Inactivation of Myostatin Delays Senescence via TREX1-SASP in Bovine Skeletal Muscle Cells
Abstract
1. Introduction
2. Results
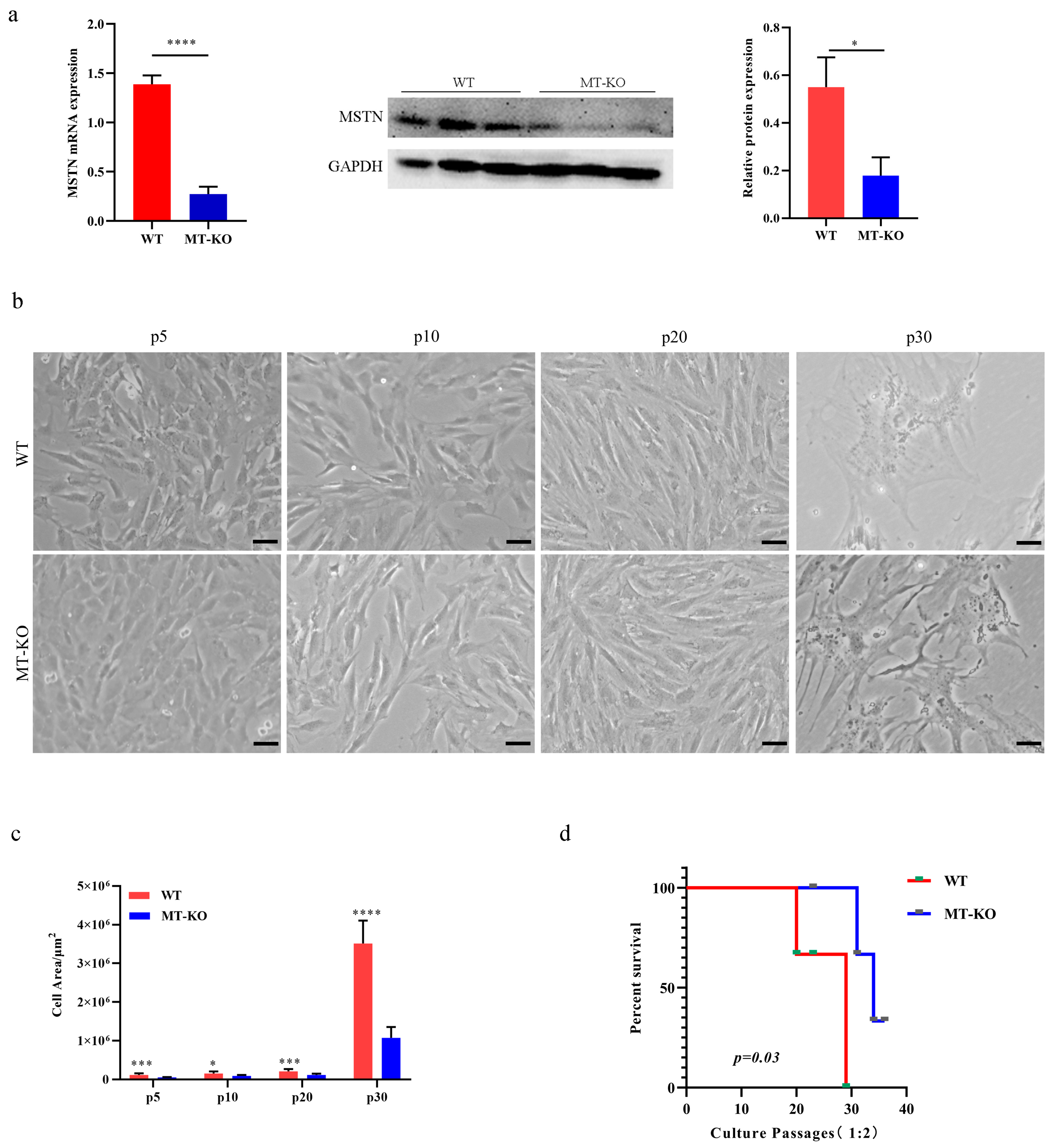
2.1. MSTN Gene Inactivation Delayed the Senescence of Long-Term Cultured bSMCs
2.2. Inactivation of the MSTN Alleviated Senescence Caused by the Long-Term Culture of bSMCs
2.3. Inactivation of the MSTN Downregulated the SASP in Skeletal Muscle Senescence Cells
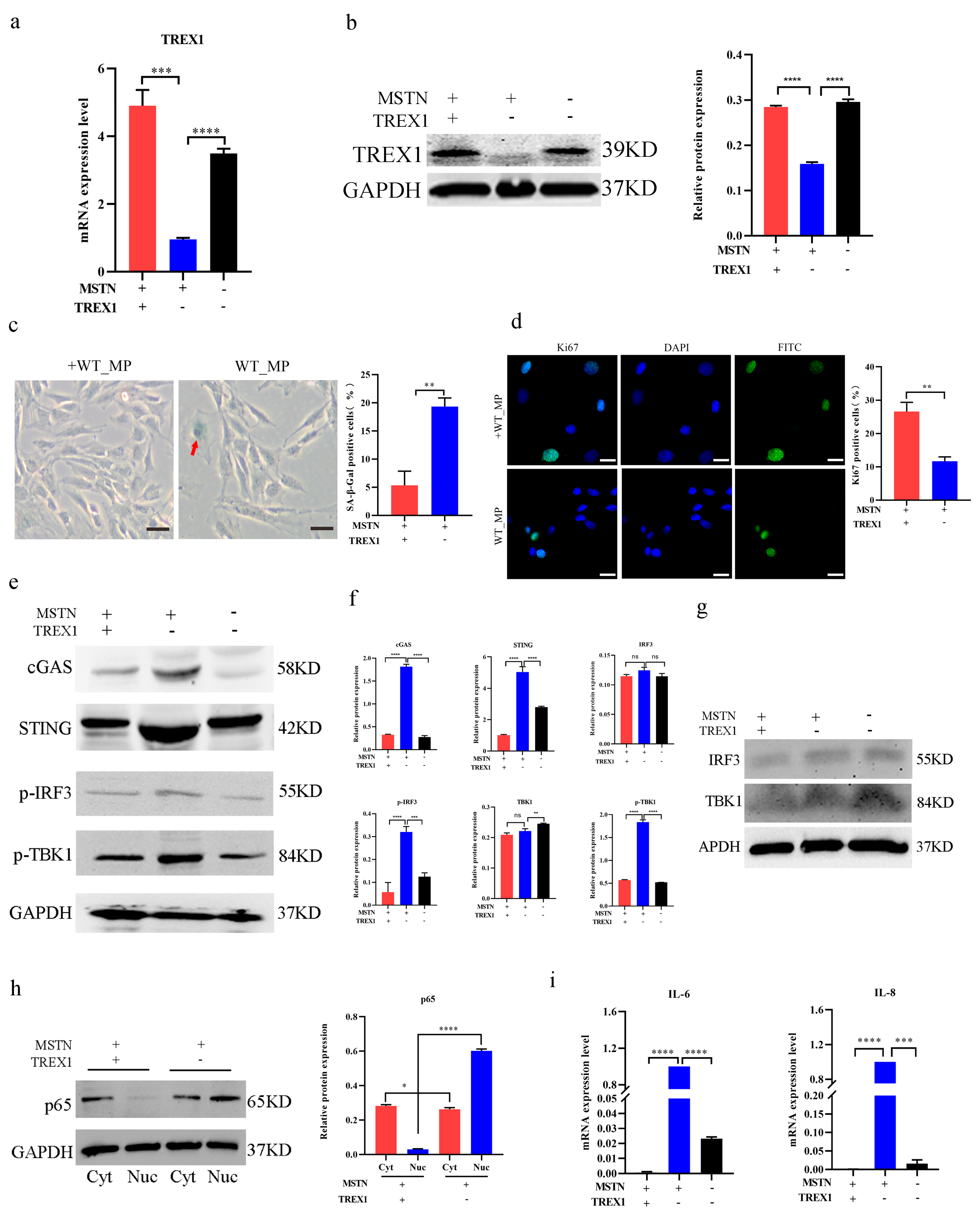
2.4. Inactivation of the MSTN Downregulated the SASP through the Upregulation of TREX1
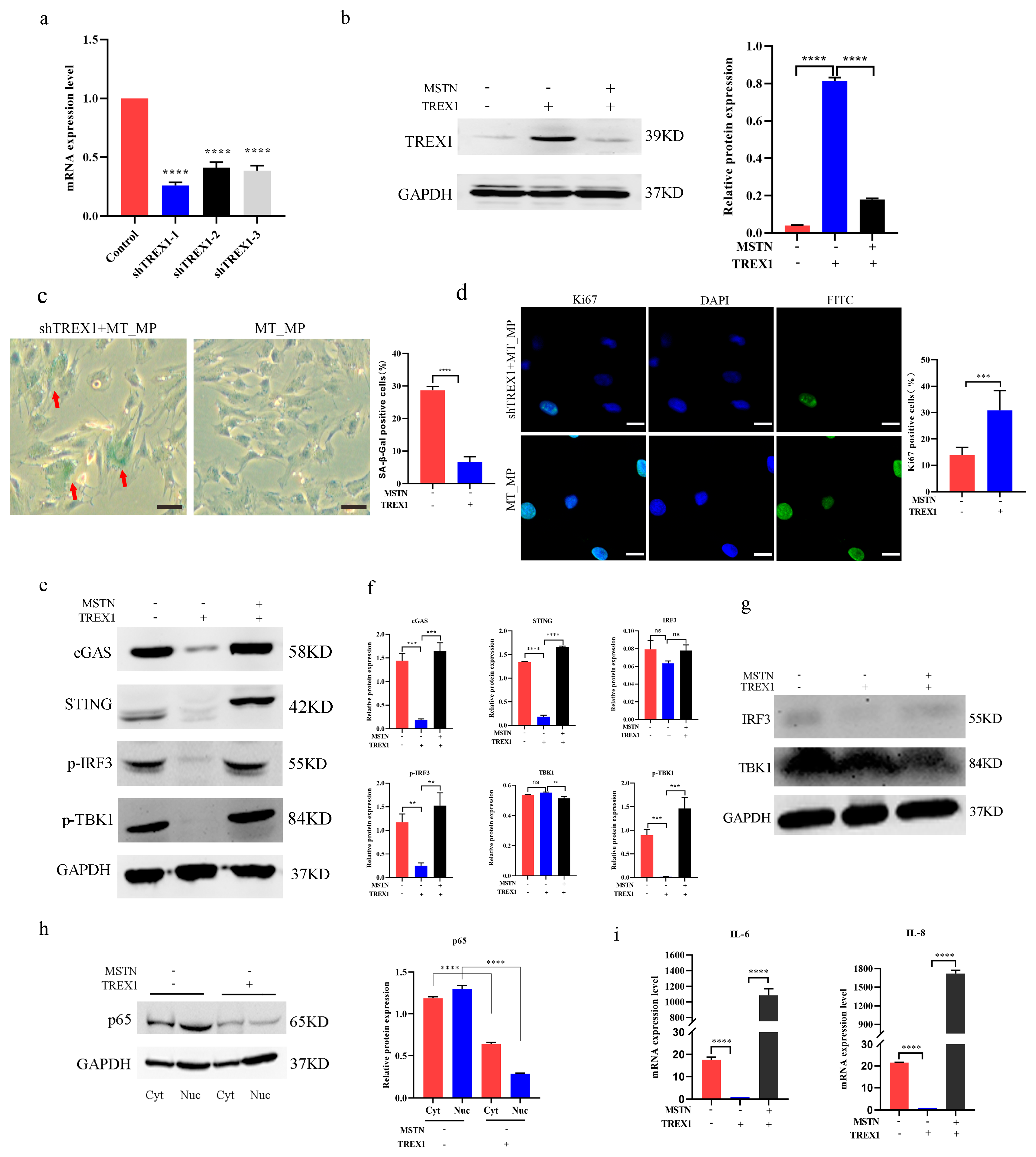
2.5. Overexpression of TREX1 Can Rejuvenate Aged bSMCs
2.6. Aging Prematurely Due to TREX1 Absence in bSMCs
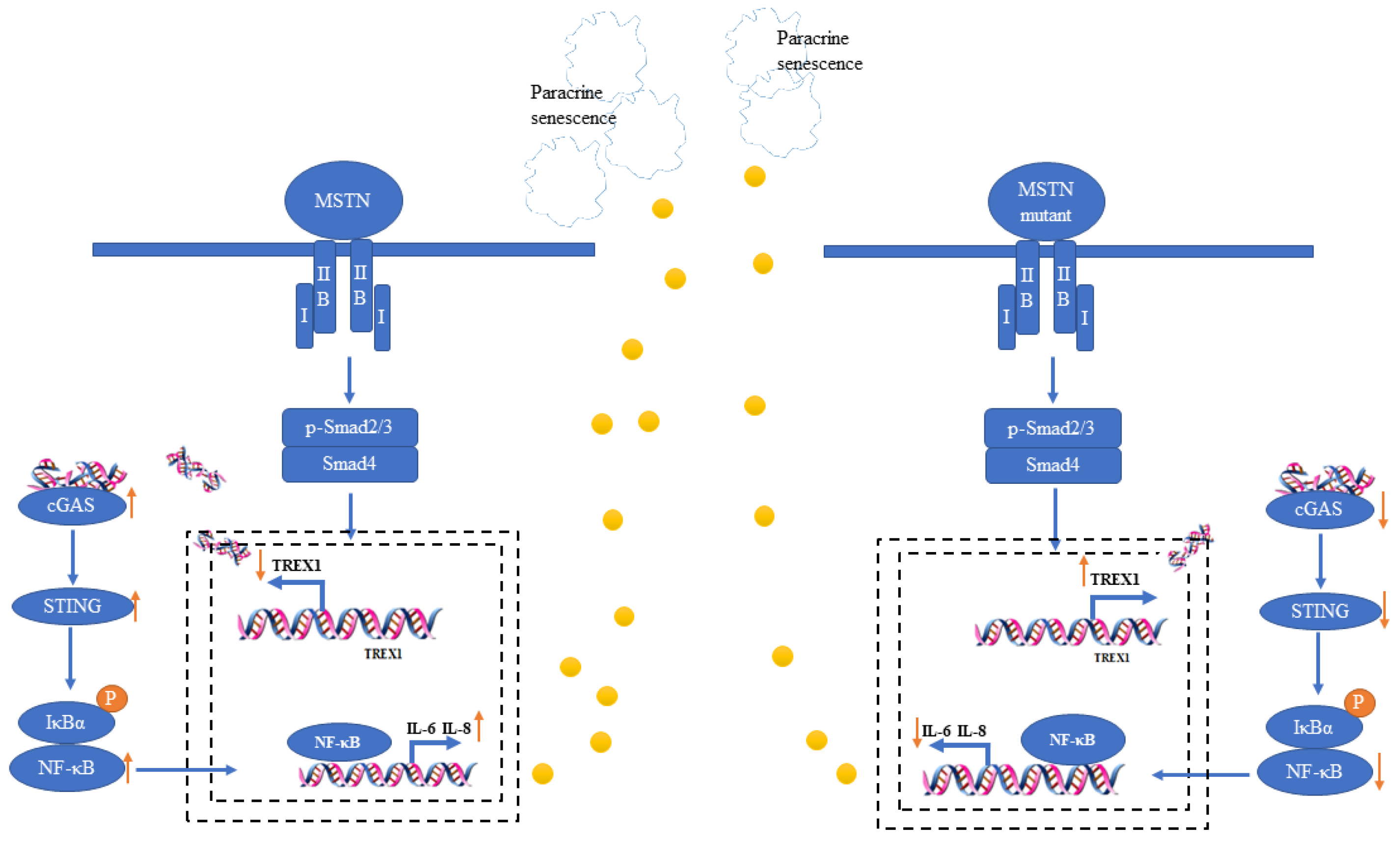
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Isolation
4.2. Telomere Length Assay
4.3. SA-β-Gal Staining
4.4. Immunofluorescent Staining
4.5. The use of Fluorescence-Activated Cell Sorting for Cell Cycle Assay (FACS)
4.6. Real-Time PCR
4.7. Western Blot
4.8. RNA-Seq Analysis
4.9. Chromatin Immunoprecipitation Assay
4.10. Luciferase Reporter
4.11. Transfection with TREX1 shRNA and Overexpression Vector
4.12. Statistics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.S. Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 2004, 351, 1030–1031. [Google Scholar]
- Paek, H.J.; Li, Z.Y.; Quan, B.H.; Yin, X.J. Application of PCR-RFLP for quick identification of MSTN mutants in MSTN mutant pig breeding. Anim. Biotechnol. 2023, 34, 2231–2239. [Google Scholar] [CrossRef]
- Gu, M.; Wang, S.; Di, A.; Wu, D.; Hai, C.; Liu, X.; Bai, C.; Su, G.; Yang, L.; Li, G. Combined Transcriptome and Metabolome Analysis of Smooth Muscle of Myostatin Knockout Cattle. Int. J. Mol. Sci. 2023, 24, 8120. [Google Scholar] [CrossRef] [PubMed]
- Song, S.Z.; He, Z.Y.; Cheng, Y.; Yu, B.L.; Zhang, T.; Li, D. MSTN modification in goat mediated by TALENs and performance analysis. Yi Chuan 2022, 44, 531–5421. [Google Scholar] [PubMed]
- Hai, C.; Bai, C.; Yang, L.; Wei, Z.; Wang, H.; Ma, H.; Ma, H.; Zhao, Y.; Su, G.; Li, G. Effects of Different Generations and Sex on Physiological, Biochemical, and Growth Parameters of Crossbred Beef Cattle by Myostatin Gene-Edited Luxi Bulls and Simmental Cows. Animal 2023, 13, 3216. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B. Editorial: Genetic engineering in farm animals. Front. Genet. 2023, 14, 1155201. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Sanes, J.R. The basement membrane/basal lamina of skeletal muscle. J. Biol. Chem. 2003, 278, 12601–12604. [Google Scholar] [CrossRef]
- Piegari, E.; Angelis, A.D.; Cappetta, D.; Russo, R.; Esposito, G.; Costantino, S.; Graiani, G.; Frati, C.; Prezioso, L.; Berrino, L. Doxorubicin induces senescence and impairs function of human cardiac progenitor cells. Basic. Res. Cardiol. 2013, 108, 334. [Google Scholar] [CrossRef]
- Barani, A.E.; Durieux, A.C.; Sabido, O.; Freyssenet, D. Age-related changes in the mitotic and metabolic characteristics of muscle-derived cells. J. Appl. Physiol. 2003, 95, 2089–2098. [Google Scholar] [CrossRef]
- Fakhari, A.; Berkland, C. Applications and emerging trends of hyaluronic acid in tissue engineering, as a dermal filler and in osteoarthritis treatment. Acta Biomater. 2013, 9, 7081–7092. [Google Scholar] [CrossRef] [PubMed]
- Stellavato, A.; Abate, L.; Vassallo, V.; Donniacuo, M.; Rinaldi, B.; Schiraldi, C. An in vitro study to assess the effect of hyaluronan-based gels on muscle-derived cells: Highlighting a new perspective in regenerative medicine. PLoS ONE 2020, 15, e0236164. [Google Scholar] [CrossRef]
- Siriett, V.; Platt, L.; Salerno, M.S.; Ling, N.; Kambadur, R.; Sharma, M. Prolonged absence of myostatin reduces sarcopenia. J. Cell Physiol. 2006, 209, 866–873. [Google Scholar] [CrossRef]
- Mendias, C.L.; Bakhurin, K.I.; Gumucio, J.P.; Shallal-Ayzin, M.V.; Davis, C.S.; Faulkner, J.A. Haploinsufficiency of myostatin protects against aging-related declines in muscle function and enhances the longevity of mice. Aging Cell 2015, 14, 704–706. [Google Scholar] [CrossRef]
- Ruiz de Galarreta, M.; Lujambio, A. DNA sensing in senescence. Nat. Cell Biol. 2017, 19, 1008–1009. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhao, J.; Chen, H.; Zhou, J.; Chen, A.; Zhang, J.; Wang, Y.; Mao, Z.; Wang, J.; Qiu, X.; et al. Bmi-1 Overexpression Improves Sarcopenia Induced by 1,25(OH)2D3 Deficiency and Downregulates GATA4-Dependent Rela Transcription. J. Bone Min. Res. 2023, 38, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Glück, S.; Ablasser, A. Innate immunosensing of DNA in cellular senescence. Curr. Opin. Immunol. 2019, 56, 31–36. [Google Scholar] [CrossRef]
- Takahashi, A.; Loo, T.M.; Okada, R.; Kamachi, F.; Watanabe, Y.; Wakita, M.; Watanabe, S.; Kawamoto, S.; Miyata, K.; BaRber, G.N. Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat. Commun. 2018, 9, 1249. [Google Scholar] [CrossRef]
- Yang, Y.G.; Lindahl, T.; Barnes, D.E. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 2007, 131, 873–886. [Google Scholar] [CrossRef]
- Atianand, M.K.; Fitzgerald, K.A. Molecular basis of DNA recognition in the immune system. J. Immunol. 2013, 190, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- McCroskery, S.; Thomas, M.; Maxwell, L.; Sharma, M.; Kambadur, R. Myostatin negatively regulates satellite cell activation and self-renewal. J. Cell Biol. 2003, 162, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.E.; Bhasin, S.; Artaza, J.; Byhower, F.; Azam, M.; Willard, D.H., Jr.; Kull, F.C., Jr.; Gonzalez-Cadavid, N. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am. J. Physiol. Endocrinol. Metab. 2001, 280, E221–E228. [Google Scholar] [CrossRef] [PubMed]
- Erdal, E.; Haider, S.; Rehwinkel, J.; Harris, A.L.; McHugh, P.J. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes. Dev. 2017, 31, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Harding, S.M.; Benci, J.L.; Irianto, J.; Discher, D.E.; Minn, A.J.; Greenberg, R.A. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017, 548, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, K.J.; Carroll, P.; Martin, C.A.; Murina, O.; Fluteau, A.; Simpson, D.J.; Olova, N.; Sutcliffe, H.; Rainger, J.K.; Leitch, A.; et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017, 548, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yang, M.; Wei, Z.; Gu, M.; Li, G. MSTN Mutant Promotes Myogenic Differentiation by Increasing Demethylase TET1 Expression via the SMAD2/SMAD3 Pathway. Int. J. Biol. Sci. 2020, 16, 1324–1334. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Glück, S.; Guey, B.; Gulen, M.F.; Wolter, K.; Kang, T.W.; Schmacke, N.A.; Bridgeman, A.; Rehwinkel, J.; Zender, L.; Ablasser, A. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017, 19, 1061–1070. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, H.Y.; Lee, J.H.; Yoon, K.H.; Chang, M.S.; Park, S.K. The age-dependent induction of apoptosis-inducing factor (AIF) in the human semitendinosus skeletal muscle. Cell. Mol. Biol. Lett. 2010, 15, 1–12. [Google Scholar] [CrossRef]
- Etienne, J.; Liu, C.; Skinner, C.M.; Conboy, M.J.; Conboy, I.M. Skeletal muscle as an experimental model of choice to study tissue aging and rejuvenation. Skelet. Muscle 2020, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Baumann, A.P.; Ibebunjo, C.; Grasser, W.A.; Paralkar, V.M. Myostatin expression in age and denervation-induced skeletal muscle atrophy. J. Musculoskelet. Neuronal Interact. 2003, 3, 8–16. [Google Scholar] [PubMed]
- Gutierrez-Salmean, G.; Ciaraldi, T.P.; Nogueira, L.; Barboza, J.; Taub, P.R.; Hogan, M.C.; Henry, R.R.; Meaney, E.; Villarreal, F.; Ceballos, G.; et al. Effects of (-)-epicatechin on molecular modulators of skeletal muscle growth and differentiation. J. Nutr. Biochem. 2014, 25, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, R.; Heintz, C. Enhancement of skeletal muscle regeneration. Dev. Dyn. 1994, 201, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965, 37, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hu, B.; Ding, Z.; Dang, Y.; Wu, J.; Di, L.; Liu, X.; Xiao, B.; Zhang, W.; Ren, R.; et al. ATF6 safeguards organelle homeostasis and cellular aging in human mesenchymal stem cells. Cell Discov. 2018, 4, 2. [Google Scholar] [CrossRef]
- Wagner, W.; Horn, P.; Castoldi, M.; Diehlmann, A.; Bork, S.; Saffrich, R.; Benes, V.; Blake, J.; Pfister, S.; Eckstein, V.; et al. Replicative senescence of mesenchymal stem cells: A continuous and organized process. PLoS ONE 2008, 3, e2213. [Google Scholar] [CrossRef] [PubMed]
- Lasry, A.; Ben-Neriah, Y. Senescence-associated inflammatory responses: Aging and cancer perspectives. Trends Immunol. 2015, 36, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Childs, B.G.; Gluscevic, M.; Baker, D.J.; Laberge, R.M.; Marquess, D.; Dananberg, J.; van Deursen, J.M. Senescent cells: An emerging target for diseases of ageing. Nat. Rev. Drug Discov. 2017, 16, 718–735. [Google Scholar] [CrossRef]
- He, S.; Sharpless, N.E. Senescence in Health and Disease. Cell 2017, 169, 1000–1011. [Google Scholar] [CrossRef]
- Salama, R.; Sadaie, M.; Hoare, M.; Narita, M. Cellular senescence and its effector programs. Genes. Dev. 2014, 28, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Kuilman, T.; Michaloglou, C.; Vredeveld, L.C.; Douma, S.; van Doorn, R.; Desmet, C.J.; Aarden, L.A.; Mooi, W.J.; Peeper, D.S. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 2008, 133, 1019–1031. [Google Scholar] [CrossRef]
- Zhixun Dou, K.G.M.G.V.J.Z. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 2017, 550, 402. [Google Scholar]
- Luo, W.D.; Wang, Y.P.; Lv, J.; Liu, Y.; Qu, Y.Q.; Xu, X.F.; Yang, L.J.; Lin, Z.C.; Wang, L.N.; Chen, R.H.; et al. Age-related self-DNA accumulation may accelerate arthritis in rats and in human rheumatoid arthritis. Nat. Commun. 2023, 14, 4394. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; LondoO, D.; Bouley, R.; Rooney, M.; Hacohen, N. Dnase2a Deficiency Uncovers Lysosomal Clearance of Damaged Nuclear DNA via Autophagy. Cell Rep. 2014, 9, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Loeser, R.F.; Collins, J.A.; Diekman, B.O. Ageing and the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016, 12, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Jang, K.M.; Park, K.K. Effects of Apamin on MPP(+)-Induced Calcium Overload and Neurotoxicity by Targeting CaMKII/ERK/p65/STAT3 Signaling Pathways in Dopaminergic Neuronal Cells. Int. J. Mol. Sci. 2022, 23, 15255. [Google Scholar] [CrossRef] [PubMed]
- Assis, S.I.S.; Amendola, L.S.; Okamoto, M.M.; Ferreira, G.D.S.; Iborra, R.T.; Santos, D.R.; Santana, M.F.M.; Santana, K.G.; Correa-Giannella, M.L.; Barbeiro, D.F.; et al. The Prolonged Activation of the p65 Subunit of the NF-Kappa-B Nuclear Factor Sustains the Persistent Effect of Advanced Glycation End Products on Inflammatory Sensitization in Macrophages. Int. J. Mol. Sci. 2024, 25, 2713. [Google Scholar] [CrossRef]
- Muñoz-Espín, D.; Serrano, M. Cellular senescence: From physiology to pathology. Nat. Rev. Mol. Cell Biol. 2014, 15, 482–496. [Google Scholar] [CrossRef]
- Riley, J.; Tait, S. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 2020, 21, e49799. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Gao, L.; Gao, Y.; Hao, Z.; Zhou, X.; Su, G.; Bai, C.; Wei, Z.; Liu, X.; Yang, L.; et al. Inactivation of Myostatin Delays Senescence via TREX1-SASP in Bovine Skeletal Muscle Cells. Int. J. Mol. Sci. 2024, 25, 5277. https://doi.org/10.3390/ijms25105277
Yang M, Gao L, Gao Y, Hao Z, Zhou X, Su G, Bai C, Wei Z, Liu X, Yang L, et al. Inactivation of Myostatin Delays Senescence via TREX1-SASP in Bovine Skeletal Muscle Cells. International Journal of Molecular Sciences. 2024; 25(10):5277. https://doi.org/10.3390/ijms25105277
Chicago/Turabian StyleYang, Miaomiao, Li Gao, Yajie Gao, Zhenting Hao, Xinyu Zhou, Guanghua Su, Chunling Bai, Zhuying Wei, Xuefei Liu, Lei Yang, and et al. 2024. "Inactivation of Myostatin Delays Senescence via TREX1-SASP in Bovine Skeletal Muscle Cells" International Journal of Molecular Sciences 25, no. 10: 5277. https://doi.org/10.3390/ijms25105277
APA StyleYang, M., Gao, L., Gao, Y., Hao, Z., Zhou, X., Su, G., Bai, C., Wei, Z., Liu, X., Yang, L., & Li, G. (2024). Inactivation of Myostatin Delays Senescence via TREX1-SASP in Bovine Skeletal Muscle Cells. International Journal of Molecular Sciences, 25(10), 5277. https://doi.org/10.3390/ijms25105277





