Chemokines as Prognostic Factor in Colorectal Cancer Patients: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Results
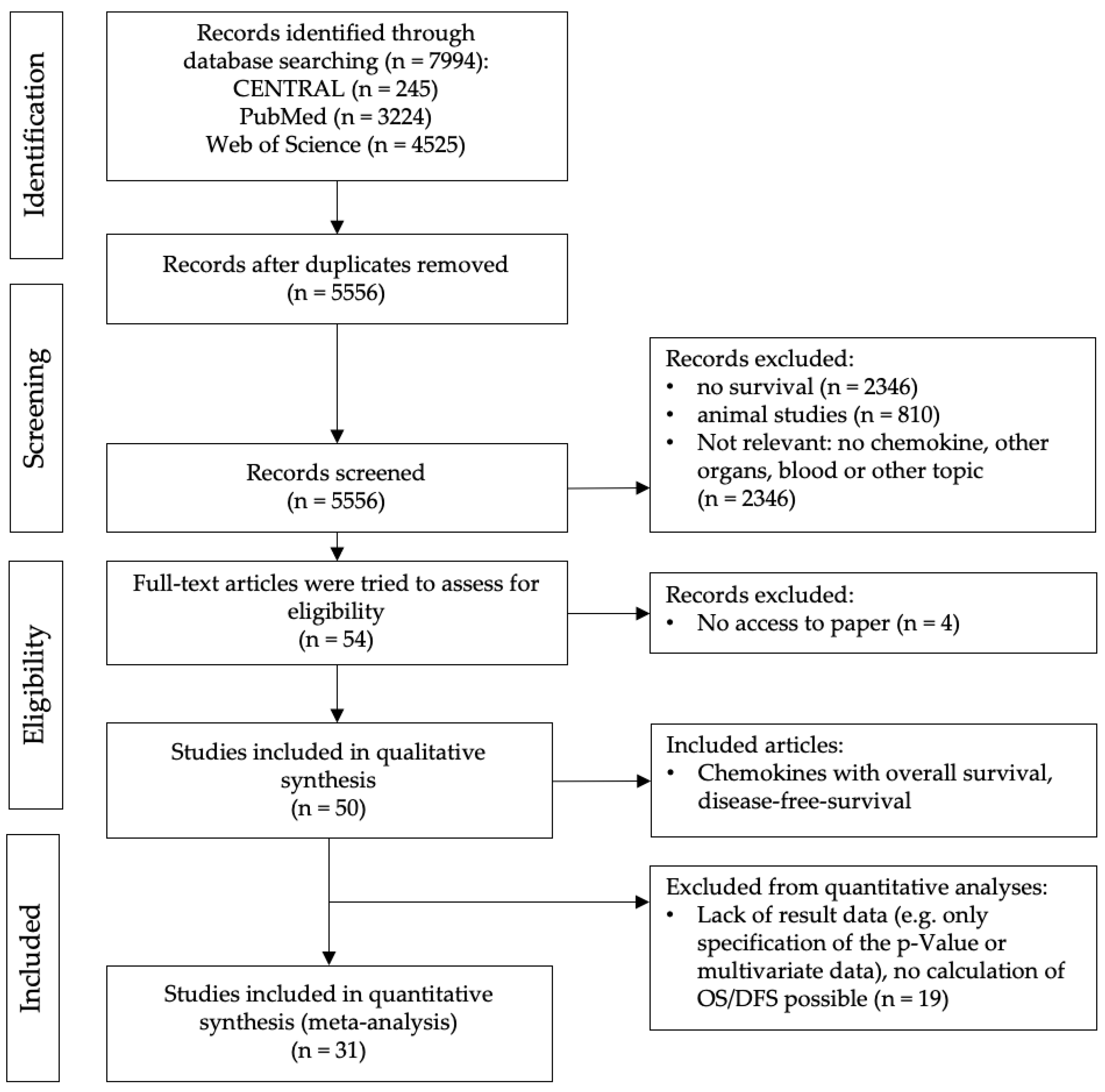
2.1. Study Selection
2.2. Qualitative Analysis
2.3. Study Characteristics
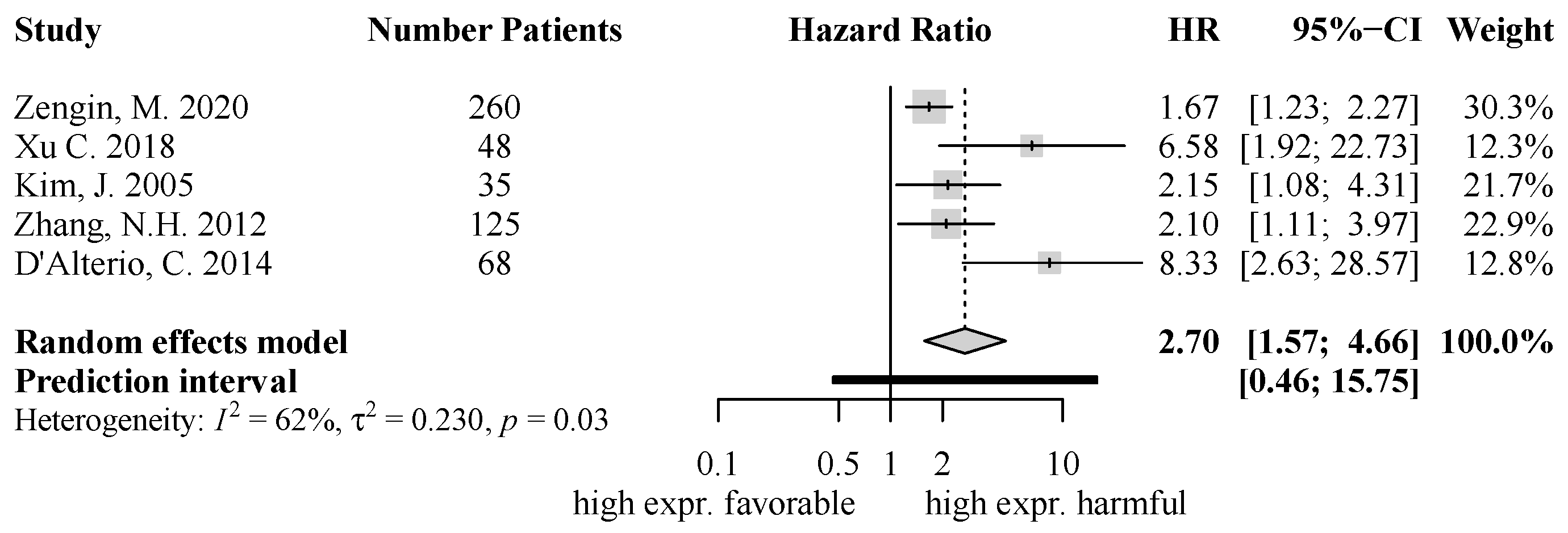
2.4. Quantitative Analysis of Chemokine Expression and Survival
2.5. Overall Risk of Bias of CXCR4
3. Discussion
4. Materials and Methods
4.1. Systematic Literature Search
4.2. Study Selection
4.3. Data Extraction
4.4. Risk of Bias—Critical Appraisal
4.5. Data Handling and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef]
- Mitchem, J.B.; Brennan, D.J.; Knolhoff, B.L.; Belt, B.A.; Zhu, Y.; Sanford, D.E.; Belaygorod, L.; Carpenter, D.; Collins, L.; Piwnica-Worms, D.; et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013, 73, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Bonapace, L.; Coissieux, M.M.; Wyckoff, J.; Mertz, K.D.; Varga, Z.; Junt, T.; Bentires-Alj, M. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature 2014, 515, 130–133. [Google Scholar] [CrossRef]
- Young, H.L.; Rowling, E.J.; Bugatti, M.; Giurisato, E.; Luheshi, N.; Arozarena, I.; Acosta, J.C.; Kamarashev, J.; Frederick, D.T.; Cooper, Z.A.; et al. An adaptive signaling network in melanoma inflammatory niches confers tolerance to MAPK signaling inhibition. J. Exp. Med. 2017, 214, 1691–1710. [Google Scholar] [CrossRef] [PubMed]
- Mollica Poeta, V.; Massara, M.; Capucetti, A.; Bonecchi, R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front. Immunol. 2019, 10, 379. [Google Scholar] [CrossRef] [PubMed]
- Halama, N.; Zoernig, I.; Berthel, A.; Kahlert, C.; Klupp, F.; Suarez-Carmona, M.; Suetterlin, T.; Brand, K.; Krauss, J.; Lasitschka, F.; et al. Tumoral Immune Cell Exploitation in Colorectal Cancer Metastases Can Be Targeted Effectively by Anti-CCR5 Therapy in Cancer Patients. Cancer Cell 2016, 29, 587–601. [Google Scholar] [CrossRef]
- Li, Y.P.; Pang, J.; Gao, S.; Bai, P.Y.; Wang, W.D.; Kong, P.; Cui, Y. Role of CXCR4 and SDF1 as prognostic factors for survival and the association with clinicopathology in colorectal cancer: A systematic meta-analysis. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2017, 39, 1010428317706206. [Google Scholar] [CrossRef]
- Braoudaki, M.; Ahmad, M.S.; Mustafov, D.; Seriah, S.; Siddiqui, M.N.; Siddiqui, S.S. Chemokines and chemokine receptors in colorectal cancer; multifarious roles and clinical impact. Semin. Cancer Biol. 2022, 86, 436–449. [Google Scholar] [CrossRef]
- Saxena, S.; Singh, R.K. Chemokines orchestrate tumor cells and the microenvironment to achieve metastatic heterogeneity. Cancer Metastasis Rev. 2021, 40, 447–476. [Google Scholar] [CrossRef]
- Singh, A.J.; Gray, J.W. Chemokine signaling in cancer-stroma communications. J. Cell Commun. Signal. 2021, 15, 361–381. [Google Scholar] [CrossRef]
- Rani, A.; Dasgupta, P.; Murphy, J.J. Prostate Cancer: The Role of Inflammation and Chemokines. Am. J. Pathol. 2019, 189, 2119–2137. [Google Scholar] [CrossRef]
- Xu, J.; Li, J.Q.; Chen, Q.L.; Shestakova, E.A.; Misyurin, V.A.; Pokrovsky, V.S.; Tchevkina, E.M.; Chen, H.B.; Song, H.; Zhang, J.Y. Advances in Research on the Effects and Mechanisms of Chemokines and Their Receptors in Cancer. Front. Pharmacol. 2022, 13, 920779. [Google Scholar] [CrossRef]
- Pang, X.; Li, K.; Wei, L.; Huang, Y.; Su, M.; Wang, L.; Cao, H.; Chen, T. IL-8 inhibits the apoptosis of MCF-7 human breast cancer cells by up-regulating Bcl-2 and down-regulating caspase-3. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2015, 31, 307–311. [Google Scholar] [PubMed]
- Miller, M.C.; Mayo, K.H. Chemokines from a Structural Perspective. Int. J. Mol. Sci. 2017, 18, 2088. [Google Scholar] [CrossRef]
- Koch, A.E.; Polverini, P.J.; Kunkel, S.L.; Harlow, L.A.; DiPietro, L.A.; Elner, V.M.; Elner, S.G.; Strieter, R.M. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science 1992, 258, 1798–1801. [Google Scholar] [CrossRef] [PubMed]
- Youn, B.S.; Mantel, C.; Broxmeyer, H.E. Chemokines, chemokine receptors and hematopoiesis. Immunol. Rev. 2000, 177, 150–174. [Google Scholar] [CrossRef]
- Lim, R.J.; Salehi-Rad, R.; Tran, L.M.; Oh, M.S.; Dumitras, C.; Crosson, W.P.; Li, R.; Patel, T.S.; Man, S.; Yean, C.E.; et al. CXCL9/10-engineered dendritic cells promote T cell activation and enhance immune checkpoint blockade for lung cancer. Cell Rep. Med. 2024, 5, 101479. [Google Scholar] [CrossRef] [PubMed]
- Kohli, K.; Pillarisetty, V.G.; Kim, T.S. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther. 2022, 29, 10–21. [Google Scholar] [CrossRef]
- Suarez-Carmona, M.; Williams, A.; Schreiber, J.; Hohmann, N.; Pruefer, U.; Krauss, J.; Jager, D.; Fromming, A.; Beyer, D.; Eulberg, D.; et al. Combined inhibition of CXCL12 and PD-1 in MSS colorectal and pancreatic cancer: Modulation of the microenvironment and clinical effects. J. Immunother. Cancer 2021, 9, e002505. [Google Scholar] [CrossRef]
- Bianchi, M.E.; Mezzapelle, R. The Chemokine Receptor CXCR4 in Cell Proliferation and Tissue Regeneration. Front. Immunol. 2020, 11, 2109. [Google Scholar] [CrossRef]
- Zengin, M.; Dursun, N.; Behzatoglu, K.; Pasaoglu, H.E.; Benek, S. Evaluation of Cxcl12 and Cxcr4 to Predict Poor Survival in Lymph Node-Positive Colorectal Cancer Patients. Pol. J. Pathol. 2020, 71, 328–338. [Google Scholar] [CrossRef]
- Hu, T.H.; Yao, Y.; Yu, S.; Han, L.L.; Wang, W.J.; Guo, H.; Tian, T.; Ruan, Z.P.; Kang, X.M.; Wang, J.; et al. SDF-1/CXCR4 promotes epithelial-mesenchymal transition and progression of colorectal cancer by activation of the Wnt/beta-catenin signaling pathway. Cancer Lett. 2014, 354, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Iwasa, S.; Yanagawa, T.; Fan, J.; Katoh, R. Expression of CXCR4 and its ligand SDF-1 in intestinal-type gastric cancer is associated with lymph node and liver metastasis. Anticancer. Res. 2009, 29, 4751–4758. [Google Scholar]
- Liang, J.J.; Zhu, S.; Bruggeman, R.; Zaino, R.J.; Evans, D.B.; Fleming, J.B.; Gomez, H.F.; Zander, D.S.; Wang, H. High levels of expression of human stromal cell-derived factor-1 are associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2598–2604. [Google Scholar] [CrossRef]
- Xie, S.; Zeng, W.; Fan, G.; Huang, J.; Kang, G.; Geng, Q.; Cheng, B.; Wang, W.; Dong, P. Effect of CXCL12/CXCR4 on increasing the metastatic potential of non-small cell lung cancer in vitro is inhibited through the downregulation of CXCR4 chemokine receptor expression. Oncol. Lett. 2014, 7, 941–947. [Google Scholar] [CrossRef][Green Version]
- Ali, S.; Lazennec, G. Chemokines: Novel targets for breast cancer metastasis. Cancer Metastasis Rev. 2007, 26, 401–420. [Google Scholar] [CrossRef]
- Darakhshan, S.; Bidmeshkipour, A.; Mansouri, K.; Saeid, H.M.; Ghanbari, A. The Effects of Tamoxifen in Combination with Tranilast on CXCL12-CXCR4 Axis and Invasion in Breast Cancer Cell Lines. Iran. J. Pharm. Res. 2014, 13, 683–693. [Google Scholar] [PubMed]
- Khare, T.; Bissonnette, M.; Khare, S. CXCL12-CXCR4/CXCR7 Axis in Colorectal Cancer: Therapeutic Target in Preclinical and Clinical Studies. Int. J. Mol. Sci. 2021, 22, 7371. [Google Scholar] [CrossRef]
- Kucia, M.; Reca, R.; Miekus, K.; Wanzeck, J.; Wojakowski, W.; Janowska-Wieczorek, A.; Ratajczak, J.; Ratajczak, M.Z. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: Pivotal role of the SDF-1-CXCR4 axis. Stem Cells 2005, 23, 879–894. [Google Scholar] [CrossRef]
- Kremer, K.N.; Peterson, K.L.; Schneider, P.A.; Meng, X.W.; Dai, H.; Hess, A.D.; Smith, B.D.; Rodriguez-Ramirez, C.; Karp, J.E.; Kaufmann, S.H.; et al. CXCR4 chemokine receptor signaling induces apoptosis in acute myeloid leukemia cells via regulation of the Bcl-2 family members Bcl-XL, Noxa, and Bak. J. Biol. Chem. 2013, 288, 22899–22914. [Google Scholar] [CrossRef]
- Zhao, H.; Ming, T.; Tang, S.; Ren, S.; Yang, H.; Liu, M.; Tao, Q.; Xu, H. Wnt signaling in colorectal cancer: Pathogenic role and therapeutic target. Mol. Cancer 2022, 21, 144. [Google Scholar] [CrossRef]
- Guo, L.; Wang, Y.; Yang, W.; Wang, C.; Guo, T.; Yang, J.; Shao, Z.; Cai, G.; Cai, S.; Zhang, L.; et al. Molecular Profiling Provides Clinical Insights Into Targeted and Immunotherapies as Well as Colorectal Cancer Prognosis. Gastroenterology 2023, 165, 414–428.e7. [Google Scholar] [CrossRef]
- Picard, E.; Verschoor, C.P.; Ma, G.W.; Pawelec, G. Relationships between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Front. Immunol. 2020, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Andre, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef] [PubMed]
- D’Alterio, C.; Avallone, A.; Tatangelo, F.; Delrio, P.; Pecori, B.; Cella, L.; Pelella, A.; D’Armiento, F.P.; Carlomagno, C.; Bianco, F.; et al. A prognostic model comprising pT stage, N status, and the chemokine receptors CXCR4 and CXCR7 powerfully predicts outcome in neoadjuvant resistant rectal cancer patients. Int. J. Cancer 2014, 135, 379–390. [Google Scholar] [CrossRef]
- Zabel, B.A.; Wang, Y.; Lewen, S.; Berahovich, R.D.; Penfold, M.E.; Zhang, P.; Powers, J.; Summers, B.C.; Miao, Z.; Zhao, B.; et al. Elucidation of CXCR7-mediated signaling events and inhibition of CXCR4-mediated tumor cell transendothelial migration by CXCR7 ligands. J. Immunol. 2009, 183, 3204–3211. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Liu, J.; Li, Z.; Zhang, W.; Wang, F.; Zhang, B. Recent Advances in CXCL12/CXCR4 Antagonists and Nano-Based Drug Delivery Systems for Cancer Therapy. Pharmaceutics 2022, 14, 1541. [Google Scholar] [CrossRef]
- Ruan, G.T.; Gong, Y.Z.; Liao, X.W.; Wang, S.; Huang, W.; Wang, X.K.; Zhu, G.Z.; Liao, C.; Gao, F. Diagnostic and prognostic values of CXC motif chemokine ligand 3 in patients with colon cancer. Oncol. Rep. 2019, 42, 1996–2008. [Google Scholar] [CrossRef]
- Li, X.; Zhong, Q.; Luo, D.; Du, Q.; Liu, W. The prognostic value of CXC subfamily ligands in stage I-III patients with colorectal cancer. PLoS ONE 2019, 14, e0214611. [Google Scholar] [CrossRef]
- Gong, Y.Z.; Ma, H.; Ruan, G.T.; Zhu, L.C.; Liao, X.W.; Wang, S.; Yan, L.; Huang, W.; Huang, K.T.; Xie, H.; et al. Diagnosis and prognostic value of C-X-C motif chemokine ligand 1 in colon adenocarcinoma based on The Cancer Genome Atlas and Guangxi cohort. J. Cancer 2021, 12, 5506–5518. [Google Scholar] [CrossRef]
- Li, L.; Jiang, K.; Li, D.; Li, D.; Fan, Z.; Dai, G.; Tu, S.; Liu, X.; Wei, G. The Chemokine CXCL7 Is Related to Angiogenesis and Associated With Poor Prognosis in Colorectal Cancer Patients. Front. Oncol. 2021, 11, 754221. [Google Scholar] [CrossRef]
- Liu, M.J.; Guo, H.; Jiang, L.L.; Jiao, M.; Wang, S.H.; Tian, T.; Fu, X.; Wang, W.J. Elevated RBP-Jκ and CXCL11 Expression in Colon Cancer is Associated with an Unfavorable Clinical Outcome. Cancer Manag. Res. 2021, 13, 3651–3661. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Dai, W.; Yang, L.; Yang, H.; Ding, L.; He, Y.; Song, X.; Cui, J. Elevated expression of CXCL16 correlates with poor prognosis in patients with colorectal cancer. Cancer Manag. Res. 2019, 11, 4691–4697. [Google Scholar] [CrossRef] [PubMed]
- Okikawa, S.; Higashijima, J.; Nishi, M.; Yoshimoto, T.; Eto, S.; Takasu, C.; Kashihara, H.; Tokunaga, T.; Yoshikawa, K.; Shimada, M. SDF-1 expression after preoperative chemoradiotherapy is associated with prognosis in patients with advanced lower rectal cancer. J. Med. Investig. JMI 2021, 68, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zheng, L.; Li, D.; Chen, G.; Gu, J.; Chen, J.; Yao, Q. CXCR4 overexpression is correlated with poor prognosis in colorectal cancer. Life Sci. 2018, 208, 333–340. [Google Scholar] [CrossRef]
- Luo, X.; Tai, J.; Zhao, Y.; Zhao, P.; Sun, D.; Wang, L. Associations of C-X-C motif chemokine ligands 1/2/8/13/14 with clinicopathological features and survival profile in patients with colorectal cancer. Oncol. Lett. 2022, 24, 348. [Google Scholar] [CrossRef]
- Kim, J.; Takeuchi, H.; Lam, S.T.; Turner, R.R.; Wang, H.J.; Kuo, C.; Foshag, L.; Bilchik, A.J.; Hoon, D.S. Chemokine receptor CXCR4 expression in colorectal cancer patients increases the risk for recurrence and for poor survival. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 2744–2753. [Google Scholar] [CrossRef]
- Terada, H.; Urano, T.; Konno, H. Association of interleukin-8 and plasminogen activator system in the progression of colorectal cancer. Eur. Surg. Res. Eur. Chir. Forschung. Rech. Chir. Eur. 2005, 37, 166–172. [Google Scholar] [CrossRef]
- Wang, S.C.; Lin, J.K.; Wang, H.S.; Yang, S.H.; Li, A.F.; Chang, S.C. Nuclear expression of CXCR4 is associated with advanced colorectal cancer. Int. J. Color. Dis. 2010, 25, 1185–1191. [Google Scholar] [CrossRef]
- Ottaiano, A.; Franco, R.; Aiello Talamanca, A.; Liguori, G.; Tatangelo, F.; Delrio, P.; Nasti, G.; Barletta, E.; Facchini, G.; Daniele, B.; et al. Overexpression of both CXC chemokine receptor 4 and vascular endothelial growth factor proteins predicts early distant relapse in stage II-III colorectal cancer patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 2795–2803. [Google Scholar] [CrossRef]
- Watanabe, H.; Miki, C.; Okugawa, Y.; Toiyama, Y.; Inoue, Y.; Kusunoki, M. Decreased expression of monocyte chemoattractant protein-1 predicts poor prognosis following curative resection of colorectal cancer. Dis. Colon Rectum 2008, 51, 1800–1805. [Google Scholar] [CrossRef]
- Akishima-Fukasawa, Y.; Nakanishi, Y.; Ino, Y.; Moriya, Y.; Kanai, Y.; Hirohashi, S. Prognostic significance of CXCL12 expression in patients with colorectal carcinoma. Am. J. Clin. Pathol. 2009, 132, 202–210, quiz 307. [Google Scholar] [CrossRef]
- Oladipo, O.; Conlon, S.; O’Grady, A.; Purcell, C.; Wilson, C.; Maxwell, P.J.; Johnston, P.G.; Stevenson, M.; Kay, E.W.; Wilson, R.H.; et al. The expression and prognostic impact of CXC-chemokines in stage II and III colorectal cancer epithelial and stromal tissue. Br. J. Cancer 2011, 104, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Han, X.; Yan, J.; Pan, Y.; Gong, J.; Di, J.; Cheng, Z.; Jin, Z.; Wang, Z.; Zheng, Q.; et al. The prognostic significance of chemokine receptor CXCR3 expression in colorectal carcinoma. Biomed. Pharmacother. 2012, 66, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.H.; Li, J.; Li, Y.; Zhang, X.T.; Liao, W.T.; Zhang, J.Y.; Li, R.; Luo, R.C. Co-expression of CXCR4 and CD133 proteins is associated with poor prognosis in stage II-III colon cancer patients. Exp. Ther. Med. 2012, 3, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Chen, Y.; He, X.; Wu, X.; Ke, J.; Zou, Y.; Cai, Z.; Zeng, Y.; Wang, L.; Wang, J.; et al. CCL18 as an independent favorable prognostic biomarker in patients with colorectal cancer. J. Surg. Res. 2013, 183, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Chen, Y.; Wu, X.; Yuan, R.; Cai, Z.; He, X.; Fan, X.; Wang, L.; Wu, X.; Lan, P. CCL21 as an independent favorable prognostic factor for stage III/IV colorectal cancer. Oncol. Rep. 2013, 30, 659–666. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, D.; Dai, T.; Xue, L.; Liu, X.; Wu, B.; Geng, J.; Mao, X.; Wang, R.; Chen, L.; Chu, X. Expression of chemokine receptor CXCR7 in colorectal carcinoma and its prognostic significance. Int. J. Clin. Exp. Pathol. 2015, 8, 13051–13058. [Google Scholar]
- Wu, Z.; Huang, X.; Han, X.; Li, Z.; Zhu, Q.; Yan, J.; Yu, S.; Jin, Z.; Wang, Z.; Zheng, Q.; et al. The chemokine CXCL9 expression is associated with better prognosis for colorectal carcinoma patients. Biomed. Pharmacother. 2016, 78, 8–13. [Google Scholar] [CrossRef]
- Yao, H.; Lv, Y.; Bai, X.; Yu, Z.; Liu, X. Prognostic value of CXCL17 and CXCR8 expression in patients with colon cancer. Oncol. Lett. 2020, 20, 2711–2720. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Yang, X.; Cheng, L.; Liu, R.; Lei, Y.; Dong, D.; Li, F.; Lau, Q.C.; Deng, L.; Nice, E.C.; et al. Chemokine CXCL14 is associated with prognosis in patients with colorectal carcinoma after curative resection. J. Transl. Med. 2013, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Zou, R.; Lin, F.; Zheng, S.; Shen, X.; Xue, X. Expression and effect of CXCL14 in colorectal carcinoma. Mol. Med. Rep. 2014, 10, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.X.; Tang, W.T.; Liu, Y.; Wang, G.H.; Liang, Z.L.; Cui, L. CCX-CKR expression in colorectal cancer and patient survival. Int. J. Biol. Markers 2014, 29, E40–E48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ou, B.; Feng, H.; Wang, P.; Yin, S.; Zhu, C.; Wang, S.; Chen, C.; Zheng, M.; Zong, Y.; et al. Overexpression of CXCR2 predicts poor prognosis in patients with colorectal cancer. Oncotarget 2017, 8, 28442–28454. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, C.; Wu, X.; Li, J.; Hu, D.; Jian, J.; Chen, C.; Zheng, X.; Yang, C. Chemokine (C-X-C motif) ligand 1 is associated with tumor progression and poor prognosis in patients with colorectal cancer. Biosci. Rep. 2018, 38, BSR20180580. [Google Scholar] [CrossRef]
- Li, R.B.; Zhang, S.Y.; Liu, G. Identification and validation of a pyroptosis-related prognostic model for colorectal cancer. Funct. Integr. Genom. 2023, 23, 21. [Google Scholar] [CrossRef] [PubMed]
- Bockorny, B.; Semenisty, V.; Macarulla, T.; Borazanci, E.; Wolpin, B.M.; Stemmer, S.M.; Golan, T.; Geva, R.; Borad, M.J.; Pedersen, K.S.; et al. BL-8040, a CXCR4 antagonist, in combination with pembrolizumab and chemotherapy for pancreatic cancer: The COMBAT trial. Nat. Med. 2020, 26, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Yue, M.; Chen, M.M.; Zhang, B.; Wang, Y.; Li, P.; Zhao, Y. The functional roles of chemokines and chemokine receptors in colorectal cancer progression. Biomed. Pharmacother. 2024, 170, 116040. [Google Scholar] [CrossRef]
- Blanchetot, C.; Verzijl, D.; Mujic-Delic, A.; Bosch, L.; Rem, L.; Leurs, R.; Verrips, C.T.; Saunders, M.; de Haard, H.; Smit, M.J. Neutralizing nanobodies targeting diverse chemokines effectively inhibit chemokine function. J. Biol. Chem. 2013, 288, 25173–25182. [Google Scholar] [CrossRef]
- Chatterjee, S.; Behnam Azad, B.; Nimmagadda, S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 2014, 124, 31–82. [Google Scholar] [CrossRef] [PubMed]
- Maganti, H.; Visram, A.; Shorr, R.; Fulcher, J.; Sabloff, M.; Allan, D.S. Plerixafor in combination with chemotherapy and/or hematopoietic cell transplantation to treat acute leukemia: A systematic review and metanalysis of preclinical and clinical studies. Leuk. Res. 2020, 97, 106442. [Google Scholar] [CrossRef] [PubMed]
- Kotb, R.M.; Ibrahim, S.S.; Mostafa, O.M.; Shahin, N.N. Potential role of CXCR4 in trastuzumab resistance in breast cancer patients. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166520. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Bmj 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.D.; Moons, K.G.M.; Snell, K.I.E.; Ensor, J.; Hooft, L.; Altman, D.G.; Hayden, J.; Collins, G.S.; Debray, T.P.A. A guide to systematic review and meta-analysis of prognostic factor studies. BMJ 2019, 364, k4597. [Google Scholar] [CrossRef] [PubMed]
- Tierney, J.F.; Stewart, L.A.; Ghersi, D.; Burdett, S.; Sydes, M.R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Balduzzi, S.; Rucker, G.; Schwarzer, G. How to perform a meta-analysis with R: A practical tutorial. Evid. Based Ment. Health 2019, 22, 153–160. [Google Scholar] [CrossRef]
- Deeks, J.J.; Higgins, J.P.T.; Altman, D.G.; on behalf of the Cochrane Statistical Methods Group. Chapter 10: Analysing Data and Undertaking Meta-Analyses. In Cochrane Handbook for Systematic Reviews of Interventions (Updated August 2023). Available online: https://training.cochrane.org/handbook/current/chapter-10#section-10-10-2 (accessed on 18 January 2024).
- Higgins, J.P.; Thompson, S.G.; Spiegelhalter, D.J. A re-evaluation of random-effects meta-analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2009, 172, 137–159. [Google Scholar] [CrossRef]

| Chemokine Receptor | Chemokine Ligand |
|---|---|
| CXCR1 | CXCL6, CXCL8 |
| CXCR2 | CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7, CXCL8 |
| CXCR3 | CXCL9, CXCL10, CXCL11, CXCL12 |
| CXCR4 | CXCL12 |
| CXCR5 | CXCL13 |
| CXCR6 | CXCL16 |
| CXCR7 | CXCL11, CXCL12 |
| Study | Number of Patients | SurvivalType | Investigated Chemokine | Study Participation | Study Attrition | Prognostic Factor Measurement | Outcome Measurement | Study Confounding | Statistical Analysis and Reporting |
|---|---|---|---|---|---|---|---|---|---|
| Ruan G. T. 2019 [39] | 212 | OS | CXCL3 | Low | Low | Low | Low | Low | Low |
| Zengin, M. 2020 [22] | 260 | OS DFS | CXCR4 CXCL12 | Low | Low | Low | Low | Low | Moderate |
| Li, X. 2019 [40] | 491 | OS DFS | CXCL9 CXCL10 CXCL11 CXCL13 | Moderate | Low | Low | Low | Low | Low |
| Gong, Y.Z. 2021 [41] | 438 | OS | CXCL1 | Moderate | Low | Low | Low | Moderate | Moderate |
| Li, L. 2021 [42] | 160 | OS | CXCL7 | Low | Low | Low | Low | Moderate | Low |
| Liu, M.J. 2021 [43] | 342 | OS | CXCL11 | Low | Low | Low | Low | Low | Low |
| Chen, Z. 2019 [44] | 142 | OS | CXCL16 | Low | Low | Low | Low | Low | Low |
| Okikawa, S. 2021 [45] | 98 | OS DFS | CXCL12 | Low | Moderate | Low | Moderate | Low | Moderate |
| Xu, C. 2018 [46] | 48 | OS | CXCR4 | Low | Moderate | Low | Low | Low | Low |
| Luo, X. 2022 [47] | 232 | OS | CXCL1 CXCL2 CXCL8 CXCL13 CXCL14 | Low | Low | Low | Low | Moderate | Moderate |
| Kim, J. 2005 [48] | 92 | OS DFS | CXCR4 | Low | Low | Low | Moderate | Moderate | Moderate |
| Terada, H. 2005 [49] | 87 | OS | CXCL8 | Low | Moderate | Low | Low | Low | Low |
| Wang, S. 2010 [50] | 388 | DFS | CXCR4 | Low | Low | Low | Low | Low | Low |
| Ottaiano, A. 2006 [51] | 72 | DFS | CXCR4 | Low | Low | Low | Low | Low | Low |
| Watanabe, H. 2008 [52] | 101 | OS | CCL2 | Low | Low | Low | Low | Moderate | Moderate |
| Akishima-Fukasawa, 2009 [53] | 165 | OS DFS | CXCL12 | Low | Low | Low | Low | Low | Low |
| Oladipo, O. 2011 [54] | 228 | OS DFS | CXCL8 | Low | Low | Low | Low | Low | Low |
| Wu, Z. 2012 [55] | 112 | OS | CXCR3 | Low | Low | Low | Low | Low | Low |
| Zhang, N.H. 2012 [56] | 125 | OS | CXCR4 | Low | Low | Low | Low | Low | Low |
| Yuan, R. 2013 [57] | 371 | OS DFS | CCL18 | Low | Low | Low | Low | Low | Low |
| Zou, Y. 2013 [58] | 143 | OS DFS | CCL21 | Low | Low | Low | Low | Low | Low |
| Yang, D. 2015 [59] | 96 | OS DFS | CXCR7 | Low | Low | Low | Low | Low | Low |
| Wu, Z. 2016 [60] | 130 | OS | CXCL9 | Low | Low | Low | Low | Low | Low |
| Yao, H. 2020 [61] | 101 | OS | CXCR8 CXCL17 | Low | Low | Low | Low | Low | Low |
| D’Alterio, 2014 [36] | 68 | OS DFS | CXCR4 CXCR7 CXCL12 | Low | Moderate | Low | Low | Moderate | Low |
| Zeng, J. 2013 [62] | 226 | OS DFS | CXCL14 | Low | Moderate | Low | Moderate | Moderate | Moderate |
| Lin, K. 2014 [63] | 40 | OS | CXCL14 | Moderate | Moderate | Low | Low | Moderate | Moderate |
| Zhu, Y.X. 2014 [64] | 136 | OS DFS | CCX-CKR | Low | Low | Low | Low | Low | Low |
| Zhao, J. 2017 [65] | 56 | OS DFS | CXCR2 | Low | Moderate | Low | Low | Low | Low |
| Zhuo, C. 2018 [66] | 276 | OS DFS | CXCL1 | Low | Moderate | Low | Low | Low | Low |
| Li, R. 2023 [67] | 643 | OS | CXCL8 | Low | Low | Low | Low | Moderate | Low |
| Study Information | Evaluation of Cxcl12 and Cxcr4 to Predict Poor Survival in Lymph Node-Positive Colorectal Cancer Patients | CXCR4 Overexpression Is Correlated with Poor Prognosis in Colorectal Cancer | Chemokine Receptor CXCR4 Expression in Colorectal Cancer Patients Increases the Risk for Recurrence and for Poor Survival | Co-Expression of CXCR4 and CD133 Proteins Is Associated with Poor Prognosis in Stage II-III Colon Cancer Patients | A Prognostic Model Comprising pT Stage, N Status, and the Chemokine Receptors CXCR4 and CXCR7 Powerfully Predicts Outcome in Neoadjuvant Resistant Rectal Cancer Patients | Nuclear Expression of CXCR4 Is Associated with Advanced Colorectal Cancer | Overexpression of both CXC Chemokine Receptor 4 and Vascular Endothelial Growth Factor Proteins Predicts Early Distant Relapse in Stage II–III Colorectal Cancer Patients |
|---|---|---|---|---|---|---|---|
| First Author | Zengin, M. | Xu, C. | Kim, J. | Zhang, N.H. | D’Alterio, C. | Wang, S. | Ottaiano, A |
| Year Publication | 2020 | 2018 | 2005 | 2012 | 2014 | 2010 | 2006 |
| Country | Turkey | China | USA | China | Italy | Taiwan | Italy |
| Number of patients | 260 | 48 | 92 | 125 | 68 | 388 | 72 |
| AJCC | - AJCC 3: 161 - AJCC 4: 99 | Not available | Primary AJCC stages: - AJCC 1: 29 - AJCC 2: 28 -AJCC 4: 35 | - AJCC 2: 61 - AJCC 3: 64 | Preoperative AJCC stage: - AJCC2: 9 - AJCC3: 59 Postoperative AJCC stage: - AJCC1: 29 - AJCC 2: 12 - AJCC 3: 27 | - AJCC 1: 52 - AJCC 2: 133 - AJCC 3: 123 - AJCC 4: 80 | - AJCC 2: 39 - AJCC 3: 33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fellhofer-Hofer, J.; Franz, C.; Vey, J.A.; Kahlert, C.; Kalkum, E.; Mehrabi, A.; Halama, N.; Probst, P.; Klupp, F. Chemokines as Prognostic Factor in Colorectal Cancer Patients: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2024, 25, 5374. https://doi.org/10.3390/ijms25105374
Fellhofer-Hofer J, Franz C, Vey JA, Kahlert C, Kalkum E, Mehrabi A, Halama N, Probst P, Klupp F. Chemokines as Prognostic Factor in Colorectal Cancer Patients: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences. 2024; 25(10):5374. https://doi.org/10.3390/ijms25105374
Chicago/Turabian StyleFellhofer-Hofer, Johanna, Clemens Franz, Johannes A. Vey, Christoph Kahlert, Eva Kalkum, Arianeb Mehrabi, Niels Halama, Pascal Probst, and Fee Klupp. 2024. "Chemokines as Prognostic Factor in Colorectal Cancer Patients: A Systematic Review and Meta-Analysis" International Journal of Molecular Sciences 25, no. 10: 5374. https://doi.org/10.3390/ijms25105374
APA StyleFellhofer-Hofer, J., Franz, C., Vey, J. A., Kahlert, C., Kalkum, E., Mehrabi, A., Halama, N., Probst, P., & Klupp, F. (2024). Chemokines as Prognostic Factor in Colorectal Cancer Patients: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences, 25(10), 5374. https://doi.org/10.3390/ijms25105374






