Abstract
The interactions with calf thymus DNA (CT-DNA) of three Schiff bases formed by the condensation of hesperetin with benzohydrazide (HHSB or L1H3), isoniazid (HIN or L2H3), or thiosemicarbazide (HTSC or L3H3) and their CuII complexes (CuHHSB, CuHIN, and CuHTSC with the general formula [CuLnH2(AcO)]) were evaluated in aqueous solution both experimentally and theoretically. UV–Vis studies indicate that the ligands and complexes exhibit hypochromism, which suggests helical ordering in the DNA helix. The intrinsic binding constants (Kb) of the Cu compounds with CT-DNA, in the range (2.3–9.2) × 106, from CuHTSC to CuHHSB, were higher than other copper-based potential drugs, suggesting that π–π stacking interaction due to the presence of the aromatic rings favors the binding. Thiazole orange (TO) assays confirmed that ligands and Cu complexes displace TO from the DNA binding site, quenching the fluorescence emission. DFT calculations allow for an assessment of the equilibrium between [Cu(LnH2)(AcO)] and [Cu(LnH2)(H2O)]+, the tautomer that binds CuII, amido (am) and not imido (im), and the coordination mode of HTSC (O−, N, S), instead of (O−, N, NH2). The docking studies indicate that the intercalative is preferred over the minor groove binding to CT-DNA with the order [Cu(L1H2am)(AcO)] > [Cu(L2H2am)(AcO)] ≈ TO ≈ L1H3 > [Cu(L3H2am)(AcO)], in line with the experimental Kb constants, obtained from the UV–Vis spectroscopy. Moreover, dockings predict that the binding strength of [Cu(L1H2am)(AcO)] is larger than [Cu(L1H2am)(H2O)]+. Overall, the results suggest that when different enantiomers, tautomers, and donor sets are possible for a metal complex, a computational approach should be recommended to predict the type and strength of binding to DNA and, in general, to macromolecules.
1. Introduction
Transition metals have a significant impact on the functioning of living organisms due to their unique properties such as the interconversion of several oxidation states and coordination geometries and the electrochemical behavior. Moreover, they are active toward organic nucleophiles, enhancing the bioactivity of many organic ligands including Schiff bases [1,2,3,4,5,6,7,8].
Copper (Cu) is a transition metal that deserves to be mentioned. Indeed, it is an essential element that is involved in many biological processes such as heme synthesis, cellular respiration, redox and oxygenation reactions, and electron transfer [9,10,11,12,13]; at relatively low intracellular concentrations, copper acts as a key catalytic cofactor in a wide range of biological processes including mitochondrial respiration, antioxidant defense, and the synthesis of various biocompounds [14]. Moreover, the interest in the potential uses of copper in medicine has increased in the last twenty years, and several compounds have been tested, both in vitro and in vivo, as potential anticancer drugs [15,16,17,18,19,20,21]. The observation that tumor growth and metastasis require a higher demand for copper has given an extra boost to the research of Cu-related diagnostics and treatments in the fight against cancer [22,23,24,25]. Today, copper is one of the most widely used metals of the first transition series to develop anticancer drugs due to its redox nature, biocompatible properties, and high effectiveness in inducing cancer cell death [22,26]. For instance, three novel Cu2+ complexes bearing N,N,O-chelating salphen-like ligands affected HeLa cells to an extent similar to cisplatin and significantly better than carboplatin [27]. The compound CuII–elesclomol, where elesclomol is N-malonyl-bis(N-methyl-N-thiobenzoyl hydrazide) in its doubly deprotonated form, has been proposed for metastatic melanoma [28], and the species Cu–triapine is active against many types of tumors [28,29]. In addition to their use as potential anticancer agents, the study of the properties of copper complexes has led to the development of non-steroidal anti-inflammatory drugs [18].
Various Cu compounds have reached clinical trials. The combination of copper or copper–gluconato with disulfiram (tetraethylthiuram disulfide) was proposed against various tumors and for newly diagnosed glioblastoma multiform (phase 2, identifier NCT03363659) [28,30], metastatic pancreatic cancer (phase 2, NCT03714555), and metastatic breast cancer (phase 2, NCT03323346). The species Cu–ATSM, with ATSM being diacetyl-bis(4-methyl-3-thiosemicarbazone), is not only under phase 2 of clinical trials for the treatment of rectal cancer (NCT03951337) [25], but has also progressed to phase 2/3 (NCT04082832) for its use against the neurodegenerative disease amyotrophic lateral sclerosis [31]. The complex 64Cu–DOTA, where DOTA is 2,2′,2″,2‴-(1,4,7,10-tetraazacyclododecane-1,4,7,10-tetrayl)tetraacetate, reached phase 1 (NCT02708511) for positron emission tomography-computed tomography use in imaging patients with ovarian and breast cancer [32]. The species Cu–histidinato is at phase 1/2 of the trials for its employment in the treatment of Menkes disease (NCT00001262). Finally, Casiopeinas®, formed by CuII, 1,10-phenanthroline or 2,2′-bipyridine or one their derivatives, plus a monoanionic non-toxic ligand like glycinate or acetylacetonate, has been proposed for the treatment of breast and colon cancer [28,33,34,35]; the compound CasIII-ia, [CuII(Me2bipy)(acac)]+, where Me2bipy is 4,4′-dimethyl-2,2′-bipyridine and acac is acetylacetonate, is currently undergoing phase 1 clinical trials in Mexico [36].
The exact mode of action of copper-based potential drugs is not always clear. They act with different mechanisms such as the inhibition of proteasome activity [37,38], telomerase activity [39], the formation of reactive oxygen species (ROS) [40,41], and, in particular, DNA interaction [42,43]. For this reason, the research on the interactions of copper complexes with DNA could be very important in biotechnology, pharmacology, and medicine for discovering and developing new potential drugs.
For a metallocompound, binding to DNA can occur in a variety of ways. Overall, they can be classified into two general categories that comprise covalent and non-covalent binding [44]; in this last case, intercalation between base pairs and minor or major DNA groove-binding interactions are involved [45,46]. Copper complexes give mostly non-covalent interactions, either by intercalation, electrostatic attraction, and/or groove binding [47]. DNA intercalation has been reported in many studies [48,49,50,51], and this results in an inhibitory action on topoisomerases [20] and in a nuclease activity with the break of the double-strand of DNA [47,48]. In particular, the capability of copper complexes to behave as artificial nuclease can be utilized to design new potential anticancer therapeutics, and numerous cases of Cu complexes with outstanding anticancer activity and a lower toxicity profile than conventional Pt drugs have been reported in the literature [17,47,52]. Such behavior is shown, for example, by the family of Casiopeines® [53]. For example, the aforementioned CasIII-ia and CasII-gly ([CuII(Me2phen)(Gly)]+ with Me2phen = 4,7-dimethyl-1,10-phenanthroline and Gly = glycinate) exhibit higher activity and lower toxicity compared to cisplatin. CasIII-Ea, with formula [CuII(Me2phen)(acac)]+, shows IC50 values of 4.9 and 2.1 μM on MCF-7 (breast cancer) and HCT-15 (colon cancer) cell lines, respectively, compared with 5.6 and 21.8 μM measured for cisplatin [54]; on the other hand, low toxicity is observed in non-tumor cells with accelerated growth like 3T3-L1 (healthy mice fibroblasts), treated with the CasIII-Ea compound [55]. As a final comment, it must be observed that binding between DNA and a metal complex can constitute hybrid catalysts in which transition metal complexes are embedded in a biomolecular scaffold represented by DNA; as a result of the formation of the new catalytic system, the reaction will proceed enantioselectively, and will optimally result in an additional acceleration [56].
Among the copper complexes, those formed by Schiff bases have been the object of extensive study. These compounds have attracted considerable interest in the scientific community due to their interesting properties, particularly those related to their biological and pharmacological action and possible use in medicine [1,2,3,4,5,6,7,8,57,58,59,60,61,62,63]. For example, hesperetin Schiff bases containing benzohydrazide (HHSB or L1H3), isoniazid (HIN or L2H3), and thiosemicarbazide (HTSC or L3H3) and their complexes with copper(II) have been investigated in analytical/spectral studies, and biological action in vitro such as cytotoxicity against human cancer cells, genotoxicity, and antimicrobial activity was ascertained by this and other groups [64,65,66]. Hesperetin (HESP) and its copper complex, CuHESP, were studied in some cases for comparison. The CuHHSB complex acts as a chemical nuclease during the cleavage of plasmid DNA in aqueous solution and is more effective than the free ligand HHSB against HeLa and K562 (human erythroleukemia) cells. In addition, among the tested bacterial strains, CuHHSB is very active toward Staphylococcus aureus [65]. All three copper complexes (CuHHSB, CuHIN, and CuHTSC) have an oxidative damaging effect greater than the ligands, with CuHIN showing the most prominent oxidative activity [66]. From our previous studies, it was found that modification of the structure of hesperetin increases its biological activity, for example, antioxidant activity, and for this reason, in the present work, we decided to perform another series of investigations to verify whether other properties of the modified ligands depended on the structural features. In this study, to allow for a better understanding of their potential biological effects, the interactions with DNA of the three hesperetin Schiff bases (HHSB, HTSC, and HIN) and their CuII compounds (CuHHSB, CuHIN, and CuHTSC) were screened in an aqueous solution both experimentally and theoretically. HESP and CuHESP were evaluated for comparison. UV–Vis and fluorescence spectra were carried out to obtain the binding characteristics; DFT methods were implemented to determine the structure of the most abundant species in solution, considering the exchange equilibrium between the monodentate equatorial ligand (AcO−) and solvent (H2O), the amido–imido tautomerism of the Schiff base ligands and the donor set (which may be (O−, N, S) or (O−, N, NH2) for L3H3); docking calculations allowed us to assess the binding to DNA and the effect of the nature of the substituents, unveiling new insights on the action of the copper complexes. The results could be useful for accelerating the development of this class of Cu-based drugs.
2. Results
2.1. Experimental Studies
2.1.1. Behavior of CuII Complexes in Aqueous Solution
The rationale for the interaction of Schiff base ligands HHSB, HTSC, and HIN with Cu2+ ions was presented in a previous publication [66]. Spectral data (FTIR, UV–Vis, EPR, ESI-MS) and electrochemical techniques showed that, in the acetate complexes, the tested Schiff bases act as neutral tridentate ligand coordinating to Cu2+ through two oxygens or oxygen/sulfur plus a nitrogen donor atom. EPR measurements indicated that, in solution, the complexes keep their structures, with the ligands remaining bound to Cu2+ ions in a tridentate fashion with the (O−, N, Oket) or (O−, N, S) donor set.
The three tridentate Schiff base chelating ligands in the fully protonated form can be indicated as LnH3, with n = 1–3. These were obtained from the functionalization of the hesperetin with three different hydrazides, benzohydrazide (HHSB or L1H3) [65], isoniazid or pyridine-4-carbohydrazide (HIN or L2H3), and thiosemicarbazide (HTSC or L3H3) [67] (Figure 1). The three titratable protons in aqueous solution were those on the OH groups in positions 5 and 7 of the ring A and 3′ of the flavonoid moiety (ring B).
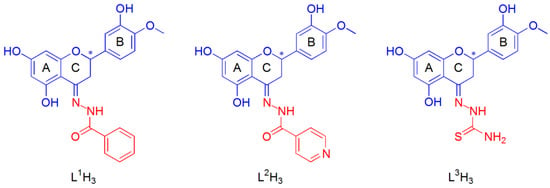
Figure 1.
Structural formula of the free hesperetin Schiff base ligands: benzohydrazide (L1H3); isoniazid (L2H3); thiosemicarbazide (L3H3). A different color is used for the molecular moiety deriving from hesperetin (blue) and hydrazides (red). The asterisk indicates the stereogenic carbon atom in position 2 of the C ring of hesperetin.
With CuII, they form different complexes in aqueous solutions, from [Cu(LH2)]+ to [Cu(LnH)], [Cu(Ln)]−, and [Cu(LnH–1)]2−. When starting from Cu(AcO)2·H2O, one solid compound is formed, [Cu(LnH2)(AcO)], around pH 6–7. They are also indicated as CuHHSB, CuHIN, and CuHTSC (Figure 2).
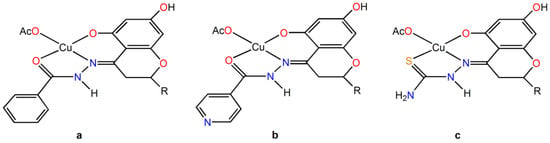
Figure 2.
Structural formula of the complexes: (a) CuHHSB, (b) CuHIN, and (c) CuHTSC. The R group is the ring B of hesperetin with OH and OCH3 substituents on the 3′ and 4′ positions.
Using EPR spectroscopy, it was demonstrated that the equilibrium [Cu(LnH2)(AcO)] + H2O/Solv ⇄ [Cu(LnH2)(H2O/Solv)]+ + AcO− is established in water-containing mixtures or in an organic solvent (Solv) like DMSO or DMF. From [Cu(LnH2)(AcO)] to [Cu(LnH2)(H2O/Solv)]+, the value of gz increases and Az(Cu) decreases by about (6–10) × 10−4 cm−1 [66]. The different steric hindrance of acetato and aqua ligand and the charge of the species could result in a different interaction with DNA.
Moreover, the coordination of LnH2− ligands can occur in the amido (am) or imido (im) form (Figure 3). In this case, the presence of a negative charge on the CO group and of NH instead of N atom in the five-membered chelate ring could also yield a different type of interaction with the double strand of DNA.

Figure 3.
Structure for the amido (a) and imido (b) coordination for the CuHHSB, CuHIN, and CuHTSC complexes. X represents a water or a AcO– ligand. The R group is the ring B of hesperetin with OH and OCH3 substituents on the 3′ and 4′ positions, while R’ stands for benzohydrazide, isoniazid, or thiosemicarbazide moieties of the HHSB, HIN, and HTSC ligands.
Finally, both R and S enantiomers of the HHSB, HIN, and HTSC ligands, due to the stereogenic carbon in position 2 of the C ring of hesperetin (indicated with an asterisk in Figure 1), could interact with DNA with significant differences in the structural complementarity, and hence in terms of energy binding.
2.1.2. UV–Vis Studies on the DNA Binding
Absorption titrations were carried out to determine the DNA binding of the ligands and their CuII complexes in tris(hydroxymethyl)aminomethane (Tris) buffer. The UV–Vis absorption spectra of compounds in the absence and presence of calf thymus DNA (CT-DNA) are shown in Figure 4.

Figure 4.
UV–Vis absorption spectra of the ligands and CuII compounds in the absence and presence of calf thymus DNA. The arrows show the changes in the absorbance after the addition of increasing amounts of CT-DNA.
If the interaction of a metal species with DNA is by intercalation, the π*- and π-orbital of the base pairs may couple, resulting in a decrease of the π–π* transition energy and giving rise to a red shift (bathochromic effect) and a decrease in absorption (hypochromic effect). The hypochromism of the π–π* transition is often employed to find the binding constant between a metal species and CT-DNA, according to Equation (1).
In Equation (1), [CT-DNA] is the base pairs concentration, εa is the apparent extinction coefficient at a given concentration, while εf and εb are the coefficients of free and fully bound metal species, respectively [68,69].
The results obtained in this study indicate that the spectral profiles of the ligands and complexes are very different. After the addition of CT-DNA, the absorption bands of HHSB, HIN, HTSC and the corresponding copper complexes exhibited hypochromism, of which the highest one was for HHSB, but to a lower extent for the HIN, HTSC, and Cu species. Red shift was not observed. Hypochromic shift in the spectra of the compounds suggests helical ordering of both ligands and Cu compounds in the DNA helix. The absorption spectra of HHSB and HTSC when titrated with CT-DNA also showed an isosbestic point. The intrinsic binding constants (Kb) of the compounds with CT-DNA are reported in Table 1. Notably, the values found for CuII complexes, in the range (2.3–9.2) × 106, were higher than other copper-based compounds, for example, those formed by amidino-O-methylurea derivatives ((0.6–1.2) × 105, ref. [70]), non-steroidal anti-inflammatory drugs piroxicam and lornoxicam ((2.7–3.4) × 104, refs. [71,72]), Schiff base salicylaldehyde or 2-hydroxy-1-naphthalidene and L-valine and plus 1,10-phenanthroline ((5.7–6.5) × 103, refs. [73,74]), and N-salicyl-β-amino alcohol Schiff bases ((0.04–2.4) × 106, ref. [75]). The possibility of π–π stacking interaction due to the presence of aromatic rings and extended electronic delocalization favors the binding of CuHHSB, CuHIN, and CuHTSC, as confirmed by the high Kb values of the ligands ((3.7–6.9) × 106, Table 1). Notably, the values determined for the ligands and Cu complexes were higher than the Kb of the adduct EB–DNA (1.2 × 105, ref. [76]), where EB is ethidium bromide, a compound known for its capability to behave as an intercalative agent in the DNA double strand.

Table 1.
Experimental data on the interaction of the HTSC, HHSB, and HIN ligands and their CuII complexes with DNA.
The results of the UV–Vis measurements and emission spectra presented in this study indicate the pro-oxidant activity of the tested compounds, which we previously demonstrated using the alkaline comet test to assess DNA breakage in HeLa cells exposed to various concentrations of HHSB, HTSC, HIN, CuHHSB, CuHIN, and CuHTSC [66]. Among the ligands, the most pronounced oxidative activity was revealed for HTSC, and, among the complexes, for CuHIN. Generally, CuII species have a greater oxidative damaging effect than the ligands. Moreover, both hesperetin azomethine derivatives and their copper complexes disclosed significantly lower impact on DNA damage compared to cisplatin [66].
The standard binding free energy G was estimated using the equation ∆G° = −RTlnKb, where R is the universal gas constant (8.314 J·mol−1·K−1) and T is the temperature in Kelvins [77]. From this equation, it follows that the higher the binding constant Kb between CT-DNA and the ligand or Cu complexes, the more negative the standard free energy. Based on the data in Table 1, all processes were spontaneous under our experimental conditions with constant pressure and temperature. The reactions with CT-DNA that occurred most spontaneously were those observed for HHSB and CuHHSB.
For the complexes but not for the ligands, a linear correlation was found between the values of the Kb binding constants and the DNA double strain breaks (DNA-DSBs) (Figure 5). This phenomenon suggests a different mechanism of interaction of Cu complexes and ligands with DNA.
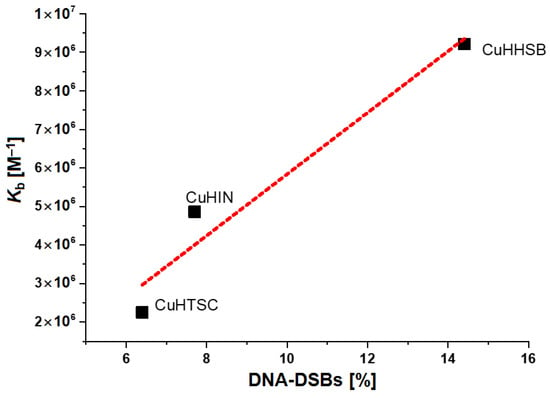
Figure 5.
Relationship between the values of Kb and DNA-DSBs (DNA double-strain breaks) for CuII complexes. Data taken from ref. [66]. The linear equation to fit the experimental points is y = a + bx, with a = (−2.14 ± 1.88) × 106 and b = (7.98 ± 1.85) × 105. The value of R2 is 0.90.
2.1.3. Thiazole Orange (TO) Displacement Assay
Thiazole orange (TO) is a good intercalating dye, and the structure of the DNA–TO intercalation complex has been studied by many authors [78,79,80]. The fluorescence of TO increases after intercalating with DNA; hence, if a compound intercalates into the helix of DNA, it would compete with TO for the DNA intercalation sites, leading to a significant decrease in fluorescence intensity [81,82].
The emission spectra of the DNA-TO system decreased after the separate addition of each of the compounds studied (Figure 6). This result indicates that they are able to replace TO in the DNA−TO adduct, resulting in the dissociation of TO and in a decrease in the emission intensity. In other words, all of the studied compounds, HHSB, HIN, HTSC, and their corresponding CuII complexes, can compete with TO for the intercalation sites of DNA, suggesting that they may intercalate into the helix of DNA. For these measurements, HESP and CuHESP were studied for comparison.
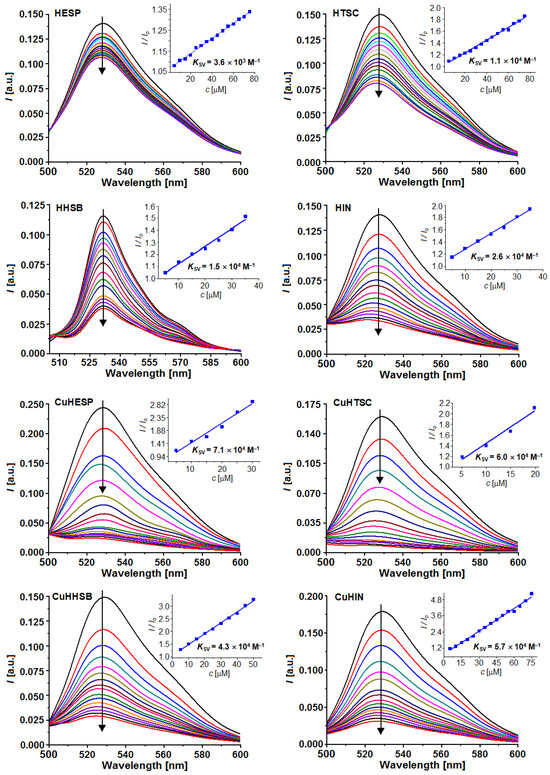
Figure 6.
Fluorescence spectra of the DNA–TO adduct upon the addition of HESP, HHSB, HIN, HTSC, and their corresponding complexes, CuHESP, CuHHSB, CuHIN, and CuHTSC. The concentration of CT-DNA was 2.5 × 10−5 M, and that of TO was 1.0 × 10−5 M, while the concentrations of the ligands and complexes were in the range (0.5–7.5) × 10−5 M. Insets represent Stern–Volmer relationships. The arrows show the changes in the fluorescence after the addition of increasing amounts of CT-DNA.
An analysis of the emission spectra of the DNA-TO system in the presence of an increasing concentration of the studied ligands and copper compounds was carried out in order to estimate the quenching % upon their addition, their quenching constant KSV, and apparent binding constant Kapp to DNA (Table 2). The data were analyzed using the Stern–Volmer equation, which puts in relationship the fluorescence with and without the quencher (F and F0), respectively, the concentration of the quencher [Q] (in this case, the hesperetin Schiff base ligands or the Cu complexes) and the Stern–Volmer quenching constant (KSV, in M−1): (F0/F) = 1 + KSV[Q]. The value of the apparent binding constant (Kapp) can be found with the equation Kapp = KTO × [TO]/[C50%], where KTO is 3.0 × 106 M−1, [TO] is the experimental concentration of TO, and [C50%] is the concentration of the quencher that produces a fluorescence decrease of 50%.

Table 2.
Thiazole orange (TO) quenching assay results for hesperetin Schiff base ligands and CuII complexes.
All of tested compounds reduced the fluorescence intensity, indicating that they are able to compete with TO for the same binding sites, or that they interact with DNA at different sites, but close to TO. Among the ligands and complexes, HIN and CuHTSC possessed the most quenching ability. From the values of KSV, quenching, and Kapp presented in Table 2, the order of increasing quenching and binding strength of the studied compounds is as follows: CuHTSC > CuHESP > CuHIN > CuHHSB > HIN > HHSB > HTSC > HESP. The order of the apparent binding constants suggests that Cu2+ ions have a distinct effect on quenching compared to the ligands themselves. This is probably related to the influence of the type of substituent inserted into the hesperetin moiety and to the interaction of the modified molecule with Cu2+. In fact, it must be observed that the electronic density of the HOMO in HESP is mainly localized over the rings A and B, while it is on ring A on the =N–NH group and the O atom of ring C in the remaining hesperetin derivatives [67]; therefore, the existence of electron-withdrawing substituents in HIN, HHSB, and HTSC enhances the electronic delocalization and conjugation, possibly influencing the process of quenching and binding of the ligands and metal species to DNA.
The apparent binding constants increased in the order opposite to DNA oxidative damage or DNA double strain breaks (DSBs) from the comet assay [66]; these observations may indicate that the free ligands and CuII species interact with slightly different mechanisms of action depending on the chemical environment. The values of Kapp (around 106) point to a preferential interaction with DNA through intercalation, but other pathways (electrostatic and/or minor/major groove) could occur.
Finally, it should be noted that the values of the Kapp constants of CuHIN and CuHHSB were very close to each other. This suggests that the isoniazid moiety affects in a negligible mode the binding strength of the CuII complexes.
2.2. Computational Studies
2.2.1. Geometry Optimization and Speciation Analysis of the Copper Complexes
Previously published spectroscopic results suggest that, in solution, [Cu(LnH2)(AcO)] has a square planar geometry around Cu2+ ion with donors (O−, N, O/S) and the fourth coordination position filled by the co-ligand (i.e., acetate AcO−) or a solvent molecule in [Cu(LnH2)(H2O)]+, with the protons on the OH groups in positions 7 and 3′ [66]. The DFT-characterized structures agree with the experimental outcomes, as shown in Figure 7.
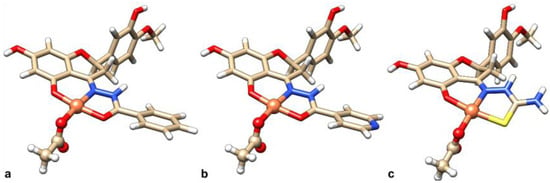
Figure 7.
DFT-optimized geometry structures of the complexes: (a) [Cu(L1H2)(AcO)], (b) [Cu(L2H2)(AcO)], and (c) [Cu(L3H2)(AcO)].
According to what was experimentally observed in solution [65,66], the three complexes can undergo ligand exchange of the acetate co-ligand with a solvent molecule and, additionally, both the amido and imido tautomers of the three ligands can be in equilibrium, each of them potentially chelating the Cu2+ ion. Therefore, a plethora of species must be considered when questioning which interacts with DNA.
In this work, the behavior of the CuII species in solution, particularly co-ligand/solvent replacement and tautomerism, were studied, aiming at the determination of the active species in the systems with DNA. For this purpose, the complex with L1H3 was selected as a model of the whole set of ligands.
The replacement of the acetato ligand by a water molecule was modeled as shown in Equation (2), and the Cartesian coordinates for the DFT-optimized structure are reported in Section S1 of the Supplementary Materials (Tables S1.1–S1.4).
[Cu(L1H2)(AcO)]aq + (H2O)8 aq ⇄ [Cu(L1H2)(H2O)]+aq + [(H2O)7·AcO]−aq
For the sake of accuracy, the two exchanging partners in this equilibrium (i.e., H2O and AcO−) were solvated using a mixed explicit/continuum method, according to Bryantsev et al. [83]. The calculated ΔGaq was 2.0 kcal mol−1, suggesting that the species are in equilibrium in solution at room temperature.
The study of the tautomerism of the Schiff bases (Figure 8) was assessed by considering the relative stability of the CuII complexes of the respective amido or imido tautomers (Equation (3)) and assuming the exchange of a proton with the solvent.
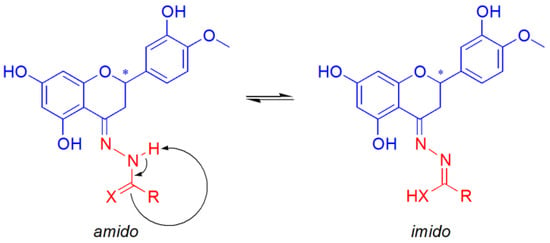
Figure 8.
Schiff base ligand tautomerism. X represents an atom of O or S. The asterisk indicates the stereogenic carbon atom in position 2 of the C ring of hesperetin.
The following reaction was studied:
[Cu(L1H2am)(H2O)]+aq + (H2O)14 aq ⇄ [Cu(L1Him)(H2O)]aq + [(H2O)14·H]+aq
The Cartesian coordinates for the species involved in Equation (3), optimized by DFT methods, are reported in Section S2 (Tables S2.1–S2.3). The calculated ΔGaq of 7.8 kcal mol−1 allowed us to exclude the presence of the imido form of the CuII complex in aqueous solution. This is in line with the characterization in the solid state and with the experimental EPR results (Table 3 and ref. [66]).

Table 3.
Calculated and experimental EPR parameters, gz and Az, of the species.
The ligand L3H3 deserves a deeper consideration compared to the other ones. Indeed, its coordination mode is ambiguous, (O−, N, S) or (O−, N, NH2), as shown in Figure 9. The computed ΔGaq for the two conformational isomers was 2.1 kcal·mol−1 with an energy barrier of 14.8 kcal·mol−1, ensuring their equilibrium at room temperature (Figure 9).
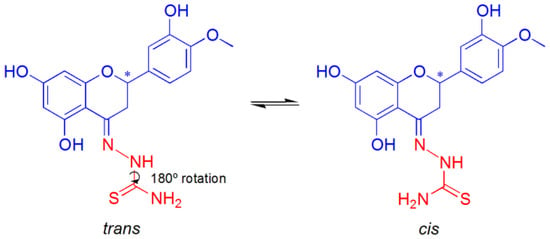
Figure 9.
Isomerization of the ligand L3H3. The asterisk indicates the stereogenic carbon atom in position 2 of the C ring of hesperetin.
The S-coordination is preferred over the N-coordination with a ΔGaq for Equation (4) of 9.3 kcal mol−1, suggesting that the equilibrium between the two linkage isomers can be considered totally shifted toward the S-coordinated species. The Cartesian coordinates for [Cu(L3H2-κS)(AcO)]aq and [Cu(L3H2-κN)(AcO)]aq are listed in Section S3 (Tables S3.1 and S3.2).
[Cu(L3H2-κS)(AcO)]aq ⇄ [Cu(L3H2-κN)(AcO)]aq
The computed spin Hamiltonian EPR parameters are in line with the experimental outcomes that suggested the S-coordination to be the favored in solution [66]. The predicted parameters are listed in Table 3.
Regarding the fourth coordination position, our DFT simulations confirmed the monodentate coordination of the acetato co-ligand with the non-coordinating oxygen perpendicular to the plane of the complex. The low energy barrier related to the flipping of the acetate, 2.9 kcal·mol−1, also ensures an equilibrium between two isoenergetic conformational isomers (Figure 10 and Figure S1). The coordinates for the two conformers are presented in Section S4 (Tables S1.1 and S4.1).
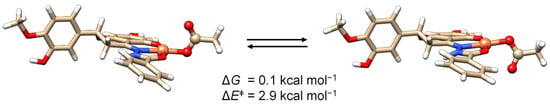
Figure 10.
Interconversion between the most stable conformers of [Cu(L3H2am)(AcO)] and the relative energies. The values of ΔG and of the energy barrier ΔE‡ for the flipping of the acetato ligand are also shown.
In general, the overall calculations allow us to discriminate which species are significantly present in solution. On the one hand, the amido–imido tautomerism of each complex and the linkage isomerism (–S or –NH2 donor) in the case of L3H3 were both significantly shifted toward only one of the two species (i.e., the amido tautomer and the –S donor, respectively). Thus, every imido tautomer as well as the coordination set (O−, N, NH2) for L3H3 can be ruled out from the docking calculations. On the other hand, the ∆G for the exchange of AcO– with solvent indicates that the two species [Cu(LnH2am)(AcO)] and [Cu(LnH2am)(H2O)]+ have comparable concentrations in solution. Torsional freedom for the Cu–OAc and Cu–OH2 bonds was accounted for.
2.2.2. Docking with DNA
For hesperetin acting as a stereogenic unit in all the complexes due to its asymmetric C2 atom on the ring C (see Figure 1), docking calculations for the L1H3 ligand and complexes were run with both the R and S enantiomers as the benchmark. Table 4 summarizes the species docked with DNA. In Section S5, the MOL2 files of each ligand structure implemented for the docking calculations are reported. In Section S5.1, the MOL2 files of each ligand structure implemented for the docking calculations are listed (from Tables S5.1–S5.11), while Section S5.2 contains the tables displaying the whole solutions and clusters for each calculation (from Tables S5.12–S5.33).

Table 4.
Species selected as ligands for the docking calculations.
For our systems, the interaction with DNA could occur in several modes: intercalative binding occurs through the insertion of a Cu complex, possibly positively charged, with aromatic ring(s) between two adjacent base pairs, stabilized by π–π stacking between the ring and the base pairs; groove binding with the reversible interaction of the copper species with a structure complementary to DNA major; and minor groove, which can vary with the size and shape of the groove [45].
We set up two different docking assays for the study of groove and intercalative binding modes. The structures of DNA crystallized with the pyrrolo[2,1-c][1,4]benzodiazepines derivative (PDB code 2K4L [84]) and intercalated by a dimeric derivative of the thiazole orange cation, TO (PDB code 108D [85]), were used as model receptors for the groove and intercalative binding assays, respectively. For the purpose of comparison, docking was additionally assessed for TO and the free ligand L1H3.
The results, gathered in Table 5, suggest that both the binding modes displayed high Fitness values, with intercalation favored for all species. The intercalative affinity order was [Cu(L1H2am)(AcO)] > [Cu(L2H2am)(AcO)] ≈ TO ≈ L1H3 > [Cu(L3H2am)(AcO)], in line with the experimental binding constants, Kb, obtained from the UV–Vis spectroscopy (Table 1).

Table 5.
Best GoldScore solutions for all of the CuII complexes and ligands with DNA for minor groove and intercalative binding. Fitness scores are sorted in order of decreasing values.
The main type of interactions found with the docking results for minor groove and intercalative binding modes were van der Waals contacts. These are more effective in the intercalative binding model because the DNA structure allows the ligand or metal species to enter the cavity between the base pairs (C3T4:G14A13 or A5G6:T12C11) to maximize these interactions, which can be described in terms of π–π contacts between the base pairs and the aromatic moieties of the hesperetin Schiff base ligands. In the groove binding model, ligands and metal moieties are not allowed to intercalate, and smaller F were observed. From the analysis of the four additive terms constituting the scoring function (see Equation (5) in Section 3.4), it is clear that the most important term among the four is Svdw_ext (i.e., the one accounting for van der Waals interactions between the ligand and CT-DNA) (Table 5 and Table S5.34). Conversely, the terms accounting for the hydrogen bonds, Shb_ext, and the intramolecular van der Waals forces plus the torsional strain energy, Sint, provides a minor contribution or are not relevant. The term that considers the intramolecular hydrogen bonds, Shb_int, is zero in all cases. From a structural point of view, the extent of the aromaticity seems to play a major role in binding stabilization, with the more extended aromatic systems (i.e., L1H3 and L2H3) being favored to the smaller (i.e., L3H3). Moreover, the R enantiomers were slightly favored in all cases.
Comparing the two AcO–-and H2O-containing complexes, the respective order of affinity was [Cu(L1H2am)(AcO)] > [Cu(L1H2am)(H2O)]+, highlighting the relevance of the co-ligand in adduct stabilization.
The formation of DNA adducts can be rationalized as a multistep process, in which minor groove binding is the first step. From this intermediate, the adduct evolves toward the more stable intercalated adduct. This is described in Figure 11.

Figure 11.
Schematized multistep binding process between DNA and [Cu(LnH2am)(AcO)], along with the Fitness values of the docking assays of the formed adducts. The symbols “⋯” and “⊃“ indicate, respectively, the minor groove and intercalative binding.
3. Materials and Methods
3.1. Synthesis
The synthesis and characterization of the CuHHSB complex have been presented in ref. [65], while the synthesis and description of the complexes CuHIN and CuHTSC were reported in ref. [66]. CuHESP was synthesized according to ref. [86]. All ligands (HHSB, HIN, and HTSC) were prepared according to the literature procedure [64]. The racemic hesperetin, benzohydrazide, 2-aminobenzohydrazide, isoniazid, thiosemicarbazide, and copper(II) acetate monohydrate (Cu(AcO)2·H2O) were purchased from Sigma-Aldrich Co. (Poznań, Poland). Thiazole orange and calf thymus DNA were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). All reagents were of analytical quality and used without further purification.
3.2. Spectroscopic Measurements
All absorption spectra were recorded at room temperature (25 °C) using matched quartz cells of 1.0 cm path length with a Varian UV–Visible Perkin-Elmer Lambda 11 spectrophotometer. Absorption spectra of 25 μM HHSB, HIN, HTSC, and complexes CuHHSB, CuHIN, CuHTSC, and the changes in their respective spectra on the subsequent addition of CT-DNA from 2.5 μM to 25 (35) μM in increasing concentration were recorded. All experiments were carried out in Tris buffer (5 mM Tris-HCl, 50 mM NaCl, pH 7.2). While measuring the absorption spectra, the solutions were allowed to incubate for 10 min before the absorption spectra were recorded, and an equal amount of CT-DNA was added to both the compound solution for the reference solution to eliminate the absorbance of CT-DNA itself. The intrinsic binding constant (Kb) of our compounds with CT-DNA was obtained using Equation (1) [68,69,87,88]. Spectrofluorimetric measurements were performed on a Hitachi spectrophotometer, model F-2000. Tris buffer containing 25 µM DNA and 25 µM TO was titrated with solutions of the tested ligands or CuII complexes (concentrations in the range of 10–300 µM were used).
The inner filter correction was included in the values of fluorescence intensity in the emission spectra, as shown in Figure 6. The inner filter correction was calculated according to the equation , where Fobs is the measured fluorescence, Fcorr is the correct fluorescence intensity that would be measured in the absence of inner-filter effects, dex and dem are the cuvette pathlength in the excitation and emission direction (1 cm), respectively, and Aex and Aem are the measured absorbance values at the excitation and emission wavelength, respectively, caused by ligand addition [89]. The emission spectra were recorded in the 500–600 nm (emission) wavelength range at an excitation wavelength λex of 430 nm. Measurements were performed in a quartz cuvette at room temperature.
3.3. DFT Calculations
Geometry optimization and vibrational frequency calculations were run by Gaussian 16 [90] using the DFT method with the B3LYP functional and Grimme’s D3 correction [91]. The basis-set 6-31G(d,p) was applied to the main group elements, while the SDD plus f-functions was employed for the metal [92]. The SMD continuum model for water was used to account for the aqueous environment [93]. Frequency calculations allowed us to obtain the thermal and entropic corrections. To determine the Gibbs free energies, these corrections were added to the potential energy obtained by single-point calculations on the optimized structures, with the basis-set def2-TZVP for the main group and def2-QZVP for the d-block elements [94]. A mixed explicit/continuum model [83] was used for aqueous water molecules, hydronium, and acetate. A dataset collection of the computational results is available in the ioChem-BD repository and can be accessed via https://doi.org/10.19061/iochem-bd-1-322 [95].
The EPR parameters were computed using the ORCA program [96,97,98]. The combination of the PBE0 [99] and 6-311G(d,p) basis-set was used to calculate the g tensor, while the B3LYP [100,101] was applied to determine the A tensor with the same basis-set, following the published results of Sciortino et al. [102].
3.4. Docking Calculations
Docking calculations were performed by GOLD 5.3 [103]. Previously, the original ligands of the DNA crystallographic structures (PDB code 2K4L [84] for the minor groove binding model; PDB code 108D [85] for the intercalation model) were removed and hydrogen atoms were added using UCSF Chimera [104].
The fluorescent dye thiazole orange (TO), used in the fluorescent quenching experiments, was optimized with the aforementioned method and used as the docking ligand for the purpose of comparison. The binding site comprised the whole structure for both models (PDB codes 2K4L [84] and 108D [85], respectively). For each ligand, 100 genetic algorithm (GA) runs were processed. Free rotation of the AcO–Cu coordination bond was considered along the simulations.
GoldScore, the scoring function used to calculate the Fitness score (F) in this study, has already been validated for docking with metal complexes as ligands (it must be noted that, in docking terminology, the species interacting with the receptor is generically named as the ligand) [105,106]. The polynomial scoring function consists of a sum of four terms (Equation (5)), accounting for hydrogen bonds between the ligand and receptor (Shb_ext), van del Waals interactions between the ligand and receptor (Svdw_ext), intramolecular hydrogen bonds of the ligand (Shb_int), and a term that summarizes the intramolecular van der Waals forces of the ligand and its torsional strain energy (Sint). Each term is weighted by the coefficients α = 1, β = 1.375, γ = 1, and δ =1.
Fitness score (F) = α Shb_ext + β Svdw_ext + γ Shb_int + δ Sint
The GA was set automatically with a minimum 1 × 105 operations. All other settings were left as the default. Finally, the docking solution were clustered by GOLD using an RMSD with a threshold of 2.5 Å.
4. Conclusions
Among the antitumor drugs, after the discovery of cisplatin and the development of its derivatives, metal complexes have gained a broad space in experimentation, and Cu-based potential therapeutics are worth being mentioned for their high activity and low toxicity. The binding studies of metal complexes with DNA are at the basis of the development of existing or new metal-based drugs. For metal complexes formed by first-row transition elements, these studies present several limitations: the possibility of exchange reactions of the ligands with water or solvent molecules, the formation of two or more isomers and of the corresponding enantiomers, the variation in the coordination number, and the possibility of the existence of tautomeric forms for the organic ligands can result in a wide variety of species able to interact with DNA. Therefore, it is not trivial to interpret the experimental results of the interaction studies, often based on UV–Vis or fluorescence spectroscopy. Some authors have noticed that the scarce knowledge of these aspects could be at the basis of the lack of interest by pharmaceutical companies for metal-based drugs compared with organic compounds, and the failed development of many of them [107]. Therefore, all approaches useful to characterize the metal–protein binding, preferably based on multiple techniques and a combined application of experimental and computational techniques, are desirable.
The systems discussed in the present study, containing a Schiff base formed by hesperetin with benzohydrazide (HHSB), isoniazid (HIN), and thiosemicarbazide (HTSC) represent a good example of what was above-mentioned. In fact, the solid compounds with the general formula [Cu(LnH2)(AcO)] can exchange the acetate ion with water, the ligands can bind the Cu centers in the amido or imido form, and HTSC can coordinate with the (O−, N, NH2) or (O−, N, S) donor set. The combined approach of the DFT and docking methods allowed us to demonstrate that: (i) an equilibrium between [Cu(LnH2)(AcO)] and [Cu(LnH2)(H2O)]+ exists in aqueous solution, and so both species can interact with DNA; (ii) the amido and not imido tautomer of HHSB, HIN, and HTSC binds Cu2+ ion; and (iii) the coordination mode of HTSC is (O−, N, S) and not (O−, N, NH2). Moreover, the results indicate that the intercalative binding is stronger than minor groove interaction with the order [Cu(L1H2am)(AcO)] > [Cu(L2H2am)(AcO)] ≈ TO ≈ L1H3 > [Cu(L3H2am)(AcO)], in agreement with the experimental binding constants to DNA (Kb) obtained from UV–Vis spectroscopy, with the aromaticity of the ligands playing a major role in binding stabilization. The computational data also suggest that the binding of [Cu(LnH2am)(AcO)] is preferred over [Cu(LnH2am)(H2O)]+, and the binding of R is favored compared to S enantiomers.
As a general conclusion, it must be highlighted that when different donor sets for the ligands, isomers, enantiomers, and tautomers for the metal complexes are possible, a computational approach should be recommended to predict the type and strength of binding to DNA, and in general, to macromolecules. To this it must be added, however, that a complete characterization of these systems is often not possible because a mixture of metal moieties can form and interact with DNA as well as because at the physiological metal concentration, not greater than a few μM, hydrolysis can lead to hydroxide species that could dominate in the solution at pH around 7.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25105283/s1.
Author Contributions
A.S., experimental investigation; F.P., computational investigation; G.S., computational investigation; F.M., supervision of computational investigation; E.L.-C., conceptualization, supervision, writing—original draft, and funding acquisition; E.G. conceptualization, supervision, writing—original draft, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by the National Science Center, Poland 2021/05/X/ST4/01006 and the Fondazione di Sardegna (grant FdS2017Garribba). G.S. also acknowledges the Spanish MICINN Juan de la Cierva Program, FJC2019-039135-I.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Material.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Khan, E.; Hanif, M.; Akhtar, M.S. Schiff bases and their metal complexes with biologically compatible metal ions; biological importance, recent trends and future hopes. Rev. Inorg. Chem. 2022, 42, 307–325. [Google Scholar] [CrossRef]
- Sinicropi, M.S.; Ceramella, J.; Iacopetta, D.; Catalano, A.; Mariconda, A.; Rosano, C.; Saturnino, C.; El-Kashef, H.; Longo, P. Metal Complexes with Schiff Bases: Data Collection and Recent Studies on Biological Activities. Int. J. Mol. Sci. 2022, 23, 14840. [Google Scholar] [CrossRef]
- Soroceanu, A.; Bargan, A. Advanced and Biomedical Applications of Schiff-Base Ligands and Their Metal Complexes: A Review. Crystals 2022, 12, 1436. [Google Scholar] [CrossRef]
- Boulechfar, C.; Ferkous, H.; Delimi, A.; Djedouani, A.; Kahlouche, A.; Boublia, A.; Darwish, A.S.; Lemaoui, T.; Verma, R.; Benguerba, Y. Schiff bases and their metal complexes: A review on the history, synthesis, and applications. Inorg. Chem. Commun. 2023, 150, 110451. [Google Scholar] [CrossRef]
- Sandhu, Q.-U.-A.; Pervaiz, M.; Majid, A.; Younas, U.; Saeed, Z.; Ashraf, A.; Khan, R.R.M.; Ullah, S.; Ali, F.; Jelani, S. Schiff base metal complexes as anti-inflammatory agents. J. Coord. Chem. 2023, 76, 1094–1118. [Google Scholar] [CrossRef]
- Singh, A.; Gogoi, H.P.; Barman, P. General Applications of Schiff Bases and Their Metal Complexes. In Schiff Base Metal Complexes: Synthesis and Applications; Wiley-VCH GmbH: Weinheim, Germany, 2023; pp. 119–128. [Google Scholar]
- Singh, P.; Yadav, P.; Kaur Sodhi, K.; Tomer, A.; Bali Mehta, S. Advancement in the synthesis of metal complexes with special emphasis on Schiff base ligands and their important biological aspects. Results Chem. 2024, 7, 101222. [Google Scholar] [CrossRef]
- Thakur, S.; Jaryal, A.; Bhalla, A. Recent advances in biological and medicinal profile of Schiff bases and their metal complexes: An updated version (2018–2023). Results Chem. 2024, 7, 101350. [Google Scholar] [CrossRef]
- Karlin, K.D.; Tyeklár, Z. Bioinorganic Chemistry of Copper; Chapman & Hall, Inc.: New York, NY, USA, 1993. [Google Scholar]
- Festa, R.A.; Thiele, D.J. Copper: An essential metal in biology. Curr. Biol. 2011, 21, R877–R883. [Google Scholar] [CrossRef]
- Maret, W.; Wedd, A. Binding, Transport and Storage of Metal Ions in Biological Cells; The Royal Society of Chemistry: London, UK, 2014. [Google Scholar]
- Rehder, D. Bioinorganic Chemistry; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Zoroddu, M.A.; Aaseth, J.; Crisponi, G.; Medici, S.; Peana, M.; Nurchi, V.M. The essential metals for humans: A brief overview. J. Inorg. Biochem. 2019, 195, 120–129. [Google Scholar] [CrossRef]
- Chen, L.; Min, J.; Wang, F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct. Target Ther. 2022, 7, 378. [Google Scholar] [CrossRef]
- Tisato, F.; Marzano, C.; Porchia, M.; Pellei, M.; Santini, C. Copper in diseases and treatments, and copper-based anticancer strategies. Med. Res. Rev. 2010, 30, 708–749. [Google Scholar] [CrossRef]
- Iakovidis, I.; Delimaris, I.; Piperakis, S.M. Copper and Its Complexes in Medicine: A Biochemical Approach. Mol. Biol. Int. 2011, 2021, 594529. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in Copper Complexes as Anticancer Agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef]
- Hussain, A.; AlAjmi, M.F.; Rehman, M.T.; Amir, S.; Husain, F.M.; Alsalme, A.; Siddiqui, M.A.; AlKhedhairy, A.A.; Khan, R.A. Copper(II) complexes as potential anticancer and Nonsteroidal anti-inflammatory agents: In Vitro and In Vivo studies. Sci. Rep. 2019, 9, 5237. [Google Scholar] [CrossRef]
- Kellett, A.; Molphy, Z.; McKee, V.; Slator, C. Recent Advances in Anticancer Copper Compounds. In Metal-Based Anticancer Agents; Casini, A., Vessières, A., Meier-Menches, S.M., Eds.; The Royal Society of Chemistry: Croydon, UK, 2019; pp. 91–119. [Google Scholar]
- Molinaro, C.; Martoriati, A.; Pelinski, L.; Cailliau, K. Copper Complexes as Anticancer Agents Targeting Topoisomerases I and II. Cancers 2020, 12, 2863. [Google Scholar] [CrossRef]
- Gu, Y.-Q.; Zhong, Y.-J.; Hu, M.-Q.; Li, H.-Q.; Yang, K.; Dong, Q.; Liang, H.; Chen, Z.-F. Terpyridine copper(II) complexes as potential anticancer agents by inhibiting cell proliferation, blocking the cell cycle and inducing apoptosis in BEL-7402 cells. Dalton Trans. 2022, 51, 1968–1978. [Google Scholar] [CrossRef]
- Ji, P.; Wang, P.; Chen, H.; Xu, Y.; Ge, J.; Tian, Z.; Yan, Z. Potential of Copper and Copper Compounds for Anticancer Applications. Pharmaceuticals 2023, 16, 234. [Google Scholar] [CrossRef]
- Rani, J.J.; Roy, S. Recent Development of Copper (II) Complexes of Polypyridyl Ligands in Chemotherapy and Photodynamic Therapy. ChemMedChem 2023, 18, e202200652. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, T.; Yuan, Y.; Li, N.; Wang, X.; Guan, J. Copper and Copper Complexes in Tumor Therapy. ChemMedChem 2024, 19, e202400060. [Google Scholar] [CrossRef] [PubMed]
- Abdolmaleki, S.; Aliabadi, A.; Khaksar, S. Unveiling the promising anticancer effect of copper-based compounds: A comprehensive review. J. Cancer Res. Clin. Oncol. 2024, 150, 213. [Google Scholar] [CrossRef] [PubMed]
- da Silva, D.A.; De Luca, A.; Squitti, R.; Rongioletti, M.; Rossi, L.; Machado, C.M.L.; Cerchiaro, G. Copper in tumors and the use of copper-based compounds in cancer treatment. J. Inorg. Biochem. 2022, 226, 111634. [Google Scholar] [CrossRef] [PubMed]
- Peña, Q.; Sciortino, G.; Maréchal, J.-D.; Bertaina, S.; Simaan, A.J.; Lorenzo, J.; Capdevila, M.; Bayón, P.; Iranzo, O.; Palacios, Ò. Copper(II) N,N,O-Chelating Complexes as Potential Anticancer Agents. Inorg. Chem. 2021, 60, 2939–2952. [Google Scholar] [CrossRef] [PubMed]
- Denoyer, D.; Clatworthy, S.A.S.; Cater, M.A. Copper Complexes in Cancer Therapy. In Metallo-Drugs: Development and Action of Anticancer Agents; Sigel, A., Sigel, H., Freisinger, E., Sigel, R.K.O., Eds.; De Gruyter: Berlin, Germany, 2018; pp. 469–506. [Google Scholar]
- Mathuber, M.; Hager, S.; Keppler, B.K.; Heffeter, P.; Kowol, C.R. Liposomal formulations of anticancer copper(II) thiosemicarbazone complexes. Dalton Trans. 2021, 50, 16053–16066. [Google Scholar] [CrossRef] [PubMed]
- Kannappan, V.; Ali, M.; Small, B.; Rajendran, G.; Elzhenni, S.; Taj, H.; Wang, W.; Dou, Q.P. Recent Advances in Repurposing Disulfiram and Disulfiram Derivatives as Copper-Dependent Anticancer Agents. Front. Mol. Biosci. 2021, 8, 741316. [Google Scholar] [CrossRef] [PubMed]
- Hilton, J.B.W.; Kysenius, K.; Liddell, J.R.; Mercer, S.W.; Paul, B.; Beckman, J.S.; McLean, C.A.; White, A.R.; Donnelly, P.S.; Bush, A.I.; et al. Evidence for disrupted copper availability in human spinal cord supports CuII(atsm) as a treatment option for sporadic cases of ALS. Sci. Rep. 2024, 14, 5929. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.; Srinivas, S.M.; Azevedo, C.; Greene, L.; Bauchet, A.-L.; Jouannot, E.; Lacoste-Bourgeacq, A.-S.; Guizon, I.; Cohen, P.; Naneix, A.-L.; et al. Two Patient Studies of a Companion Diagnostic Immuno-Positron Emission Tomography (PET) Tracer for Measuring Human CA6 Expression in Cancer for Antibody Drug Conjugate (ADC) Therapy. Mol. Imaging 2020, 19, 1536012120939398. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Azuara, L. Preparation of New Mixed Copper Aminoacidate Complexes from Phenylate Phenathrolines to Be Used as “Anticancerigenic” Agents. U.S. Patent 07/628,628: Re 35,458, 18 February 1992. [Google Scholar]
- Ruiz-Azuara, L. Process to Obtain New Mixed Copper Aminoacidate Complexes from Phenylatephenanthroline to Be Used as Anticancerigenic Agents. U.S. Patent 07/628,843: RE 35,458, 18 February 1992. [Google Scholar]
- Ruiz-Azuara, L. Copper Amino Acidate Diimine Nitrate Compounds and Their Methyl Derivatives and a Process for Preparing Them. U.S. Patent 5,576,326, 19 November 1996. [Google Scholar]
- Figueroa-DePaz, Y.; Resendiz-Acevedo, K.; Dávila-Manzanilla, S.G.; García-Ramos, J.C.; Ortiz-Frade, L.; Serment-Guerrero, J.; Ruiz-Azuara, L. DNA, a target of mixed chelate copper(II) compounds (Casiopeinas®) studied by electrophoresis, UV–vis and circular dichroism techniques. J. Inorg. Biochem. 2022, 231, 111772. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, X.; Chen, J.; Yang, Q.; Yang, L.; Xu, D.; Zhang, P.; Wang, X.; Liu, J. Hinokitiol copper complex inhibits proteasomal deubiquitination and induces paraptosis-like cell death in human cancer cells. Eur. J. Pharmacol. 2017, 815, 147–155. [Google Scholar] [CrossRef]
- Chen, X.; Dou, Q.P.; Liu, J.; Tang, D. Targeting Ubiquitin–Proteasome System with Copper Complexes for Cancer Therapy. Front. Mol. Biosci. 2021, 8, 649151. [Google Scholar] [CrossRef]
- Qin, Q.-P.; Meng, T.; Tan, M.-X.; Liu, Y.-C.; Luo, X.-J.; Zou, B.-Q.; Liang, H. Synthesis, crystal structure and biological evaluation of a new dasatinib copper(II) complex as telomerase inhibitor. Eur. J. Med. Chem. 2018, 143, 1597–1603. [Google Scholar] [CrossRef]
- Rivera-Guevara, C.; Bravo-Gómez, M.E.; Ruiz-Azuara, L. Chemotherapy and Design of New Antineoplastic Compounds. In Molecular Oncology: Principles and Recent Advances; Camacho, J., Ed.; Bentham: Sharjah, United Arab Emirates, 2012; pp. 172–191. [Google Scholar]
- Fatfat, M.; Merhi, R.A.; Rahal, O.; Stoyanovsky, D.A.; Zaki, A.; Haidar, H.; Kagan, V.E.; Gali-Muhtasib, H.; Machaca, K. Copper chelation selectively kills colon cancer cells through redox cycling and generation of reactive oxygen species. BMC Cancer 2014, 14, 527. [Google Scholar] [CrossRef]
- Becco, L.; García-Ramos, J.C.; Azuara, L.R.; Gambino, D.; Garat, B. Analysis of the DNA Interaction of Copper Compounds Belonging to the Casiopeínas® Antitumoral Series. Biol. Trace Elem. Res. 2014, 161, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Erxleben, A. Interactions of copper complexes with nucleic acids. Coord. Chem. Rev. 2018, 360, 92–121. [Google Scholar] [CrossRef]
- Gianferrara, T.; Bratsos, I.; Alessio, E. A categorization of metal anticancer compounds based on their mode of action. Dalton Trans. 2009, 7588–7598. [Google Scholar] [CrossRef]
- Pages, B.J.; Ang, D.L.; Wright, E.P.; Aldrich-Wright, J.R. Metal complex interactions with DNA. Dalton Trans. 2015, 44, 3505–3526. [Google Scholar] [CrossRef] [PubMed]
- Boros, E.; Dyson, P.J.; Gasser, G. Classification of Metal-Based Drugs according to Their Mechanisms of Action. Chem 2020, 6, 41–60. [Google Scholar] [CrossRef] [PubMed]
- McGivern, T.J.P.; Afsharpour, S.; Marmion, C.J. Copper complexes as artificial DNA metallonucleases: From Sigman’s reagent to next generation anti-cancer agent? Inorg. Chim. Acta 2018, 472, 12–39. [Google Scholar] [CrossRef]
- Gama, S.; Rodrigues, I.; Marques, F.; Palma, E.; Correia, I.; Carvalho, M.F.N.N.; Pessoa, J.C.; Cruz, A.; Mendo, S.; Santos, I.C.; et al. New ternary bipyridine–terpyridine copper(II) complexes as self-activating chemical nucleases. RSC Adv. 2014, 4, 61363–61377. [Google Scholar] [CrossRef]
- Galindo-Murillo, R.; García-Ramos, J.C.; Ruiz-Azuara, L.; Cheatham, T.E., III.; Cortés-Guzmán, F. Intercalation processes of copper complexes in DNA. Nucleic Acids Res. 2015, 43, 5364–5376. [Google Scholar] [CrossRef]
- Sangeetha, S.; Murali, M. Non-covalent DNA binding, protein interaction, DNA cleavage and cytotoxicity of [Cu(quamol)Cl]·H2O. Int. J. Biol. Macromol. 2018, 107, 2501–2511. [Google Scholar] [CrossRef]
- Romo, A.I.B.; Carepo, M.P.; Levín, P.; Nascimento, O.R.; Díaz, D.E.; Rodríguez-López, J.; León, I.E.; Bezerra, L.F.; Lemus, L.; Diógenes, I.C.N. Synergy of DNA intercalation and catalytic activity of a copper complex towards improved polymerase inhibition and cancer cell cytotoxicity. Dalton Trans. 2021, 50, 11931–11940. [Google Scholar] [CrossRef]
- Krasnovskaya, O.; Naumov, A.; Guk, D.; Gorelkin, P.; Erofeev, A.; Beloglazkina, E.; Majouga, A. Copper Coordination Compounds as Biologically Active Agents. Int. J. Mol. Sci. 2020, 21, 3965. [Google Scholar] [CrossRef] [PubMed]
- Maciel-Flores, C.E.; Lozano-Alvarez, J.A.; Bivián-Castro, E.Y. Recently Reported Biological Activities and Action Targets of Pt(II)- and Cu(II)-Based Complexes. Molecules 2024, 29, 1066. [Google Scholar] [CrossRef]
- Bravo-Gómez, M.E.; García-Ramos, J.C.; Gracia-Mora, I.; Ruiz-Azuara, L. Antiproliferative activity and QSAR study of copper(II) mixed chelate [Cu(N–N)(acetylacetonato)]NO3 and [Cu(N–N)(glycinato)]NO3 complexes, (Casiopeínas®). J. Inorg. Biochem. 2009, 103, 299–309. [Google Scholar] [CrossRef]
- Aguilar-Jiménez, Z.; Espinoza-Guillén, A.; Resendiz-Acevedo, K.; Fuentes-Noriega, I.; Mejía, C.; Ruiz-Azuara, L. The Importance of Being Casiopeina as Polypharmacologycal Profile (Mixed Chelate–Copper (II) Complexes and Their In Vitro and In Vivo Activities). Inorganics 2023, 11, 394. [Google Scholar] [CrossRef]
- Rioz-Martínez, A.; Roelfes, G. DNA-based hybrid catalysis. Curr. Opin. Chem. Biol. 2015, 25, 80–87. [Google Scholar] [CrossRef]
- Singh, A.; Gogoi, H.P.; Barman, P. Schiff Base Metal Complexes. In Schiff Base Metal Complexes: Synthesis and Applications; Barman, P., Singh, A., Eds.; Wiley-VCH GmbH: Weinheim, Germany, 2023; pp. 29–36. [Google Scholar]
- More, M.S.; Joshi, P.G.; Mishra, Y.K.; Khanna, P.K. Metal complexes driven from Schiff bases and semicarbazones for biomedical and allied applications: A review. Mater. Today Chem. 2019, 14, 100195. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Ahmed, S.S.; Alam, S.M.R. Biomedical applications of Schiff base metal complexes. J. Coord. Chem. 2020, 73, 3109–3149. [Google Scholar] [CrossRef]
- Khan, T.; Zehra, S.; Alvi, A.; Fatima, U.; Lawrence, A.I. Medicinal Utility of Some Schiff Bases and their Complexes with First Transition Series Metals: A Review. Orient. J. Chem. 2021, 37, 1051–1061. [Google Scholar] [CrossRef]
- Raju, S.K.; Settu, A.; Thiyagarajan, A.; Rama, D.; Sekar, P.; Kumar, S. Biological applications of Schiff bases: An overview. GSC Biol. Pharm. Sci. 2022, 21, 203–215. [Google Scholar] [CrossRef]
- Kumari, P.; Choudhary, M.; Kumar, A.; Yadav, P.; Singh, B.; Kataria, R.; Kumar, V. Copper(II) Schiff base complexes: Synthetic and medicinal perspective. Inorg. Chem. Commun. 2023, 158, 111409. [Google Scholar] [CrossRef]
- Mushtaq, I.; Ahmad, M.; Saleem, M.; Ahmed, A. Pharmaceutical significance of Schiff bases: An overview. Future J. Pharm. Sci. 2024, 10, 16. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Z.-y. DNA binding affinity and antioxidative activity of copper(II) and zinc(II) complexes with a novel hesperetin Schiff base ligand. Inorg. Chim. Acta 2009, 362, 4823–4831. [Google Scholar] [CrossRef]
- Lodyga-Chruscinska, E.; Symonowicz, M.; Sykula, A.; Bujacz, A.; Garribba, E.; Rowinska-Zyrek, M.; Oldziej, S.; Klewicka, E.; Janicka, M.; Krolewska, K.; et al. Chelating ability and biological activity of hesperetin Schiff base. J. Inorg. Biochem. 2015, 143, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Sykuła, A.; Nowak, A.; Garribba, E.; Dzeikala, A.; Rowińska-Żyrek, M.; Czerwińska, J.; Maniukiewicz, W.; Łodyga-Chruścińska, E. Spectroscopic Characterization and Biological Activity of Hesperetin Schiff Bases and Their Cu(II) Complexes. Int. J. Mol. Sci. 2023, 24, 761. [Google Scholar] [CrossRef]
- Sykula, A.; Kowalska-Baron, A.; Dzeikala, A.; Bodzioch, A.; Lodyga-Chruscinska, E. An experimental and DFT study on free radical scavenging activity of hesperetin Schiff bases. Chem. Phys. 2019, 517, 91–103. [Google Scholar] [CrossRef]
- Wolfe, A.; Shimer, G.H., Jr.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef] [PubMed]
- Pyle, A.M.; Rehmann, J.P.; Meshoyrer, R.; Kumar, C.V.; Turro, N.J.; Barton, J.K. Mixed-ligand complexes of ruthenium(II): Factors governing binding to DNA. J. Am. Chem. Soc. 1989, 111, 3051–3058. [Google Scholar] [CrossRef]
- Chaveerach, U.; Meenongwa, A.; Trongpanich, Y.; Soikum, C.; Chaveerach, P. DNA binding and cleavage behaviors of copper(II) complexes with amidino-O-methylurea and N-methylphenyl-amidino-O-methylurea, and their antibacterial activities. Polyhedron 2010, 29, 731–738. [Google Scholar] [CrossRef]
- Roy, S.; Banerjee, R.; Sarkar, M. Direct binding of Cu(II)-complexes of oxicam NSAIDs with DNA backbone. J. Inorg. Biochem. 2006, 100, 1320–1331. [Google Scholar] [CrossRef]
- Goswami, S.; Ray, S.; Sarkar, M. Spectroscopic studies on the interaction of DNA with the copper complexes of NSAIDs lornoxicam and isoxicam. Int. J. Biol. Macromol. 2016, 93, 47–56. [Google Scholar] [CrossRef]
- Dong, J.; Li, L.; Liu, G.; Xu, T.; Wang, D. Synthesis, crystal structure and DNA-binding properties of a new copper(II) complex with L-valine Schiff base and 1,10-phenanthroline. J. Mol. Struct. 2011, 986, 57–63. [Google Scholar] [CrossRef]
- Li, J.; Dong, J.; Cui, H.; Xu, T.; Li, L. A copper(II) complex of the Schiff base from l-valine and 2-hydroxy-1-naphthalidene plus 1,10-phenanthroline: Synthesis, crystal structure, and DNA interaction. Transit. Met. Chem. 2012, 37, 175–182. [Google Scholar] [CrossRef]
- Tunç, T.; Demirel, N.; Emir, M.; Günel, A.; Çolak, M.; Karacan, N. DNA Binding and Cleavage Activity of Three New Copper(II) Complexes of Chiral N-salicyl-β-amino Alcohol Schiff Bases. J. Mex. Chem. Soc. 2018, 62, 51–66. [Google Scholar] [CrossRef]
- Dimitrakopoulou, A.; Dendrinou-Samara, C.; Pantazaki, A.A.; Alexiou, M.; Nordlander, E.; Kessissoglou, D.P. Synthesis, structure and interactions with DNA of novel tetranuclear, [Mn4(II/II/II/IV)] mixed valence complexes. J. Inorg. Biochem. 2008, 102, 618–628. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Netzel, T.L.; Nafisi, K.; Zhao, M.; Lenhard, J.R.; Johnson, I. Base-Content Dependence of Emission Enhancements, Quantum Yields, and Lifetimes for Cyanine Dyes Bound to Double-Strand DNA: Photophysical Properties of Monomeric and Bichromomphoric DNA Stains. J. Phys. Chem. 1995, 99, 17936–17947. [Google Scholar] [CrossRef]
- Boger, D.L.; Tse, W.C. Thiazole orange as the fluorescent intercalator in a high resolution fid assay for determining DNA binding affinity and sequence selectivity of small molecules. Bioorg. Med. Chem. 2001, 9, 2511–2518. [Google Scholar] [CrossRef] [PubMed]
- Ihmels, H.; Otto, D. Intercalation of Organic Dye Molecules into Double-Stranded DNA—General Principles and Recent Developments. In Supermolecular Dye Chemistry; Würthner, F., Ed.; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2005; Volume 258, pp. 161–204. [Google Scholar]
- Ribeiro, N.; Roy, S.; Butenko, N.; Cavaco, I.; Pinheiro, T.; Alho, I.; Marques, F.; Avecilla, F.; Costa Pessoa, J.; Correia, I. New Cu(II) complexes with pyrazolyl derived Schiff base ligands: Synthesis and biological evaluation. J. Inorg. Biochem. 2017, 174, 63–75. [Google Scholar] [CrossRef]
- Brodowska, K.; Correia, I.; Garribba, E.; Marques, F.; Klewicka, E.; Łodyga-Chruscińska, E.; Costa Pessoa, J.; Dzeikala, A.; Chrusciński, L. Coordination ability and biological activity of a naringenin thiosemicarbazone. J. Inorg. Biochem. 2016, 165, 36–48. [Google Scholar] [CrossRef]
- Bryantsev, V.S.; Diallo, M.S.; Goddard, W.A., III. Calculation of Solvation Free Energies of Charged Solutes Using Mixed Cluster/Continuum Models. J. Phys. Chem. B 2008, 112, 9709–9719. [Google Scholar] [CrossRef] [PubMed]
- Antonow, D.; Barata, T.; Jenkins, T.C.; Parkinson, G.N.; Howard, P.W.; Thurston, D.E.; Zloh, M. Solution Structure of a 2:1 C2-(2-Naphthyl) Pyrrolo[2,1-c][1,4]benzodiazepine DNA Adduct: Molecular Basis for Unexpectedly High DNA Helix Stabilization. Biochemistry 2008, 47, 11818–11829. [Google Scholar] [CrossRef] [PubMed]
- Spielmann, H.P.; Wemmer, D.E.; Jacobsen, J.P. Solution Structure of a DNA Complex with the Fluorescent Bis-Intercalator TOTO Determined by NMR Spectroscopy. Biochemistry 1995, 34, 8542–8553. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, L.V.; Gouvea, L.R.; Sousa, A.C.; Albuquerque, R.M.; Teixeira, S.F.; de Azevedo, R.A.; Louro, S.R.W.; Ferreira, A.K.; Beraldo, H. Copper(II) complexes with naringenin and hesperetin: Cytotoxic activity against A 549 human lung adenocarcinoma cells and investigation on the mode of action. BioMetals 2016, 29, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, P.; Sathyadevi, P.; Cowley, A.H.; Butorac, R.R.; Dharmaraj, N. Evaluation of DNA binding, DNA cleavage, protein binding and in vitro cytotoxic activities of bivalent transition metal hydrazone complexes. Eur. J. Med. Chem. 2011, 46, 3376–3387. [Google Scholar] [CrossRef] [PubMed]
- Brodowska, K.; Sykuła, A.; Garribba, E.; Łodyga-Chruścińska, E.; Sójka, M. Naringenin Schiff base: Antioxidant activity, acid–base profile, and interactions with DNA. Transit. Met. Chem. 2016, 41, 179–189. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, A.W.; Böhme, M.; Dapprich, S.; Gobbi, A.; Höllwarth, A.; Jonas, V.; Köhler, K.F.; Stegmann, R.; Veldkamp, A.; Frenking, G. A set of f-polarization functions for pseudo-potential basis sets of the transition metals Sc-Cu, Y-Ag and La-Au. Chem. Phys. Lett. 1993, 208, 111–114. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Weigend, F.; Furche, F.; Ahlrichs, R. Gaussian basis sets of quadruple zeta valence quality for atoms H–Kr. J. Chem. Phys. 2003, 119, 12753–12762. [Google Scholar] [CrossRef]
- Álvarez-Moreno, M.; de Graaf, C.; López, N.; Maseras, F.; Poblet, J.M.; Bo, C. Managing the Computational Chemistry Big Data Problem: The ioChem-BD Platform. J. Chem. Inf. Model. 2015, 55, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Neese, F. ORCA—An Ab Initio, DFT and Semiempirical Program Package, Version 5.0; Max-Planck-Institute for Chemical Energy Conversion: Mülheim, Germany, 2021. [Google Scholar]
- Neese, F. Software update: The ORCA program system—Version 5.0. WIREs Comput. Mol. Sci. 2022, 12, e1606. [Google Scholar] [CrossRef]
- Neese, F.; Wennmohs, F.; Becker, U.; Riplinger, C. The ORCA quantum chemistry program package. J. Chem. Phys. 2020, 152, 224108. [Google Scholar] [CrossRef] [PubMed]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868, Erratum in Phys. Rev. Lett. 1997, 78, 1396–1396. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Sciortino, G.; Lubinu, G.; Maréchal, J.-D.; Garribba, E. DFT Protocol for EPR Prediction of Paramagnetic Cu(II) Complexes and Application to Protein Binding Sites. Magnetochemistry 2018, 4, 55. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Sciortino, G.; Sanna, D.; Ugone, V.; Lledós, A.; Maréchal, J.-D.; Garribba, E. Decoding Surface Interaction of VIVO Metallodrug Candidates with Lysozyme. Inorg. Chem. 2018, 57, 4456–4469. [Google Scholar] [CrossRef]
- Sciortino, G.; Sanna, D.; Ugone, V.; Micera, G.; Lledós, A.; Maréchal, J.-D.; Garribba, E. Elucidation of Binding Site and Chiral Specificity of Oxidovanadium Drugs with Lysozyme through Theoretical Calculations. Inorg. Chem. 2017, 56, 12938–12951. [Google Scholar] [CrossRef] [PubMed]
- Scior, T.; Guevara-Garcia, J.A.; Do, Q.T.; Bernard, P.; Laufer, S. Why Antidiabetic Vanadium Complexes are Not in the Pipeline of “Big Pharma” Drug Research? A Critical Review. Curr. Med. Chem. 2016, 23, 2874–2891. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).