Runx2 and Polycystins in Bone Mechanotransduction: Challenges for Therapeutic Opportunities
Abstract
1. Introduction
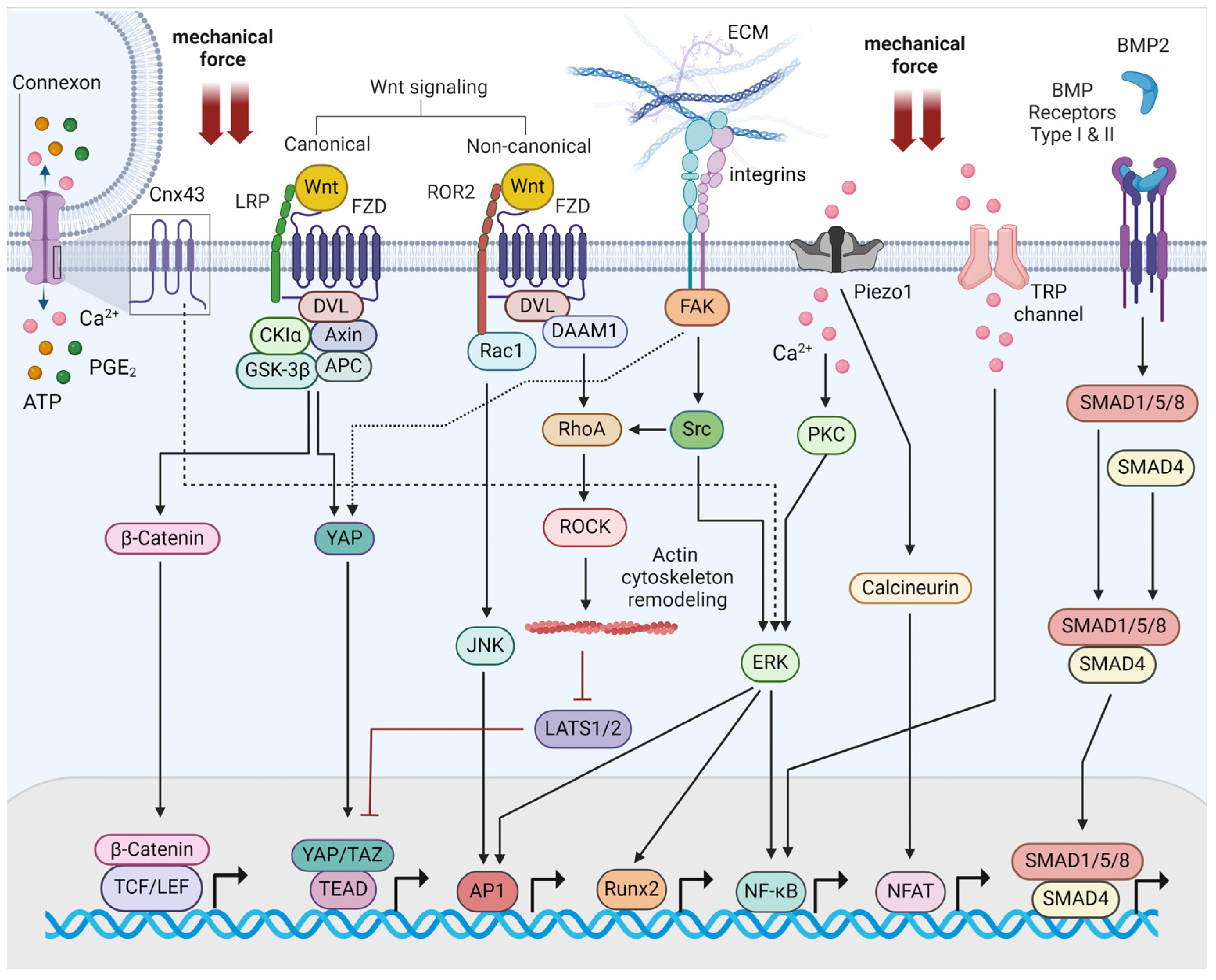
2. Molecular Mechanisms of Bone Mechanotransduction
3. Runx2 as an Effector of Bone Mechanotransduction
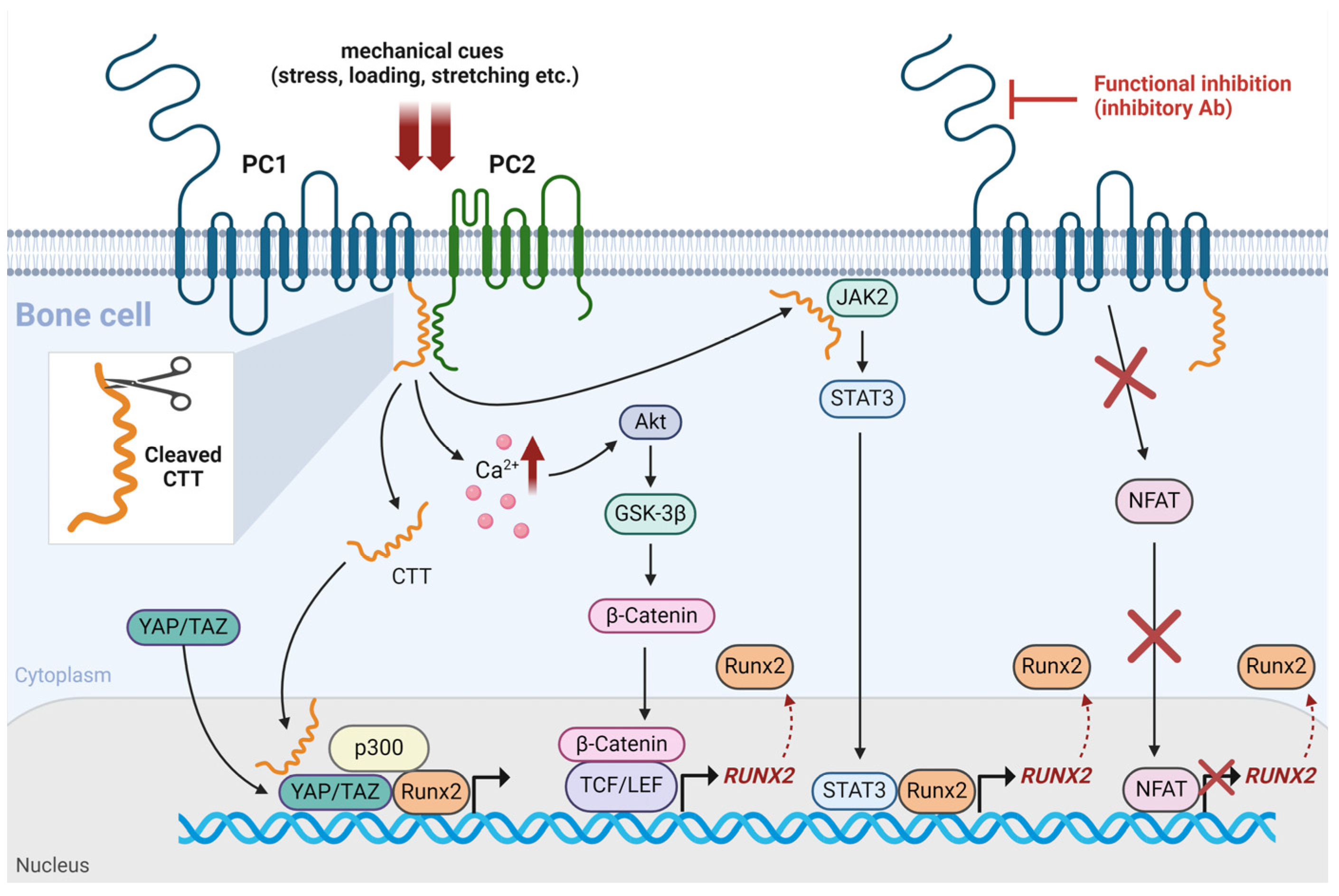
4. Mechanosensitive Polycystins and their Interaction with Runx2
5. Therapeutic Opportunities
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Takata, S.; Yasui, N. Disuse osteoporosis. J. Med. Investig. 2001, 48, 147–156. [Google Scholar]
- Wang, L.; You, X.; Zhang, L.; Zhang, C.; Zou, W. Mechanical regulation of bone remodeling. Bone Res. 2022, 10, 16. [Google Scholar] [CrossRef]
- Ma, Q.; Miri, Z.; Haugen, H.J.; Moghanian, A.; Loca, D. Significance of mechanical loading in bone fracture healing, bone regeneration, and vascularization. J. Tissue Eng. 2023, 14, 20417314231172573. [Google Scholar] [CrossRef]
- Chen, F.; Ouyang, Y.; Ye, T.; Ni, B.; Chen, A. Estrogen inhibits RANKL-induced osteoclastic differentiation by increasing the expression of TRPV5 channel. J. Cell. Biochem. 2014, 115, 651–658. [Google Scholar] [CrossRef]
- Santos, R.V.; Viana, V.A.; Boscolo, R.A.; Marques, V.G.; Santana, M.G.; Lira, F.S.; Tufik, S.; de Mello, M.T. Moderate exercise training modulates cytokine profile and sleep in elderly people. Cytokine 2012, 60, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Chen, X.; Zhang, S.; Huang, M.; Shen, X.; Xu, J.; Zou, J. The Effect of Exercise on the Prevention of Osteoporosis and Bone Angiogenesis. Biomed. Res. Int. 2019, 2019, 8171897. [Google Scholar] [CrossRef] [PubMed]
- Vignes, H.; Vagena-Pantoula, C.; Vermot, J. Mechanical control of tissue shape: Cell-extrinsic and -intrinsic mechanisms join forces to regulate morphogenesis. Semin. Cell Dev. Biol. 2022, 130, 45–55. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, L.; Zheng, Y.; Li, W. Bone remodeling induced by mechanical forces is regulated by miRNAs. Biosci. Rep. 2018, 38, BSR20180448. [Google Scholar] [CrossRef] [PubMed]
- Young, M.N.; Sindoni, M.J.; Lewis, A.H.; Zauscher, S.; Grandl, J. The energetics of rapid cellular mechanotransduction. Proc. Natl. Acad. Sci. USA 2023, 120, e2215747120. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Liu, W.; Cao, H.; Xiao, G. Molecular mechanosensors in osteocytes. Bone Res. 2020, 8, 23. [Google Scholar] [CrossRef]
- Stewart, S.; Darwood, A.; Masouros, S.; Higgins, C.; Ramasamy, A. Mechanotransduction in osteogenesis. Bone Jt. Res. 2020, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M.A.; Gu, S.; Hua, R.; Jiang, J.X. Mechanotransduction via the coordinated actions of integrins, PI3K signaling and Connexin hemichannels. Bone Res. 2021, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, Y.X.; Li, J. The role of mechanosensor Piezo1 in bone homeostasis and mechanobiology. Dev. Biol. 2023, 493, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Choi, R.B.; Robling, A.G. The Wnt pathway: An important control mechanism in bone’s response to mechanical loading. Bone 2021, 153, 116087. [Google Scholar] [CrossRef] [PubMed]
- Park, H.W.; Kim, Y.C.; Yu, B.; Moroishi, T.; Mo, J.S.; Plouffe, S.W.; Meng, Z.; Lin, K.C.; Yu, F.X.; Alexander, C.M.; et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell 2015, 162, 780–794. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Chen, J.; Liu, Y. LRP5 and LRP6 in Wnt Signaling: Similarity and Divergence. Front. Cell Dev. Biol. 2021, 9, 670960. [Google Scholar] [CrossRef] [PubMed]
- Lojk, J.; Marc, J. Roles of Non-Canonical Wnt Signalling Pathways in Bone Biology. Int. J. Mol. Sci. 2021, 22, 10840. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.L.; Chen, Z.H.; Teng, Y.Y.; Liu, S.Y.; Jia, Y.; Zhang, K.W.; Sun, Z.L.; Wu, J.J.; Yuan, Z.D.; Feng, Y.; et al. The Smad Dependent TGF-beta and BMP Signaling Pathway in Bone Remodeling and Therapies. Front. Mol. Biosci. 2021, 8, 593310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, X.; Wang, L.; Liu, Z.; Yi, Q.; Geng, B.; Chen, X.; Yu, D.; Xia, Y. The mechanosensory and mechanotransductive processes mediated by ion channels and the impact on bone metabolism: A systematic review. Arch. Biochem. Biophys. 2021, 711, 109020. [Google Scholar] [CrossRef]
- Xiao, Z.; Quarles, L.D. Physiological mechanisms and therapeutic potential of bone mechanosensing. Rev. Endocr. Metab. Disord. 2015, 16, 115–129. [Google Scholar] [CrossRef]
- Mevel, R.; Draper, J.E.; Lie, A.L.M.; Kouskoff, V.; Lacaud, G. RUNX transcription factors: Orchestrators of development. Development 2019, 146, dev148296. [Google Scholar] [CrossRef] [PubMed]
- Ziros, P.G.; Basdra, E.K.; Papavassiliou, A.G. Runx2: Of bone and stretch. Int. J. Biochem. Cell Biol. 2008, 40, 1659–1663. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Molecular Mechanism of Runx2-Dependent Bone Development. Mol. Cells 2020, 43, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Ziros, P.G.; Gil, A.P.; Georgakopoulos, T.; Habeos, I.; Kletsas, D.; Basdra, E.K.; Papavassiliou, A.G. The bone-specific transcriptional regulator Cbfa1 is a target of mechanical signals in osteoblastic cells. J. Biol. Chem. 2002, 277, 23934–23941. [Google Scholar] [CrossRef]
- Gomathi, K.; Akshaya, N.; Srinaath, N.; Moorthi, A.; Selvamurugan, N. Regulation of Runx2 by post-translational modifications in osteoblast differentiation. Life Sci. 2020, 245, 117389. [Google Scholar] [CrossRef] [PubMed]
- Jeon, E.J.; Lee, K.Y.; Choi, N.S.; Lee, M.H.; Kim, H.N.; Jin, Y.H.; Ryoo, H.M.; Choi, J.Y.; Yoshida, M.; Nishino, N.; et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 2006, 281, 16502–16511. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, J.W.; Lee, K.H.; Yoon, H.; Shin, D.H.; Ju, U.I.; Seok, S.H.; Lim, S.H.; Lee, Z.H.; Kim, H.H.; et al. Plant homeodomain finger protein 2 promotes bone formation by demethylating and activating Runx2 for osteoblast differentiation. Cell Res. 2014, 24, 1231–1249. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Okamura, H.; Nakashima, Y.; Haneji, T. Histone demethylase Jmjd3 regulates osteoblast differentiation via transcription factors Runx2 and osterix. J. Biol. Chem. 2013, 288, 33530–33541. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, A.; Srinaath, N.; Rohini, M.; Selvamurugan, N. Regulation of Runx2 by MicroRNAs in osteoblast differentiation. Life Sci. 2019, 232, 116676. [Google Scholar] [CrossRef]
- Yoon, D.S.; Kim, E.J.; Cho, S.; Jung, S.; Lee, K.M.; Park, K.H.; Lee, J.W.; Kim, S.H. RUNX2 stabilization by long non-coding RNAs contributes to hypertrophic changes in human chondrocytes. Int. J. Biol. Sci. 2023, 19, 13–33. [Google Scholar] [CrossRef]
- Lakhia, R.; Ramalingam, H.; Chang, C.M.; Cobo-Stark, P.; Biggers, L.; Flaten, A.; Alvarez, J.; Valencia, T.; Wallace, D.P.; Lee, E.C.; et al. PKD1 and PKD2 mRNA cis-inhibition drives polycystic kidney disease progression. Nat. Commun. 2022, 13, 4765. [Google Scholar] [CrossRef]
- Sun, H.; Li, Q.W.; Lv, X.Y.; Ai, J.Z.; Yang, Q.T.; Duan, J.J.; Bian, G.H.; Xiao, Y.; Wang, Y.D.; Zhang, Z.; et al. MicroRNA-17 post-transcriptionally regulates polycystic kidney disease-2 gene and promotes cell proliferation. Mol. Biol. Rep. 2010, 37, 2951–2958. [Google Scholar] [CrossRef]
- Patel, V.; Williams, D.; Hajarnis, S.; Hunter, R.; Pontoglio, M.; Somlo, S.; Igarashi, P. miR-17~92 miRNA cluster promotes kidney cyst growth in polycystic kidney disease. Proc. Natl. Acad. Sci. USA 2013, 110, 10765–10770. [Google Scholar] [CrossRef]
- Ding, H.; Li, L.X.; Harris, P.C.; Yang, J.; Li, X. Extracellular vesicles and exosomes generated from cystic renal epithelial cells promote cyst growth in autosomal dominant polycystic kidney disease. Nat. Commun. 2021, 12, 4548. [Google Scholar] [CrossRef]
- Hajarnis, S.S.; Patel, V.; Aboudehen, K.; Attanasio, M.; Cobo-Stark, P.; Pontoglio, M.; Igarashi, P. Transcription Factor Hepatocyte Nuclear Factor-1beta (HNF-1beta) Regulates MicroRNA-200 Expression through a Long Noncoding RNA. J. Biol. Chem. 2015, 290, 24793–24805. [Google Scholar] [CrossRef]
- Patel, V.; Hajarnis, S.; Williams, D.; Hunter, R.; Huynh, D.; Igarashi, P. MicroRNAs regulate renal tubule maturation through modulation of Pkd1. J. Am. Soc. Nephrol. 2012, 23, 1941–1948. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Z.; Zhang, R.; Huang, L.; Zhao, Z.; Yang, Y.; Cui, L.; Zhang, S. MiR-4787-5p Regulates Vascular Smooth Muscle Cell Apoptosis by Targeting PKD1 and Inhibiting the PI3K/Akt/FKHR Pathway. J. Cardiovasc. Pharmacol. 2021, 78, 288–296. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, S.; Xu, Z.; Zhang, J.; Li, L.; Zhao, G. The diagnostic value of microRNA-4787-5p and microRNA-4306 in patients with acute aortic dissection. Am. J. Transl. Res. 2017, 9, 5138–5149. [Google Scholar]
- Zhang, L.; Li, L.X.; Zhou, J.X.; Harris, P.C.; Calvet, J.P.; Li, X. RNA helicase p68 inhibits the transcription and post-transcription of Pkd1 in ADPKD. Theranostics 2020, 10, 8281–8297. [Google Scholar] [CrossRef]
- Qu, Z.; Quan, Z.; Zhang, Q.; Wang, Z.; Song, Q.; Zhuang, X.; Fu, C.; Xu, F.; Liu, Y.; Wang, Y.; et al. Comprehensive evaluation of differential lncRNA and gene expression in patients with intervertebral disc degeneration. Mol. Med. Rep. 2018, 18, 1504–1512. [Google Scholar] [CrossRef]
- Duplomb, L.; Droin, N.; Bouchot, O.; Thauvin-Robinet, C.; Bruel, A.L.; Thevenon, J.; Callier, P.; Meurice, G.; Pata-Merci, N.; Loffroy, R.; et al. A constitutive BCL2 down-regulation aggravates the phenotype of PKD1-mutant-induced polycystic kidney disease. Hum. Mol. Genet. 2017, 26, 4680–4688. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Qin, X.; Chen, T.; Zhou, L.; Xu, X.; Feng, J. MicroRNA-106b-5p regulates cisplatin chemosensitivity by targeting polycystic kidney disease-2 in non-small-cell lung cancer. Anticancer. Drugs 2017, 28, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Pippin, J.A.; Chang, S.; Redmond, J.; Chesi, A.; Wells, A.D.; Maerz, T.; Grant, S.F.A.; Coleman, R.M.; Hankenson, K.D.; et al. Osteoporosis GWAS-implicated DNM3 locus contextually regulates osteoblastic and chondrogenic fate of mesenchymal stem/progenitor cells through oscillating miR-199a-5p levels. JBMR Plus 2024, 8, ziae051. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Fu, X.; Li, X.; Zhao, W. Upregulation of circ_0076684 in osteosarcoma facilitates malignant processes by mediating miRNAs/CUX1. J. Orthop. Surg. Res. 2024, 19, 260. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, S.; Wei, R.; Liu, Y.; Xu, T.; Liu, Z.; Tan, Z.; Xie, Y.; Yang, D.; Liang, Z.; et al. Circular RNA circ-3626 promotes bone formation by modulating the miR-338-3p/Runx2 axis. Jt. Bone Spine 2024, 91, 105669. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.J.; Shi, X.J.; Fu, Q.; Li, Y.C.; Zhu, L.; Lu, N. MicroRNA-584-5p/RUNX family transcription factor 2 axis mediates hypoxia-induced osteogenic differentiation of periosteal stem cells. World J. Stem Cells 2023, 15, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Jiang, B.; Luo, B.; Jiang, X.; Zhang, Y.; Wang, Q. Circular RNA-FK501 binding protein 51 boosts bone marrow mesenchymal stem cell proliferation and osteogenic differentiation via modulating microRNA-205-5p/Runt-associated transcription factor 2 axis. J. Orthop. Surg. Res. 2023, 18, 782. [Google Scholar] [CrossRef] [PubMed]
- Kaur, T.; John, A.A.; Sharma, C.; Vashisht, N.K.; Singh, D.; Kapila, R.; Kapila, S. miR300 intervenes Smad3/beta-catenin/RunX2 crosstalk for therapy with an alternate function as indicative biomarker in osteoporosis. Bone 2021, 143, 115603. [Google Scholar] [CrossRef]
- Chen, C.; Tang, Z.; Song, Q.; Yang, M.; Shi, Q.; Weng, Y. Downregulated microRNA-23b promotes BMP9-mediated osteogenesis in C2C12 myoblast cells by targeting Runx2. Mol. Med. Rep. 2016, 13, 2492–2498. [Google Scholar] [CrossRef]
- Li, X.; Guo, L.; Liu, Y.; Su, Y.; Xie, Y.; Du, J.; Zhou, J.; Ding, G.; Wang, H.; Bai, Y.; et al. MicroRNA-21 promotes osteogenesis of bone marrow mesenchymal stem cells via the Smad7-Smad1/5/8-Runx2 pathway. Biochem. Biophys. Res. Commun. 2017, 493, 928–933. [Google Scholar] [CrossRef]
- Li, W.; Chen, Z.; Cai, C.; Li, G.; Wang, X.; Shi, Z. MicroRNA-505 is involved in the regulation of osteogenic differentiation of MC3T3-E1 cells partially by targeting RUNX2. J. Orthop. Surg. Res. 2020, 15, 143. [Google Scholar] [CrossRef]
- Hu, R.; Liu, W.; Li, H.; Yang, L.; Chen, C.; Xia, Z.Y.; Guo, L.J.; Xie, H.; Zhou, H.D.; Wu, X.P.; et al. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J. Biol. Chem. 2011, 286, 12328–12339. [Google Scholar] [CrossRef]
- Kureel, J.; John, A.A.; Dixit, M.; Singh, D. MicroRNA-467g inhibits new bone regeneration by targeting Ihh/Runx-2 signaling. Int. J. Biochem. Cell Biol. 2017, 85, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, L.; Xing, L.; Chen, D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 2010, 28, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, R.L.; Croce, C.M.; Stein, J.L.; Lian, J.B.; van Wijnen, A.J.; Stein, G.S. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc. Natl. Acad. Sci. USA 2011, 108, 9863–9868. [Google Scholar] [CrossRef] [PubMed]
- Wildman, B.J.; Godfrey, T.C.; Rehan, M.; Chen, Y.; Afreen, L.H.; Hassan, Q. MICROmanagement of Runx2 Function in Skeletal Cells. Curr. Mol. Biol. Rep. 2019, 5, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, H.; Enomoto-Iwamoto, M.; Iwamoto, M.; Nomura, S.; Himeno, M.; Kitamura, Y.; Kishimoto, T.; Komori, T. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J. Biol. Chem. 2000, 275, 8695–8702. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Bonnamy, J.P.; Owen, M.J.; Ducy, P.; Karsenty, G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes. Dev. 2001, 15, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, C.A.; Yamamoto, H.; Fujita, T.; Furuichi, T.; Ito, K.; Inoue, K.; Yamana, K.; Zanma, A.; Takada, K.; Ito, Y.; et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes. Dev. 2004, 18, 952–963. [Google Scholar] [CrossRef]
- Komori, T. Whole Aspect of Runx2 Functions in Skeletal Development. Int. J. Mol. Sci. 2022, 23, 5776. [Google Scholar] [CrossRef]
- Yang, L.; Tsang, K.Y.; Tang, H.C.; Chan, D.; Cheah, K.S. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. USA 2014, 111, 12097–12102. [Google Scholar] [CrossRef]
- Gao, S.; Chen, B.; Gao, M.; Xu, Y.; Yang, X.; Yang, C.; Pan, S. Substrate Stiffness of Bone Microenvironment Controls Functions of Pre-Osteoblasts and Fibroblasts In Vitro. Biomimetics 2023, 8, 344. [Google Scholar] [CrossRef]
- Sun, M.; Chi, G.; Xu, J.; Tan, Y.; Xu, J.; Lv, S.; Xu, Z.; Xia, Y.; Li, L.; Li, Y. Extracellular matrix stiffness controls osteogenic differentiation of mesenchymal stem cells mediated by integrin alpha5. Stem Cell Res. Ther. 2018, 9, 52. [Google Scholar] [CrossRef]
- Song, J.; Liu, L.; Lv, L.; Hu, S.; Tariq, A.; Wang, W.; Dang, X. Fluid shear stress induces Runx-2 expression via upregulation of PIEZO1 in MC3T3-E1 cells. Cell Biol. Int. 2020, 44, 1491–1502. [Google Scholar] [CrossRef]
- Jiang, Y.; Guan, Y.; Lan, Y.; Chen, S.; Li, T.; Zou, S.; Hu, Z.; Ye, Q. Mechanosensitive Piezo1 in Periodontal Ligament Cells Promotes Alveolar Bone Remodeling During Orthodontic Tooth Movement. Front. Physiol. 2021, 12, 767136. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, W.; Xing, Z.; Qian, J.; Chen, L.; Gu, R.; Guo, W.; Lai, X.; Zhao, W.; Li, S.; et al. A Conditional Knockout Mouse Model Reveals a Critical Role of PKD1 in Osteoblast Differentiation and Bone Development. Sci. Rep. 2017, 7, 40505. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.W.; Grant, A.D.; Parker, S.S.; Hill, S.; Whalen, M.B.; Chakrabarti, J.; Harman, M.W.; Roman, M.R.; Forte, B.L.; Gowan, C.C.; et al. Breast tumor stiffness instructs bone metastasis via maintenance of mechanical conditioning. Cell Rep. 2021, 35, 109293. [Google Scholar] [CrossRef]
- Xing, Y.; Song, L.; Zhang, Y. Autophagy Is Possibly Involved in Osteoblast Responses to Mechanical Loadings. Curr. Issues Mol. Biol. 2022, 44, 3611–3620. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Cai, J.; Zeng, Z.; Gao, X.; Shao, X.; Ding, Y.; Feng, X.; Jing, D. The interactions between mTOR and NF-kappaB: A novel mechanism mediating mechanical stretch-stimulated osteoblast differentiation. J. Cell. Physiol. 2020, 236, 4592–4603. [Google Scholar] [CrossRef]
- Esarte Palomero, O.; Larmore, M.; DeCaen, P.G. Polycystin Channel Complexes. Annu. Rev. Physiol. 2023, 85, 425–448. [Google Scholar] [CrossRef]
- Luo, L.; Roy, S.; Li, L.; Ma, M. Polycystic kidney disease: Novel insights into polycystin function. Trends Mol. Med. 2023, 29, 268–281. [Google Scholar] [CrossRef]
- Maser, R.L.; Calvet, J.P.; Parnell, S.C. The GPCR properties of polycystin-1- A new paradigm. Front. Mol. Biosci. 2022, 9, 1035507. [Google Scholar] [CrossRef] [PubMed]
- Cantero, M.D.R.; Cantiello, H.F. Polycystin-2 (TRPP2): Ion channel properties and regulation. Gene 2022, 827, 146313. [Google Scholar] [CrossRef] [PubMed]
- Gargalionis, A.N.; Korkolopoulou, P.; Farmaki, E.; Piperi, C.; Dalagiorgou, G.; Adamopoulos, C.; Levidou, G.; Saetta, A.; Fragkou, P.; Tsioli, P.; et al. Polycystin-1 and polycystin-2 are involved in the acquisition of aggressive phenotypes in colorectal cancer. Int. J. Cancer 2015, 136, 1515–1527. [Google Scholar] [CrossRef]
- Gargalionis, A.N.; Malakou, L.S.; Adamopoulos, C.; Piperi, C.; Theohari, I.; Nokhbehsaim, M.; Deschner, J.; Kokkalis, G.; Korkolopoulou, P.; Papadavid, E.; et al. Polycystin-1 downregulation induces ERK-dependent mTOR pathway activation in a cellular model of psoriasis. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3468–3476. [Google Scholar] [CrossRef] [PubMed]
- Gargalionis, A.N.; Sarlani, E.; Stofas, A.; Malakou, L.S.; Adamopoulos, C.; Bamias, A.; Boutati, E.; Constantinides, C.A.; Stravodimos, K.G.; Piperi, C.; et al. Polycystin-1 induces activation of the PI3K/AKT/mTOR pathway and promotes angiogenesis in renal cell carcinoma. Cancer Lett. 2020, 489, 135–143. [Google Scholar] [CrossRef]
- Papavassiliou, K.A.; Zoi, I.; Gargalionis, A.N.; Koutsilieris, M. Polycystin-1 affects cancer cell behaviour and interacts with mTOR and Jak signalling pathways in cancer cell lines. J. Cell. Mol. Med. 2019, 23, 6215–6227. [Google Scholar] [CrossRef] [PubMed]
- Zoi, I.; Gargalionis, A.N.; Papavassiliou, K.A.; Nasiri-Ansari, N.; Piperi, C.; Basdra, E.K.; Papavassiliou, A.G. Polycystin-1 and hydrostatic pressure are implicated in glioblastoma pathogenesis in vitro. J. Cell. Mol. Med. 2022, 26, 1699–1709. [Google Scholar] [CrossRef]
- Lu, W.; Shen, X.; Pavlova, A.; Lakkis, M.; Ward, C.J.; Pritchard, L.; Harris, P.C.; Genest, D.R.; Perez-Atayde, A.R.; Zhou, J. Comparison of Pkd1-targeted mutants reveals that loss of polycystin-1 causes cystogenesis and bone defects. Hum. Mol. Genet. 2001, 10, 2385–2396. [Google Scholar] [CrossRef]
- Merrick, D.; Mistry, K.; Wu, J.; Gresko, N.; Baggs, J.E.; Hogenesch, J.B.; Sun, Z.; Caplan, M.J. Polycystin-1 regulates bone development through an interaction with the transcriptional coactivator TAZ. Hum. Mol. Genet. 2019, 28, 16–30. [Google Scholar] [CrossRef]
- Xiao, Z.; Cao, L.; Smith, M.D.; Li, H.; Li, W.; Smith, J.C.; Quarles, L.D. Genetic interactions between polycystin-1 and Wwtr1 in osteoblasts define a novel mechanosensing mechanism regulating bone formation in mice. Bone Res. 2023, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Dalagiorgou, G.; Piperi, C.; Adamopoulos, C.; Georgopoulou, U.; Gargalionis, A.N.; Spyropoulou, A.; Zoi, I.; Nokhbehsaim, M.; Damanaki, A.; Deschner, J.; et al. Mechanosensor polycystin-1 potentiates differentiation of human osteoblastic cells by upregulating Runx2 expression via induction of JAK2/STAT3 signaling axis. Cell. Mol. Life Sci. 2017, 74, 921–936. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, W.; Ma, J.; Pan, Y.; Wang, L.; Zhang, W. Polycystin-1 mediates mechanical strain-induced osteoblastic mechanoresponses via potentiation of intracellular calcium and Akt/beta-catenin pathway. PLoS ONE 2014, 9, e91730. [Google Scholar] [CrossRef] [PubMed]
- Dalagiorgou, G.; Piperi, C.; Georgopoulou, U.; Adamopoulos, C.; Basdra, E.K.; Papavassiliou, A.G. Mechanical stimulation of polycystin-1 induces human osteoblastic gene expression via potentiation of the calcineurin/NFAT signaling axis. Cell. Mol. Life Sci. 2013, 70, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Chekroun, A.; Pujo-Menjouet, L.; Falcoz, S.; Tsuen, K.; Yueh-Hsun Yang, K.; Berteau, J.P. Theoretical evidence of osteoblast self-inhibition after activation of the genetic regulatory network controlling mineralization. J. Theor. Biol. 2022, 537, 111005. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Zhang, S.; Cao, L.; Qiu, N.; David, V.; Quarles, L.D. Conditional disruption of Pkd1 in osteoblasts results in osteopenia due to direct impairment of bone formation. J. Biol. Chem. 2010, 285, 1177–1187. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Zhang, S.; Mahlios, J.; Zhou, G.; Magenheimer, B.S.; Guo, D.; Dallas, S.L.; Maser, R.; Calvet, J.P.; Bonewald, L.; et al. Cilia-like structures and polycystin-1 in osteoblasts/osteocytes and associated abnormalities in skeletogenesis and Runx2 expression. J. Biol. Chem. 2006, 281, 30884–30895. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Dallas, M.; Qiu, N.; Nicolella, D.; Cao, L.; Johnson, M.; Bonewald, L.; Quarles, L.D. Conditional deletion of Pkd1 in osteocytes disrupts skeletal mechanosensing in mice. FASEB J. 2011, 25, 2418–2432. [Google Scholar] [CrossRef]
- Qiu, N.; Zhou, H.; Xiao, Z. Downregulation of PKD1 by shRNA results in defective osteogenic differentiation via cAMP/PKA pathway in human MG-63 cells. J. Cell. Biochem. 2012, 113, 967–976. [Google Scholar] [CrossRef]
- Kolpakova-Hart, E.; McBratney-Owen, B.; Hou, B.; Fukai, N.; Nicolae, C.; Zhou, J.; Olsen, B.R. Growth of cranial synchondroses and sutures requires polycystin-1. Dev. Biol. 2008, 321, 407–419. [Google Scholar] [CrossRef]
- Katsianou, M.A.; Papavassiliou, K.A.; Gargalionis, A.N.; Agrogiannis, G.; Korkolopoulou, P.; Panagopoulos, D.; Themistocleous, M.S.; Piperi, C.; Basdra, E.K.; Papavassiliou, A.G. Polycystin-1 regulates cell proliferation and migration through AKT/mTORC2 pathway in a human craniosynostosis cell model. J. Cell. Mol. Med. 2022, 26, 2428–2437. [Google Scholar] [CrossRef] [PubMed]
- Katsianou, M.; Papavassiliou, K.A.; Zoi, I.; Gargalionis, A.N.; Panagopoulos, D.; Themistocleous, M.S.; Piperi, C.; Papavassiliou, A.G.; Basdra, E.K. Polycystin-1 modulates RUNX2 activation and osteocalcin gene expression via ERK signalling in a human craniosynostosis cell model. J. Cell. Mol. Med. 2021, 25, 3216–3225. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Cao, L.; Liang, Y.; Huang, J.; Stern, A.R.; Dallas, M.; Johnson, M.; Quarles, L.D. Osteoblast-specific deletion of Pkd2 leads to low-turnover osteopenia and reduced bone marrow adiposity. PLoS ONE 2014, 9, e114198. [Google Scholar] [CrossRef] [PubMed]
- Khonsari, R.H.; Ohazama, A.; Raouf, R.; Kawasaki, M.; Kawasaki, K.; Porntaveetus, T.; Ghafoor, S.; Hammond, P.; Suttie, M.; Odri, G.A.; et al. Multiple postnatal craniofacial anomalies are characterized by conditional loss of polycystic kidney disease 2 (Pkd2). Hum. Mol. Genet. 2013, 22, 1873–1885. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Baudry, J.; Cao, L.; Huang, J.; Chen, H.; Yates, C.R.; Li, W.; Dong, B.; Waters, C.M.; Smith, J.C.; et al. Polycystin-1 interacts with TAZ to stimulate osteoblastogenesis and inhibit adipogenesis. J. Clin. Investig. 2018, 128, 157–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lu, Y.; Cai, G.; Chen, H.; Li, G.; Liu, L.; Sun, L.; Guan, Z.; Sun, W.; Zhao, C.; et al. Polycystin-2 mediates mechanical tension-induced osteogenic differentiation of human adipose-derived stem cells by activating transcriptional co-activator with PDZ-binding motif. Front. Physiol. 2022, 13, 917510. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Wang, D.; Yan, Z.; Ding, Y.; Zhang, J.; Liu, J.; Shao, X.; Liu, X.; Wang, L.; Luo, E.; et al. Bone Deterioration in Response to Chronic High-Altitude Hypoxia Is Attenuated by a Pulsed Electromagnetic Field Via the Primary Cilium/HIF-1alpha Axis. J. Bone Miner. Res. 2023, 38, 597–614. [Google Scholar] [CrossRef] [PubMed]
- Estevez, M.; Cicuendez, M.; Colilla, M.; Vallet-Regi, M.; Gonzalez, B.; Izquierdo-Barba, I. Magnetic colloidal nanoformulations to remotely trigger mechanotransduction for osteogenic differentiation. J. Colloid Interface Sci. 2024, 664, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Hajiali, H.; Rotherham, M.; El Haj, A.J. Remote Activation of Mechanotransduction via Integrin Alpha-5 via Aptamer-Conjugated Magnetic Nanoparticles Promotes Osteogenesis. Pharmaceutics 2023, 16, 21. [Google Scholar] [CrossRef]
- Zhou, Q.; Ren, X.; Oberoi, M.K.; Bedar, M.; Caprini, R.M.; Dewey, M.J.; Kolliopoulos, V.; Yamaguchi, D.T.; Harley, B.A.C.; Lee, J.C. beta-Catenin Limits Osteogenesis on Regenerative Materials in a Stiffness-Dependent Manner. Adv. Healthc. Mater. 2021, 10, e2101467. [Google Scholar] [CrossRef]
- Lee, P.S.; Heinemann, C.; Zheng, K.; Appali, R.; Alt, F.; Krieghoff, J.; Bernhardt, A.; Boccaccini, A.R.; van Rienen, U.; Hintze, V. The interplay of collagen/bioactive glass nanoparticle coatings and electrical stimulation regimes distinctly enhanced osteogenic differentiation of human mesenchymal stem cells. Acta Biomater. 2022, 149, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Gao, X.; Li, Y.; Sun, W.; Xu, Y.; Tan, Y.; Du, R.; Zhong, G.; Zhao, D.; Liu, Z.; et al. The mechanosensitive lncRNA Neat1 promotes osteoblast function through paraspeckle-dependent Smurf1 mRNA retention. Bone Res. 2022, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Zeng, C.; Qian, Y.; Yuan, S.; Ye, Z.; Zhao, S.; Li, R. Tensile strain promotes osteogenic differentiation of bone marrow mesenchymal stem cells through upregulating lncRNA-MEG3. Histol. Histopathol. 2021, 36, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Shin, H.L.; Kim, B.S.; Kim, H.J.; Ryoo, H.M. RUNX2-modifying enzymes: Therapeutic targets for bone diseases. Exp. Mol. Med. 2020, 52, 1178–1184. [Google Scholar] [CrossRef]
- Xu, H.; Wang, W.; Liu, X.; Huang, W.; Zhu, C.; Xu, Y.; Yang, H.; Bai, J.; Geng, D. Targeting strategies for bone diseases: Signaling pathways and clinical studies. Signal Transduct. Target. Ther. 2023, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.G.; Cerchio, K.; Stoch, S.A.; Gottesdiener, K.; Wu, M.; Recker, R.; Group, L.S. Effect of L-000845704, an alphaVbeta3 integrin antagonist, on markers of bone turnover and bone mineral density in postmenopausal osteoporotic women. J. Clin. Endocrinol. Metab. 2005, 90, 2022–2028. [Google Scholar] [CrossRef]
- Gerwin, N.; Scotti, C.; Halleux, C.; Fornaro, M.; Elliott, J.; Zhang, Y.; Johnson, K.; Shi, J.; Walter, S.; Li, Y.; et al. Angiopoietin-like 3-derivative LNA043 for cartilage regeneration in osteoarthritis: A randomized phase 1 trial. Nat. Med. 2022, 28, 2633–2645. [Google Scholar] [CrossRef]
| MicroRNA | Mechanism | Tissue | Reference |
|---|---|---|---|
| miR-17 | Polycystic Kidney Disease 1 (PKD1) and Polycystic Kidney Disease 2 (PKD2) mRNAs suppressed via their 3’-UTR miR-17-binding motif Targets PKD2 | Autosomal dominant polycystic kidney disease (ADPKD) mouse models HEK 293T | [31,32] |
| miR-17∼92 | Upregulated and produces renal cysts in mice | Polycystic kidney disease (PKD) mouse models | [33] |
| miR-200 family (miR-200b, miR-200c, and miR-429) | Downregulation of PKD1 by miRNAs secreted by exosomes Regulated by hepatocyte nuclear factor-1 (HNF-1) and targets PKD1 miR-200 downregulation by Dicer inactivation and concomitant PKD1 upregulation | Exosomes secreted from cystic epithelial cells and urine exosomes | [34,35,36] |
| miR-4787-5p | Targets PKD1 mRNA | Vascular smooth muscle cells (aortic dissection) | [37,38] |
| miR-17, miR-200c, and miR-182 | p68 induces the expression of miR-17, miR-200c, and miR-182 targeting PKD1 mRNA | Renal epithelial cells | [39] |
| lncRNAs OIP5-AS1 and UGDH-AS1 | Target PKD1 | Bioinformatics for intervertebral disc degeneration | [40] |
| miR-181a | Targets B-cell lymphoma 2 (Bcl-2) and promotes PKD phenotype | Plasma of PKD patient | [41] |
| miR-106b-5p | Targets PKD2 and sensitizes A549 cells to cisplatin | Non-small-cell lung cancer | [42] |
| miR-199a-5p | Targets Runx2 through SMAD1/5/9 | Human mesenchymal stem/progenitor cells | [43] |
| circ_0076684/miR-370-3p, miR-140-3p, and miR-193a-5p | CBX4-mediated Runx2 and circ_0076684 upregulation | Osteosarcoma | [44] |
| miR-338-3p | Targets Runx2 from upstream circ-3626 regulation | Mouse bone marrow-derived mesenchymal stem cells | [45] |
| miR-584-5p | Hypoxia-induced osteogenic differentiation through Runx2 | Primary mouse periosteal stem cells | [46] |
| miR-205-5p | Targets Runx2 by upstream circ-FK501 binding protein 51 induction | Bone marrow mesenchymal stem cells | [47] |
| miR-224-5p | Targets Runx2 for osteoblast differentiation | C2C12 myoblast cells | |
| miR-300 | Osteoblast differentiation through Smad3/β-catenin/RunX2 | Primary rat osteoblast cells, human osteoblast culture | [48] |
| miR-23b | Targets Runx2 induced by bone morphogenetic protein 9 | C2C12 myoblast cells | [49] |
| miR-21 | Osteogenic differentiation through Smad7-Smad1/5/8-Runx2 | Bone marrow mesenchymal stem cells | [50] |
| miR-505 | Osteogenic differentiation through Runx2 | MC3T3-E1 cells | [51] |
| miR-3960/miR2861 | BMP-2/miR-3960/homeobox A2 (Hoxa2) Runx2/miR-3960/miR2861 feedback loop | Primary mouse calvarial osteoblasts | [52] |
| miR-467g | Osteoblast differntiation through IHH/Runx2 targeting | Primary mouse calvarial osteoblasts, | [53] |
| human mesenchymal (skeletal) stem cells | |||
| miR-204 | Targets Runx2 promoting adipocyte differentiation | Mesenchymal progenitor cells and bone marrow stromal cells | [54] |
| miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-217, and miR-338 | Target Runx2 suppressing osteoblast differentiation | Mouse MC3T3-E1 osteoblasts, Mouse ATDC5 chondrocytes | [55] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gargalionis, A.N.; Adamopoulos, C.; Vottis, C.T.; Papavassiliou, A.G.; Basdra, E.K. Runx2 and Polycystins in Bone Mechanotransduction: Challenges for Therapeutic Opportunities. Int. J. Mol. Sci. 2024, 25, 5291. https://doi.org/10.3390/ijms25105291
Gargalionis AN, Adamopoulos C, Vottis CT, Papavassiliou AG, Basdra EK. Runx2 and Polycystins in Bone Mechanotransduction: Challenges for Therapeutic Opportunities. International Journal of Molecular Sciences. 2024; 25(10):5291. https://doi.org/10.3390/ijms25105291
Chicago/Turabian StyleGargalionis, Antonios N., Christos Adamopoulos, Christos T. Vottis, Athanasios G. Papavassiliou, and Efthimia K. Basdra. 2024. "Runx2 and Polycystins in Bone Mechanotransduction: Challenges for Therapeutic Opportunities" International Journal of Molecular Sciences 25, no. 10: 5291. https://doi.org/10.3390/ijms25105291
APA StyleGargalionis, A. N., Adamopoulos, C., Vottis, C. T., Papavassiliou, A. G., & Basdra, E. K. (2024). Runx2 and Polycystins in Bone Mechanotransduction: Challenges for Therapeutic Opportunities. International Journal of Molecular Sciences, 25(10), 5291. https://doi.org/10.3390/ijms25105291










