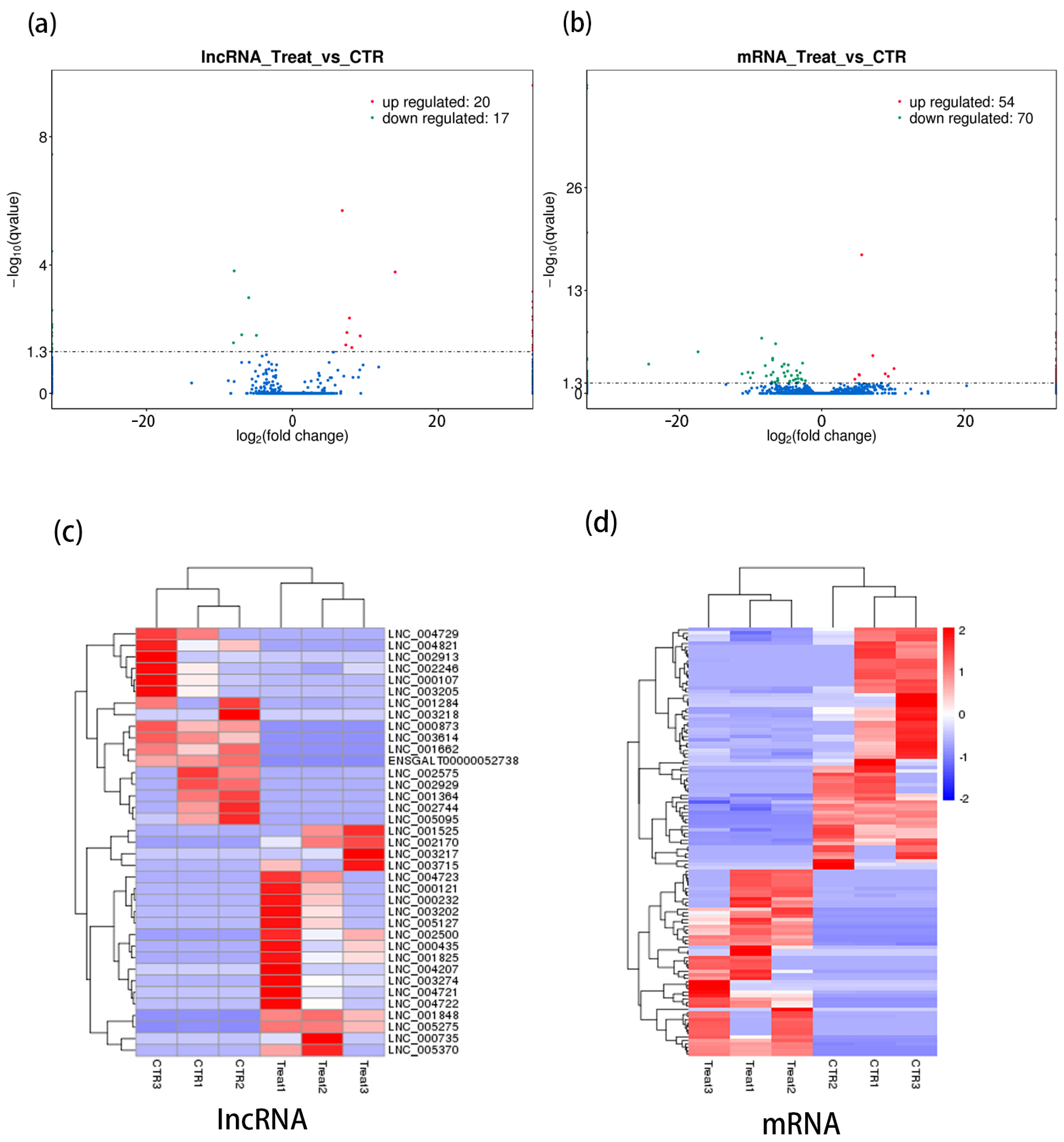
2.5. The Differentially Expressed of lncRNA and mRNA
2.6. GO Enrichment Analysis
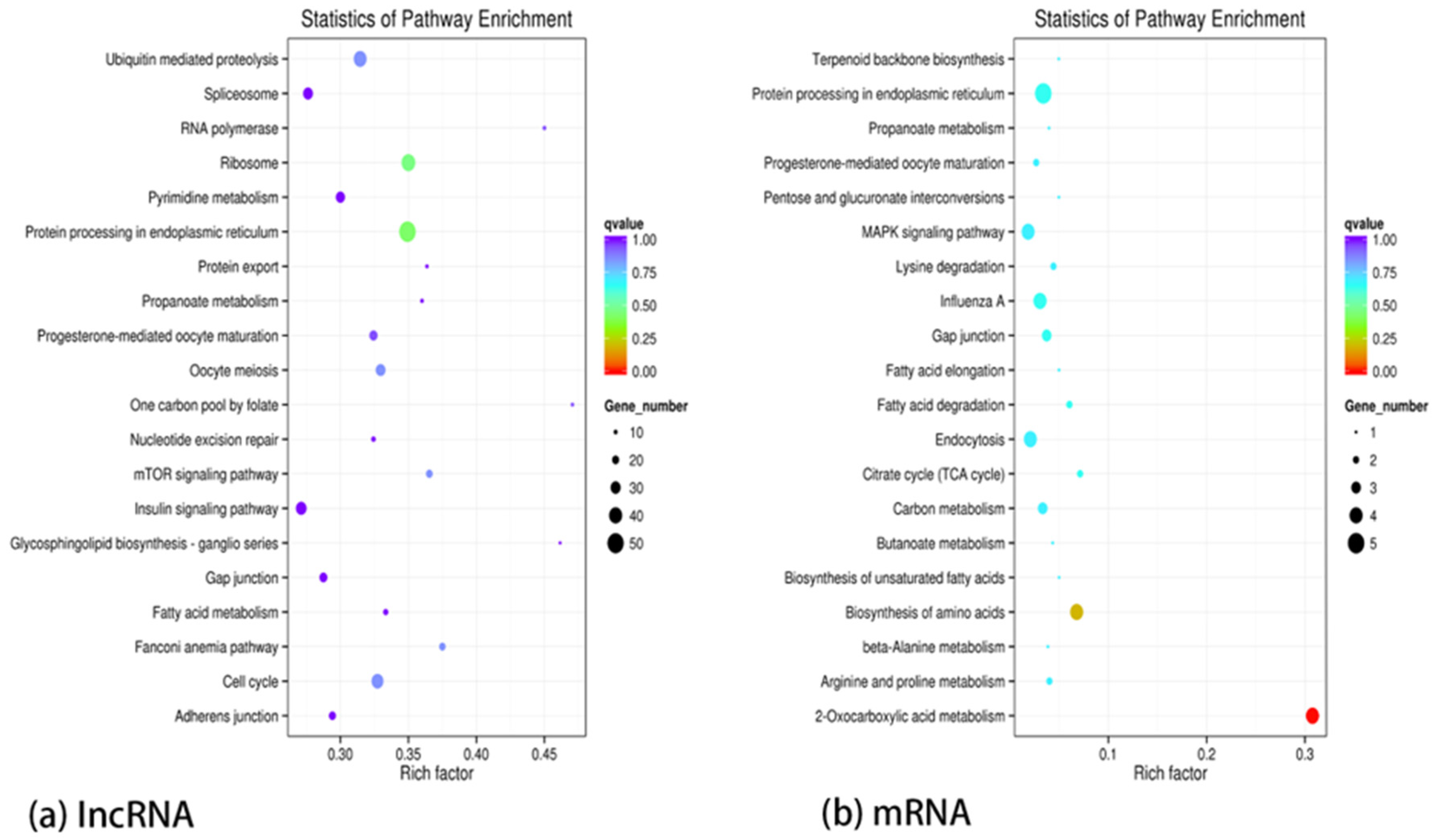
2.7. KEGG Signaling Pathway Enrichment Analysis
2.8. Expression Correlation Prediction of lncRNA and mRNA
2.9. qPCR Validation
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Sample Collect
4.2. RNA Extraction, Library Construction, Quality Control, and RNA-seq
4.3. Reads Alignments and Assembled Transcripts
4.4. LncRNA Filtration and Inter-Sample Correlation Detection
4.5. Identification of Differential Expressed Genes and Analysis of GO Category and KEGG Pathway
4.6. Quantitative Real-Time PCR (qRT-PCR) Validation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, D.-H.; Lee, K.-W. An Update on Heat Stress in Laying Hens. World’s Poult. Sci. J. 2023, 79, 689–712. [Google Scholar] [CrossRef]
- Altan, O.; Pabuçcuoğlu, A.; Altan, A.; Konyalioğlu, S.; Bayraktar, H. Effect of Heat Stress on Oxidative Stress, Lipid Peroxidation and Some Stress Parameters in Broilers. Br. Poult. Sci. 2003, 44, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Decuypere, E.; Buyse, J. Acute Heat Stress Induces Oxidative Stress in Broiler Chickens. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2006, 144, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Shini, S.; Huff, G.R.; Shini, A.; Kaiser, P. Understanding Stress-Induced Immunosuppression: Exploration of Cytokine and Chemokine Gene Profiles in Chicken Peripheral Leukocytes. Poult. Sci. 2010, 89, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Sahin, K.; Sahin, N.; Kucuk, O.; Hayirli, A.; Prasad, A.S. Role of Dietary Zinc in Heat-Stressed Poultry: A Review. Poult. Sci. 2009, 88, 2176–2183. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Jiao, H.C.; Buyse, J.; Decuypere, E. Strategies for Preventing Heat Stress in Poultry. World’s Poult. Sci. J. 2006, 62, 71–86. [Google Scholar] [CrossRef]
- Roushdy, E.M.; Zaglool, A.W.; El-Tarabany, M.S. Effects of Chronic Thermal Stress on Growth Performance, Carcass Traits, Antioxidant Indices and the Expression of HSP70, Growth Hormone and Superoxide Dismutase Genes in Two Broiler Strains. J. Therm. Biol. 2018, 74, 337–343. [Google Scholar] [CrossRef]
- Kim, D.Y.; Lim, B.; Kim, J.M.; Kil, D.Y. Integrated Transcriptome Analysis for the Hepatic and Jejunal Mucosa Tissues of Broiler Chickens Raised under Heat Stress Conditions. J. Anim. Sci. Biotechnol. 2022, 13, 79. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Acharya, A.; Ray, R.S.; Agrawal, R.; Raghuwanshi, R.; Jain, P. Cellular and Molecular Mechanisms of Curcumin in Prevention and Treatment of Disease. Crit. Rev. Food Sci. Nutr. 2020, 60, 887–939. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Gupta, S.C.; Sung, B. Curcumin: An Orally Bioavailable Blocker of TNF and Other pro-Inflammatory Biomarkers. Br. J. Pharmacol. 2013, 169, 1672–1692. [Google Scholar] [CrossRef]
- Nawab, A.; Li, G.; An, L.; Wu, J.; Chao, L.; Xiao, M.; Zhao, Y.; Birmani, M.W.; Ghani, M.W. Effect of Curcumin Supplementation on TLR4 Mediated Non-Specific Immune Responses in Liver of Laying Hens under High-Temperature Conditions. J. Therm. Biol. 2019, 84, 384–397. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jiang, W.; Liu, X.; Du, Y.; Liu, L.; Ordovas, J.M.; Lai, C.-Q.; Shen, L. Curcumin Supplementation Improves Heat-Stress-Induced Cardiac Injury of Mice: Physiological and Molecular Mechanisms. J. Nutr. Biochem. 2020, 78, 108331. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Lu, Y.; Gao, P.; Xie, X.; Li, D.; Yu, D.; Yu, M. Effect of Curcumin on Laying Performance, Egg Quality, Endocrine Hormones, and Immune Activity in Heat-Stressed Hens. Poult. Sci. 2020, 99, 2196–2202. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, X.; Liu, L.; Deng, H.; Zhang, J.; Xu, Q.; Cen, B.; Ji, A. Regulation of lncRNA Expression. Cell. Mol. Biol. Lett. 2014, 19, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Muret, K.; Klopp, C.; Wucher, V.; Esquerré, D.; Legeai, F.; Lecerf, F.; Désert, C.; Boutin, M.; Jehl, F.; Acloque, H.; et al. Long Noncoding RNA Repertoire in Chicken Liver and Adipose Tissue. Genet. Sel. Evol. 2017, 49, 6. [Google Scholar] [CrossRef]
- Li, Q.; Qiao, J.; Zhang, Z.; Shang, X.; Chu, Z.; Fu, Y.; Chu, M. Identification and Analysis of Differentially Expressed Long Non-Coding RNAs of Chinese Holstein Cattle Responses to Heat Stress. Anim. Biotechnol. 2020, 31, 9–16. [Google Scholar] [CrossRef]
- Dou, J.; Schenkel, F.; Hu, L.; Khan, A.; Khan, M.Z.; Yu, Y.; Wang, Y.; Wang, Y. Genome-Wide Identification and Functional Prediction of Long Non-Coding RNAs in Sprague-Dawley Rats during Heat Stress. BMC Genom. 2021, 22, 122. [Google Scholar] [CrossRef]
- Onagbesan, O.M.; Uyanga, V.A.; Oso, O.; Tona, K.; Oke, O.E. Alleviating Heat Stress Effects in Poultry: Updates on Methods and Mechanisms of Actions. Front. Vet. Sci. 2023, 10, 1255520. [Google Scholar] [CrossRef]
- Rozenboim, I.; Tako, E.; Gal-Garber, O.; Proudman, J.A.; Uni, Z. The Effect of Heat Stress on Ovarian Function of Laying Hens. Poult. Sci. 2007, 86, 1760–1765. [Google Scholar] [CrossRef]
- Ebeid, T.A.; Suzuki, T.; Sugiyama, T. High Ambient Temperature Influences Eggshell Quality and Calbindin-D28k Localization of Eggshell Gland and All Intestinal Segments of Laying Hens. Poult. Sci. 2012, 91, 2282–2287. [Google Scholar] [CrossRef]
- Lara, L.J.; Rostagno, M.H. Impact of Heat Stress on Poultry Production. Animals 2013, 3, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Ahmed-Farid, O.; Salah, A.S.; Nassan, M.A.; El-Tarabany, M.S. Effects of Chronic Thermal Stress on Performance, Energy Metabolism, Antioxidant Activity, Brain Serotonin, and Blood Biochemical Indices of Broiler Chickens. Animals 2021, 11, 2554. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.-P.; Liu, Y.-L.; Zhang, J.-X.; Ding, K.-N.; Lu, M.-H.; He, Y.-M. Heat Stress in Broilers of Liver Injury Effects of Heat Stress on Oxidative Stress and Autophagy in Liver of Broilers. Poult. Sci. 2022, 101, 102085. [Google Scholar] [CrossRef]
- Yan, L.; Hu, M.; Gu, L.; Lei, M.; Chen, Z.; Zhu, H.; Chen, R. Effect of Heat Stress on Egg Production, Steroid Hormone Synthesis, and Related Gene Expression in Chicken Preovulatory Follicular Granulosa Cells. Animals 2022, 12, 1467. [Google Scholar] [CrossRef]
- Ma, B.; Xing, T.; Li, J.; Zhang, L.; Jiang, Y.; Gao, F. Chronic Heat Stress Causes Liver Damage via Endoplasmic Reticulum Stress-Induced Apoptosis in Broilers. Poult. Sci. 2022, 101, 102063. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-M.; Liu, L.-P.; Yin, B.; Liu, Y.-Y.; Dong, W.-W.; Gong, S.; Zhang, J.; Tan, J.-H. Heat Stress Decreases Egg Production of Laying Hens by Inducing Apoptosis of Follicular Cells via Activating the FasL/Fas and TNF-α Systems. Poult. Sci. 2020, 99, 6084–6093. [Google Scholar] [CrossRef]
- Moniruzzaman, M.; Min, T. Curcumin, Curcumin Nanoparticles and Curcumin Nanospheres: A Review on Their Pharmacodynamics Based on Monogastric Farm Animal, Poultry and Fish Nutrition. Pharmaceutics 2020, 12, 447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bai, K.W.; He, J.; Niu, Y.; Lu, Y.; Zhang, L.; Wang, T. Curcumin Attenuates Hepatic Mitochondrial Dysfunction through the Maintenance of Thiol Pool, Inhibition of mtDNA Damage, and Stimulation of the Mitochondrial Thioredoxin System in Heat-Stressed Broilers. J. Anim. Sci. 2018, 96, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Sahin, K.; Orhan, C.; Tuzcu, Z.; Tuzcu, M.; Sahin, N. Curcumin Ameloriates Heat Stress via Inhibition of Oxidative Stress and Modulation of Nrf2/HO-1 Pathway in Quail. Food Chem. Toxicol. 2012, 50, 4035–4041. [Google Scholar] [CrossRef]
- Nawab, A.; Li, G.; Liu, W.; Lan, R.; Wu, J.; Zhao, Y.; Kang, K.; Kieser, B.; Sun, C.; Tang, S.; et al. Effect of Dietary Curcumin On the Antioxidant Status of Laying Hens Under High-Temperature Conditions. Chem. Chem. 2019, 86. [Google Scholar] [CrossRef]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The Liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Hao, Y.; Li, J.; Bao, W.; Li, G.; Gao, Y.; Gu, X. Chronic Heat Stress Induces Immune Response, Oxidative Stress Response, and Apoptosis of Finishing Pig Liver: A Proteomic Approach. Int. J. Mol. Sci. 2016, 17, 393. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fang, L.; Wu, C. Alternative Splicing and Isoforms: From Mechanisms to Diseases. Genes 2022, 13, 401. [Google Scholar] [CrossRef] [PubMed]
- Takechi, H.; Hosokawa, N.; Hirayoshi, K.; Nagata, K. Alternative 5′ Splice Site Selection Induced by Heat Shock. Mol. Cell. Biol. 1994, 14, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Nevo, Y.; Kamhi, E.; Jacob-Hirsch, J.; Amariglio, N.; Rechavi, G.; Sperling, J.; Sperling, R. Genome-Wide Activation of Latent Donor Splice Sites in Stress and Disease. Nucleic Acids Res. 2012, 40, 10980–10994. [Google Scholar] [CrossRef] [PubMed]
- Herman, A.B.; Tsitsipatis, D.; Gorospe, M. Integrated lncRNA Function upon Genomic and Epigenomic Regulation. Mol. Cell 2022, 82, 2252–2266. [Google Scholar] [CrossRef]
- Mao, Y.; Wen, C.; Yang, Z. Construction of a Co-Expression Network for lncRNAs and mRNAs Related to Urothelial Carcinoma of the Bladder Progression. Front. Oncol. 2022, 12, 835074. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chen, X.; Wang, X.; Qing, J.; Li, J.; Pan, Y. Construction of lncRNA and mRNA Co-Expression Network Associated with Nasopharyngeal Carcinoma Progression. Front. Oncol. 2022, 12, 965088. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Luo, J.; Liang, C.; Li, G.; Cai, J.; Ding, P.; Liu, Y. Identifying lncRNA and mRNA Co-Expression Modules from Matched Expression Data in Ovarian Cancer. IEEE/ACM Trans. Comput. Biol. Bioinf. 2020, 17, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Hu, E.; Cai, Y.; Zhu, W.; Chen, Q.; Li, T.; Li, Z.; Wang, Y.; Tang, T. mRNA and lncRNA Co-Expression Network in Mice of Acute Intracerebral Hemorrhage. Front. Mol. Neurosci. 2023, 16, 1166875. [Google Scholar] [CrossRef]
- Doyle, S.M.; Hoskins, J.R.; Kravats, A.N.; Heffner, A.L.; Garikapati, S.; Wickner, S. Intermolecular Interactions between Hsp90 and Hsp70. J. Mol. Biol. 2019, 431, 2729–2746. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Jin, S.; Shou, C.; Han, Z. Hsp70 Gene Family in Sebastiscus Marmoratus: The Genome-Wide Identification and Transcriptome Analysis under Thermal Stress. Genes 2023, 14, 1779. [Google Scholar] [CrossRef] [PubMed]
- Lou, S.-L.; Zhang, X.-Y.; Wang, D.-H. HSP70 Plays a Role in the Defense of Acute and Chronic Heat Stress in Mongolian Gerbils (Meriones Unguiculatus). J. Therm. Biol. 2019, 86, 102452. [Google Scholar] [CrossRef] [PubMed]
- Al-Zghoul, M.B. Thermal Manipulation during Broiler Chicken Embryogenesis Increases Basal mRNA Levels and Alters Production Dynamics of Heat Shock Proteins 70 and 60 and Heat Shock Factors 3 and 4 during Thermal Stress. Poult. Sci. 2018, 97, 3661–3670. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tang, S.; Song, E.; Yin, B.; Wu, D.; Bao, E. Hsp70 Expression Induced by Co-Enzyme Q10 Protected Chicken Myocardial Cells from Damage and Apoptosis under in Vitro Heat Stress. Poult. Sci. 2017, 96, 1426–1437. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular Chaperones in Protein Folding and Proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In Vivo Aspects of Protein Folding and Quality Control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef]
- Bonam, S.R.; Ruff, M.; Muller, S. HSPA8/HSC70 in Immune Disorders: A Molecular Rheostat That Adjusts Chaperone-Mediated Autophagy Substrates. Cells 2019, 8, 849. [Google Scholar] [CrossRef]
- Wu, E.; He, W.; Wu, C.; Chen, Z.; Zhou, S.; Wu, X.; Hu, Z.; Jia, K.; Pan, J.; Wang, L.; et al. HSPA8 Acts as an Amyloidase to Suppress Necroptosis by Inhibiting and Reversing Functional Amyloid Formation. Cell Res. 2023, 33, 851–866. [Google Scholar] [CrossRef]
- Miyazaki, M.; Nakatsura, T.; Yokomine, K.; Senju, S.; Monji, M.; Hosaka, S.; Komori, H.; Yoshitake, Y.; Motomura, Y.; Minohara, M.; et al. DNA Vaccination of HSP105 Leads to Tumor Rejection of Colorectal Cancer and Melanoma in Mice through Activation of Both CD4 T Cells and CD8 T Cells. Cancer Sci. 2005, 96, 695–705. [Google Scholar] [CrossRef]
- Shimizu, Y.; Yoshikawa, T.; Kojima, T.; Shoda, K.; Nosaka, K.; Mizuno, S.; Wada, S.; Fujimoto, Y.; Sasada, T.; Kohashi, K.; et al. Heat Shock Protein 105 Peptide Vaccine Could Induce Antitumor Immune Reactions in a Phase I Clinical Trial. Cancer Sci. 2019, 110, 3049–3060. [Google Scholar] [CrossRef]
- Scieglinska, D.; Sojka, D.R.; Gogler-Pigłowska, A.; Chumak, V.; Krawczyk, Z. Various Anti-HSPA2 Antibodies Yield Different Results in Studies on Cancer-Related Functions of Heat Shock Protein A2. Int. J. Mol. Sci. 2020, 21, 4296. [Google Scholar] [CrossRef]
- Liu, R.-J.; Niu, X.-L.; Yuan, J.-P.; Chen, H.-D.; Gao, X.-H.; Qi, R.-Q. DnaJA4 Is Involved in Responses to Hyperthermia by Regulating the Expression of F-Actin in HaCaT Cells. Chin. Med. J. 2020, 134, 456–462. [Google Scholar] [CrossRef]
- Robichon, C.; Varret, M.; Le Liepvre, X.; Lasnier, F.; Hajduch, E.; Ferré, P.; Dugail, I. DnaJA4 Is a SREBP-Regulated Chaperone Involved in the Cholesterol Biosynthesis Pathway. Biochim. Biophys. Acta 2006, 1761, 1107–1113. [Google Scholar] [CrossRef]
- Li, Y.; Kong, L.; Deng, M.; Lian, Z.; Han, Y.; Sun, B.; Guo, Y.; Liu, G.; Liu, D. Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep. Genes 2019, 10, 395. [Google Scholar] [CrossRef]
- Li, G.; Yu, X.; Portela Fontoura, A.B.; Javaid, A.; De La Maza-Escolà, V.S.; Salandy, N.S.; Fubini, S.L.; Grilli, E.; McFadden, J.W.; Duan, J.E. Transcriptomic Regulations of Heat Stress Response in the Liver of Lactating Dairy Cows. BMC Genom. 2023, 24, 410. [Google Scholar] [CrossRef]
- Naphade, S.; Bhatnagar, R.; Hanson-Smith, V.; Choi, I.; Zhang, A. Systematic Comparative Analysis of Strand-Specific RNA-Seq Library Preparation Methods for Low Input Samples. Sci. Rep. 2022, 12, 1789. [Google Scholar] [CrossRef]
- Mills, J.D.; Kawahara, Y.; Janitz, M. Strand-Specific RNA-Seq Provides Greater Resolution of Transcriptome Profiling. Curr. Genom. 2013, 14, 173–181. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Lu, Z.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and Flexible Detection of Differential Alternative Splicing from Replicate RNA-Seq Data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Lin, M.F.; Jungreis, I.; Kellis, M. PhyloCSF: A Comparative Genomics Method to Distinguish Protein Coding and Non-Coding Regions. Bioinformatics 2011, 27, i275–i282. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing Sequence Intrinsic Composition to Classify Protein-Coding and Long Non-Coding Transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for Linking Genomes to Life and the Environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Sample Name | Raw Reads | Clean Reads | Clean Bases | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|
| CTR1 | 141,354,366 | 138,329,490 | 20.75 G | 97.07 | 92.86 | 48.87 |
| CTR2 | 123,877,456 | 121,554,156 | 18.23 G | 97.09 | 92.91 | 51.66 |
| CTR3 | 144,998,210 | 142,024,464 | 21.3 G | 97.18 | 93.11 | 48.06 |
| Treat1 | 126,852,994 | 125,635,340 | 18.85 G | 97.55 | 93.61 | 49.68 |
| Treat2 | 205,193,728 | 201,015,402 | 30.15 G | 97.19 | 93.13 | 50.07 |
| Treat3 | 119,451,560 | 117,319,078 | 17.6 G | 97.29 | 93.34 | 49.88 |
| Sample Name | Total Reads | Total Mapped | Multiple Mapped | Uniquely Mapped |
|---|---|---|---|---|
| CTR1 | 138,329,490 | 129,787,959 (93.83%) | 14,303,032 (10.34%) | 115,484,927 (83.49%) |
| CTR2 | 121,554,156 | 113,126,334 (93.07%) | 15,063,522 (12.39%) | 98,062,812 (80.67%) |
| CTR3 | 142,024,464 | 134,005,785 (94.35%) | 13,746,535 (9.68%) | 120,259,250 (84.68%) |
| Treat1 | 60,167,536 | 56,707,140 (94.25%) | 7,168,028 (11.91%) | 49,539,112 (82.34%) |
| Treat2 | 201,015,402 | 188,897,443 (93.97%) | 22,901,129 (11.39%) | 165,996,314 (82.58%) |
| Treat3 | 117,319,078 | 109,761,781 (93.56%) | 14,668,105 (12.5%) | 95,093,676 (81.06%) |
| KEGG Pathway ID | Term | Cg | Bg | Gene Name |
|---|---|---|---|---|
| gga04141 | Protein processing in endoplasmic reticulum | 5 | 146 | |HSPA2, HSP70|HSPH1|HSP90AA1|HSPA8, HSC70| |
| gga03010 | Ribosome | 1 | 120 | RPL7A |
| gga04120 | Ubiquitin mediated proteolysis | 1 | 124 | CUL5 |
| gga04114 | Oocyte meiosis | 1 | 88 | ADCY1 |
| KEGG Pathway ID | Term | Cg | Bg | Gene Name |
|---|---|---|---|---|
| gga01210 | 2-Oxocarboxylic acid metabolism | 4 | 13 | |ACY1|IDH3B|IDH1|ACY1| |
| gga01230 | Biosynthesis of amino acids | 4 | 59 | |ACY1|IDH3B|IDH1|ACY1| |
| gga04141 | Protein processing in endoplasmic reticulum | 5 | 146 | |HSPA2, HSP70|HSPH1|HSP90AA1|HSPA8, HSC70| |
| gga00020 | Citrate cycle (TCA cycle) | 2 | 28 | |IDH1|IDH3B| |
| gga00071 | Fatty acid degradation | 2 | 33 | |HADHA|ECI1, DCI| |
| mRNA Gene Symbol | mRNA Gene ID | LncRNA Gene ID | Mrna Gene Description | Control Group vs. Curcumin Addition Group |
|---|---|---|---|---|
| HSPA8 | ENSGALG00000006512 | XLOC_053511 XLOC_037987 XLOC_010450 XLOC_100318 XLOC_061207 | Heat shock protein 70 family||Heat shock protein 70 kD, peptide-binding domain||Heat shock protein 70, conserved site||Heat shock protein 70 kD, C-terminal domain | Down |
| HSPH1 | ENSGALG00000017077 | XLOC_061207 XLOC_053511 XLOC_037987 XLOC_010450 | Heat shock protein 70 family||Heat shock protein 70 kD, C-terminal domain||Heat shock protein 70, conserved site||Heat shock protein 70 kD, peptide-binding domain | Down |
| HSPA2 | ENSGALG00000011715 | XLOC_100318 XLOC_061207 XLOC_037987 | Heat shock protein 70 kD, C-terminal domain||Heat shock protein 70 kD, peptide-binding domain||Heat shock protein 70, conserved site||Heat shock protein 70 family | Down |
| DNAJA4 | ENSGALG00000036850 | XLOC_061207 XLOC_037987 XLOC_053511 | HSP40/DnaJ peptide-binding||Chaperone DnaJ, C-terminal||DnaJ domain||DnaJ domain, conserved site||Chaperone DnaJ||Heat shock protein DnaJ, cysteine-rich domain | Down |
| Gene ID | Transcript ID | Primer Sequence | Product Length (bp) |
|---|---|---|---|
| IDH1 | ENSGALT00000014333 | F: CCCCATTGCCTCCATCTTTG R: CAAGCAGCAAGGTCCTTTGT | 153 |
| HSPA8 | ENSGALT00000086458 | F: AGCGACAGGGCACAAAAGAT R: GGTTCCTTTCAGCACCAACTTTC | 123 |
| BAG3 | ENSGALT00000088658 | F: TTCGGTCACCTACCGAGAAC R: CCCAGTTTCTTCAGGAGCAG | 139 |
| HSP90AA1 | ENSGALT00000081765 | F: CAAGCCTATTTGGACCAGGA R: GCTCTGAATTCCAGCTGACC | 138 |
| XLOC_015361 | LNC_001282 | F: CCGCTTTTCGCCCCAAAATGAG R: CAGACCCATAGATTGACCCATGAGG | 130 |
| XLOC_027753 | LNC_001848 | F: AGGCACACTCTAGAAAGGCAC R: CATCCCCCAACTTCTCGTTGA | 122 |
| XLOC_097982 | LNC_004633 | F: GCATTAGAGCTGCACCAACA R: TTGCCATGGTTCATTTCTCA | 150 |
| XLOC_037987 | LNC_002246 | F: TGTGATGGTTTACTGCCACC R: AGCCCGTGTAAGGTCTAAAGTG | 127 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Du, X.; Pian, H.; Yu, D. Effect of Curcumin on Hepatic mRNA and lncRNA Co-Expression in Heat-Stressed Laying Hens. Int. J. Mol. Sci. 2024, 25, 5393. https://doi.org/10.3390/ijms25105393
Wu X, Du X, Pian H, Yu D. Effect of Curcumin on Hepatic mRNA and lncRNA Co-Expression in Heat-Stressed Laying Hens. International Journal of Molecular Sciences. 2024; 25(10):5393. https://doi.org/10.3390/ijms25105393
Chicago/Turabian StyleWu, Xinyue, Xubin Du, Huifang Pian, and Debing Yu. 2024. "Effect of Curcumin on Hepatic mRNA and lncRNA Co-Expression in Heat-Stressed Laying Hens" International Journal of Molecular Sciences 25, no. 10: 5393. https://doi.org/10.3390/ijms25105393
APA StyleWu, X., Du, X., Pian, H., & Yu, D. (2024). Effect of Curcumin on Hepatic mRNA and lncRNA Co-Expression in Heat-Stressed Laying Hens. International Journal of Molecular Sciences, 25(10), 5393. https://doi.org/10.3390/ijms25105393






