Machine Learning and Single-Cell Analysis Identify Molecular Features of IPF-Associated Fibroblast Subtypes and Their Implications on IPF Prognosis
Abstract
:1. Introduction
2. Results
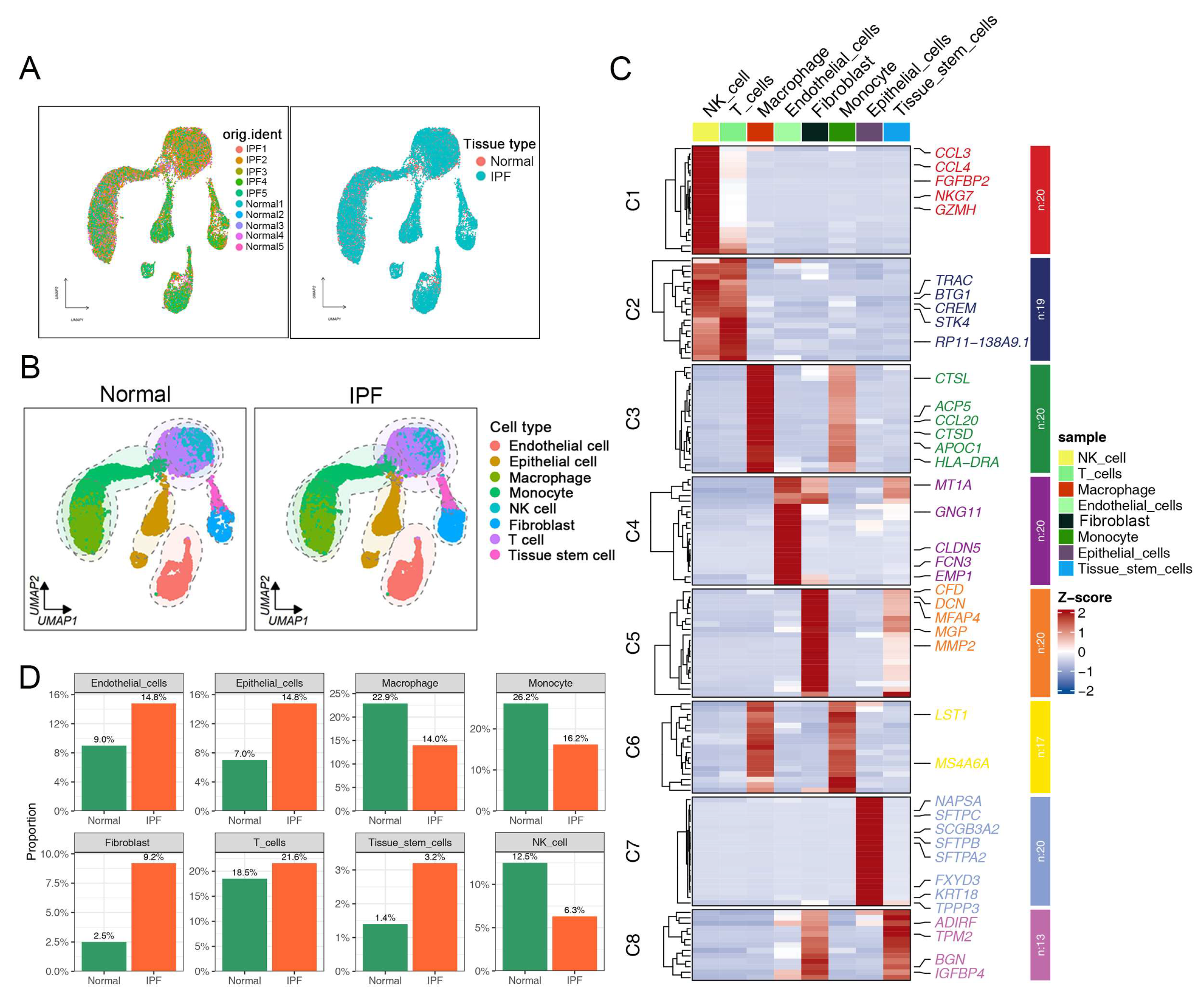
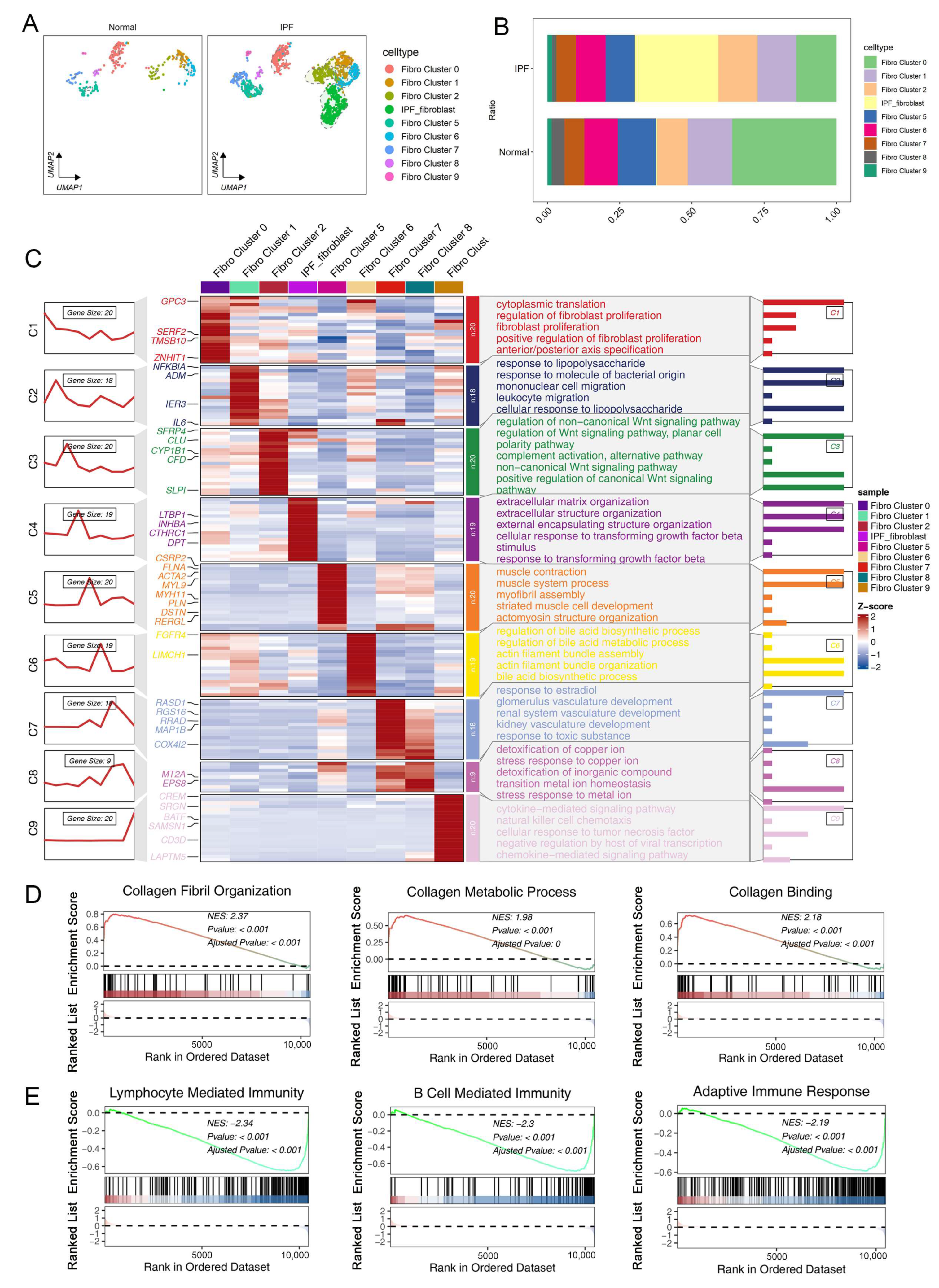
2.1. Single-Cell RNA Sequencing Reveals the Cellular Heterogeneity of Fibroblasts in Normal and Fibrotic Lung Tissues
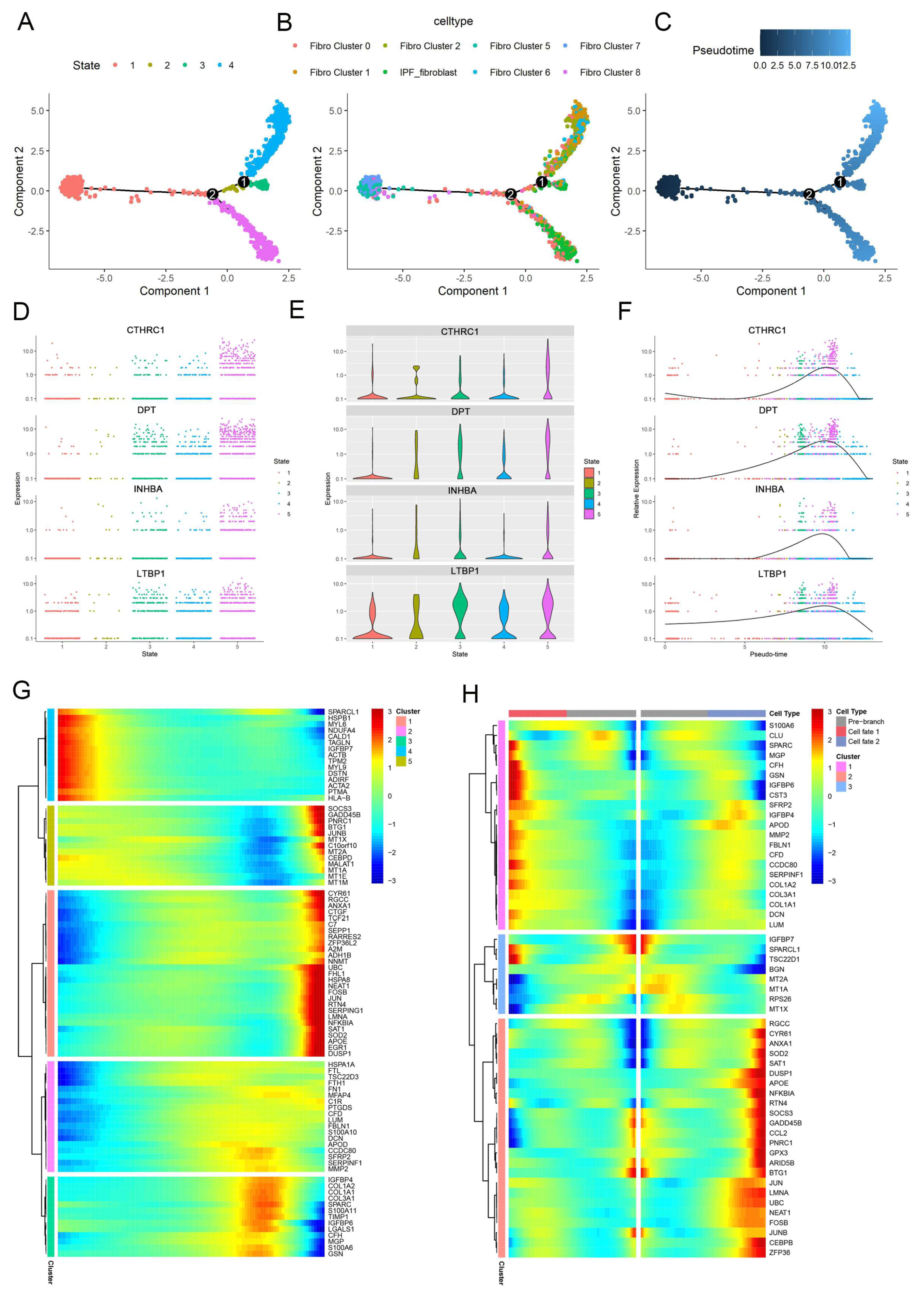
2.2. The Pseudotime Trajectory Analysis of Pathogenic Fibroblast Subtypes during Fibrogenesis
2.3. Cell–Cell Communications Analyses in Lung Fibroblast Subpopulations
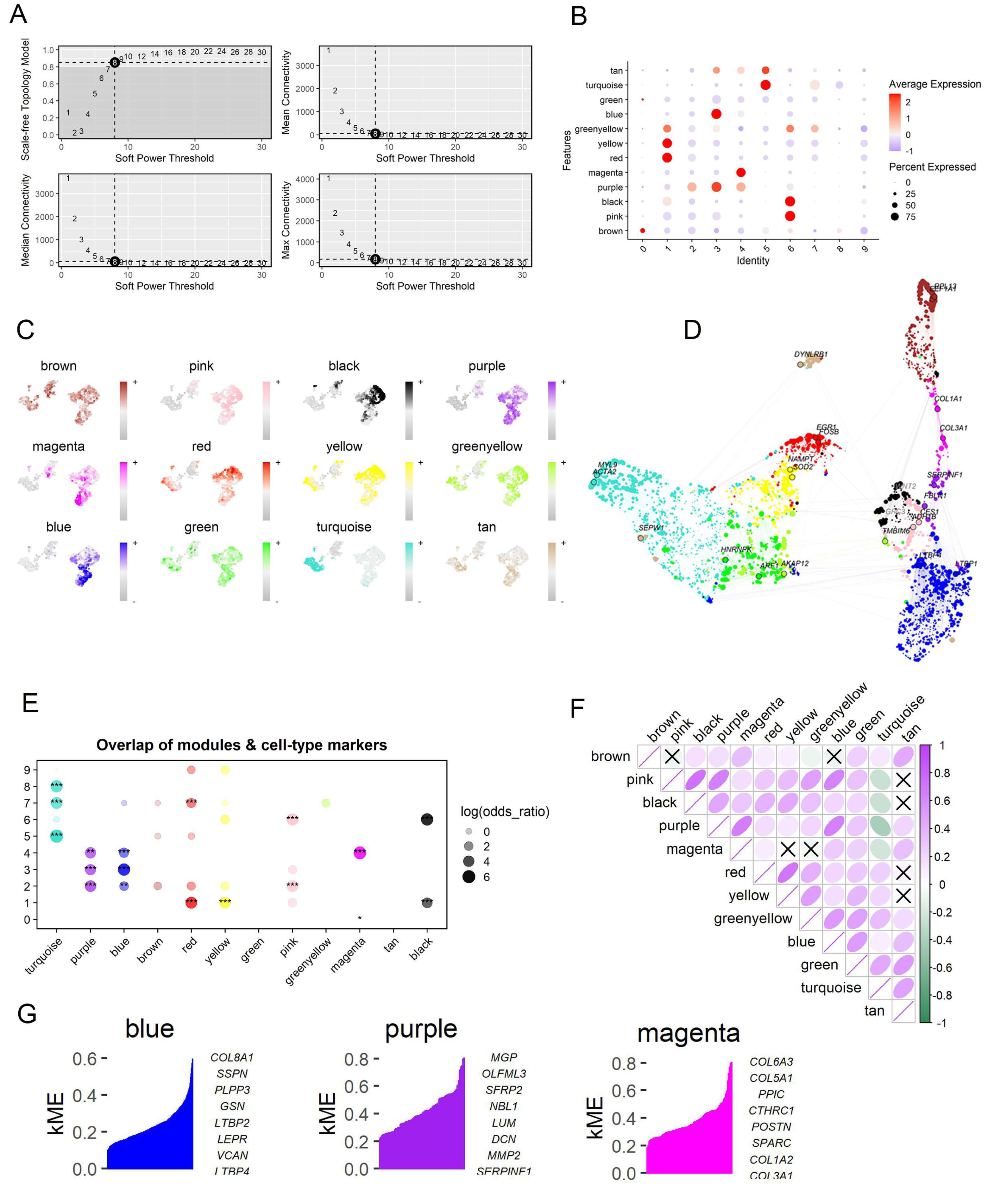
2.4. Identification of the Co-Expressed Gene Modules Associated with IPF-Fibroblasts by Using hdWGCNA
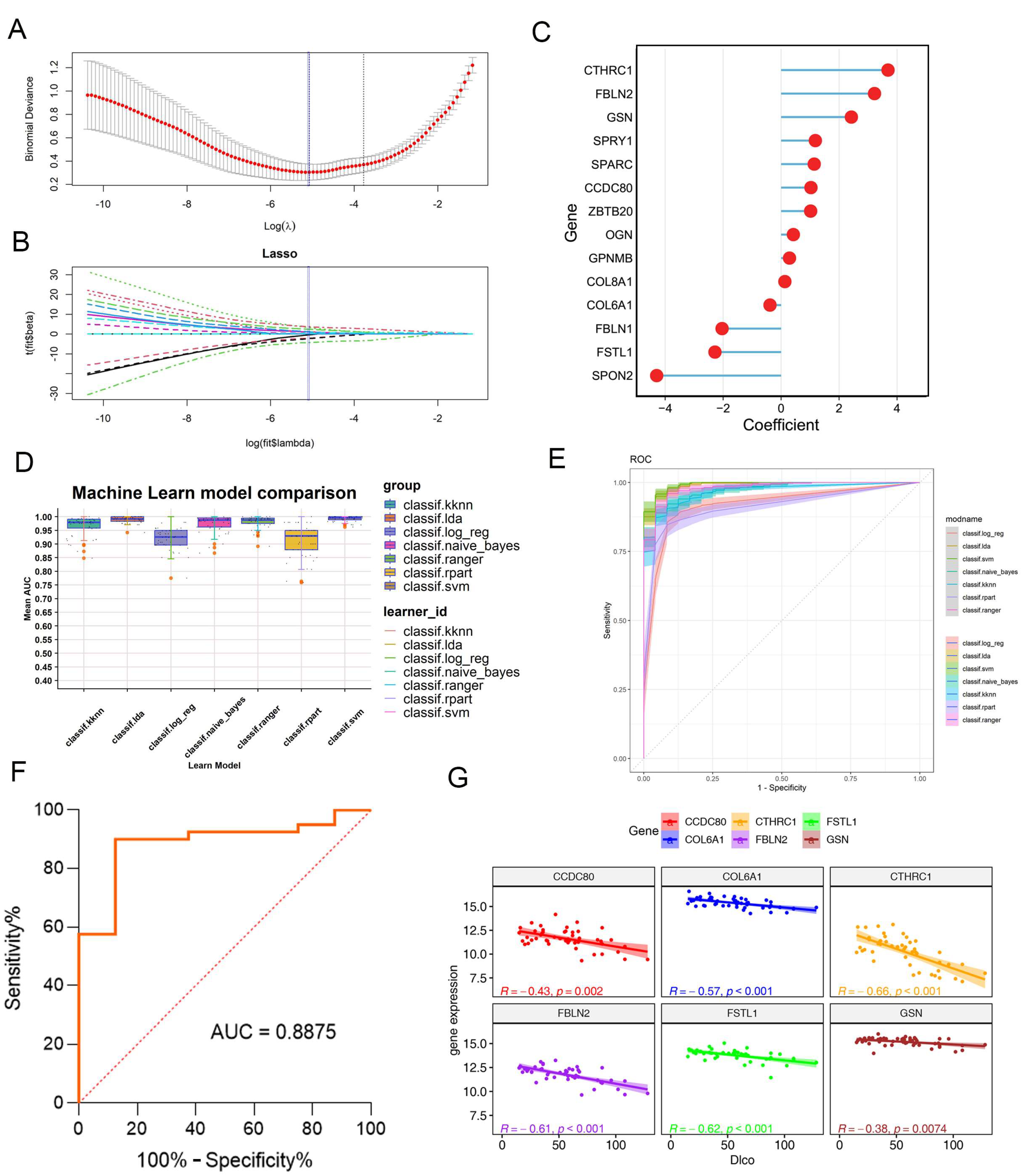
2.5. Machine Learning-Based Construction of the IPF-Fibroblast-Related Predictive Model for IPF
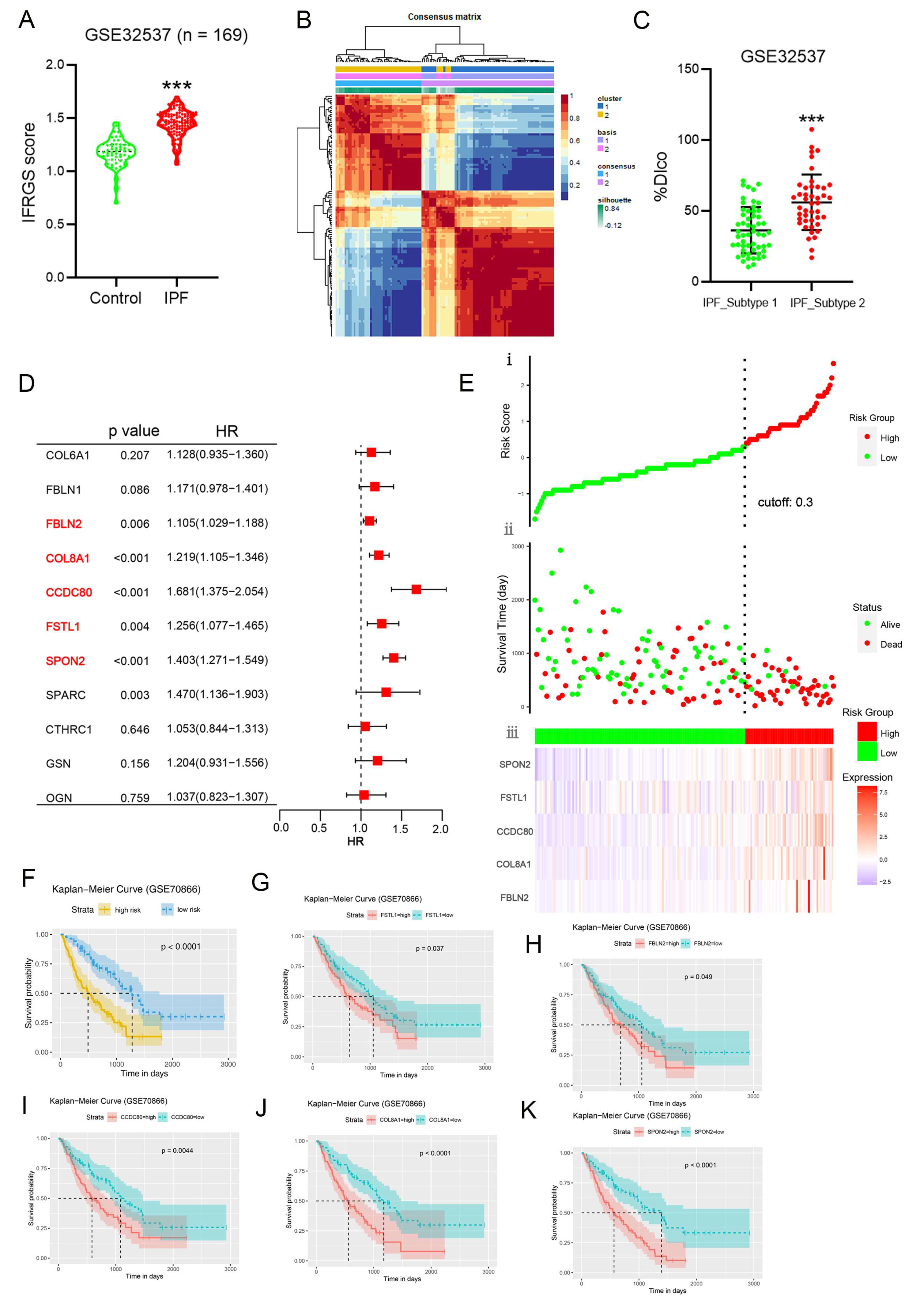
2.6. External Validation of an IPF-Fibroblast-Related Prognostic Signature
3. Discussion
4. Materials and Methods
4.1. Data Acquisition
4.2. scRNA-seq Data Processing
4.3. Trajectory and Cell–Cell Communication Analysis
4.4. Enrichment Analysis
4.5. High Dimensional Weighted Gene Co-Expression Network Analysis (hdWGCNA)
4.6. Machine Learning-Based Construction of an IPF-Fibroblast-Related Predictive Model
4.7. Non–Negative Matrix Factorization (NMF) of IPF by IFRGs and Survival Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spagnolo, P.; Kropski, J.A.; Jones, M.G.; Lee, J.S.; Rossi, G.; Karampitsakos, T.; Maher, T.M.; Tzouvelekis, A.; Ryerson, C.J. Idiopathic pulmonary fibrosis: Disease mechanisms and drug development. Pharmacol. Ther. 2021, 222, 107798. [Google Scholar] [CrossRef] [PubMed]
- Moss, B.; Ryter, S.; Rosas, I. Pathogenic Mechanisms Underlying Idiopathic Pulmonary Fibrosis. Annu. Rev. Pathol. 2022, 17, 515–546. [Google Scholar] [CrossRef] [PubMed]
- Sgalla, G.; Biffi, A.; Richeldi, L. Idiopathic pulmonary fibrosis: Diagnosis, epidemiology and natural history. Respirology 2016, 21, 427–437. [Google Scholar] [CrossRef] [PubMed]
- King, T.E., Jr.; Tooze, J.A.; Schwarz, M.I.; Brown, K.R.; Cherniack, R.M. Predicting survival in idiopathic pulmonary fibrosis: Scoring system and survival model. Am. J. Respir. Crit. Care Med. 2001, 164, 1171–1181. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.; Liu, Z.; Zuo, H.; Yang, Z.; Qu, J. Idiopathic Pulmonary Fibrosis: An Update on Pathogenesis. Front. Pharmacol. 2021, 12, 797292. [Google Scholar] [CrossRef] [PubMed]
- Dean, C.H.; Lloyd, C.M. Lung Alveolar Repair: Not All Cells Are Equal. Trends Mol. Med. 2017, 23, 871–873. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.; Linn, G.; Chow, Y.H.; Kobayashi, A.; Mittelsteadt, K.; Altemeier, W.A.; Gharib, S.A.; Schnapp, L.M.; Duffield, J.S. Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2013, 188, 820–830. [Google Scholar] [CrossRef]
- Bamberg, A.; Redente, E.F.; Groshong, S.D.; Tuder, R.M.; Cool, C.D.; Keith, R.C.; Edelman, B.L.; Black, B.P.; Cosgrove, G.P.; Wynes, M.W.; et al. Protein Tyrosine Phosphatase-N13 Promotes Myofibroblast Resistance to Apoptosis in Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2018, 198, 914–927. [Google Scholar] [CrossRef]
- Lederer, D.J.; Martinez, F.J. Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 2018, 378, 1811–1823. [Google Scholar] [CrossRef]
- Günther, A.; Korfei, M.; Mahavadi, P.; von der Beck, D.; Ruppert, C.; Markart, P. Unravelling the progressive pathophysiology of idiopathic pulmonary fibrosis. Eur. Respir. Rev. 2012, 21, 152–160. [Google Scholar] [CrossRef]
- Liu, G.; Philp, A.M.; Corte, T.; Travis, M.A.; Schilter, H.; Hansbro, N.G.; Burns, C.J.; Eapen, M.S.; Sohal, S.S.; Burgess, J.K.; et al. Therapeutic targets in lung tissue remodelling and fibrosis. Pharmacol. Ther. 2021, 225, 107839. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.D.; Watt, F.M. Fibroblast heterogeneity: Implications for human disease. J. Clin. Investig. 2018, 128, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Valenzi, E.; Bulik, M.; Tabib, T.; Morse, C.; Sembrat, J.; Trejo Bittar, H.; Rojas, M.; Lafyatis, R. Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann. Rheum. Dis. 2019, 78, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Li, Q.; Shi, J.; Wei, J.; Li, P.; Chang, C.H.; Shultz, L.D.; Ren, G. Lung fibroblasts facilitate pre-metastatic niche formation by remodeling the local immune microenvironment. Immunity 2022, 55, 1483–1500.e9. [Google Scholar] [CrossRef] [PubMed]
- Hanley, C.J.; Waise, S.; Ellis, M.J.; Lopez, M.A.; Pun, W.Y.; Taylor, J.; Parker, R.; Kimbley, L.M.; Chee, S.J.; Shaw, E.C.; et al. Single-cell analysis reveals prognostic fibroblast subpopulations linked to molecular and immunological subtypes of lung cancer. Nat. Commun. 2023, 14, 387. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Dobie, R.; Wilson-Kanamori, J.R.; Dora, E.F.; Henderson, B.E.P.; Luu, N.T.; Portman, J.R.; Matchett, K.P.; Brice, M.; Marwick, J.A.; et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019, 575, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Kuppe, C.; Ibrahim, M.M.; Kranz, J.; Zhang, X.; Ziegler, S.; Perales-Patón, J.; Jansen, J.; Reimer, K.C.; Smith, J.R.; Dobie, R.; et al. Decoding myofibroblast origins in human kidney fibrosis. Nature 2021, 589, 281–286. [Google Scholar] [CrossRef]
- Reyfman, P.A.; Walter, J.M.; Joshi, N.; Anekalla, K.R.; McQuattie-Pimentel, A.C.; Chiu, S.; Fernandez, R.; Akbarpour, M.; Chen, C.I.; Ren, Z.; et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 199, 1517–1536. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021, 25, 1315–1360. [Google Scholar] [CrossRef]
- Maddali, M.V.; Kalra, A.; Muelly, M.; Reicher, J.J. Development and validation of a CT-based deep learning algorithm to augment non-invasive diagnosis of idiopathic pulmonary fibrosis. Respir. Med. 2023, 219, 107428. [Google Scholar] [CrossRef] [PubMed]
- El-Hasnony, I.M.; Elzeki, O.M.; Alshehri, A.; Salem, H. Multi-Label Active Learning-Based Machine Learning Model for Heart Disease Prediction. Sensors 2022, 22, 1184. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Chen, H.; Ke, S.; Mo, L.; Qiu, M.; Zhu, G.; Zhu, W.; Liu, L. Identifying potential biomarkers of idiopathic pulmonary fibrosis through machine learning analysis. Sci. Rep. 2023, 13, 16559. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.; Wu, E.; Zhang, A.; Alizadeh, A.A.; Zou, J. From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell 2023, 186, 1772–1791. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Hofmanninger, J.; Nenning, K.H.; Prayer, F.; Röhrich, S.; Sverzellati, N.; Poletti, V.; Tomassetti, S.; Weber, M.; Prosch, H.; et al. Unsupervised machine learning identifies predictive progression markers of IPF. Eur. Radiol. 2023, 33, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.D.; Li, F.; Chen, J.Y.; Zhang, J.; Qian, L.L.; Wang, R.X. Analysis of potential genetic biomarkers using machine learning methods and immune infiltration regulatory mechanisms underlying atrial fibrillation. BMC Med. Genom. 2022, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, L.H.; de Andrade, J.A.; Zibrak, J.D.; Padilla, M.L.; Albera, C.; Nathan, S.D.; Wijsenbeek, M.S.; Stauffer, J.L.; Kirchgaessler, K.U.; Costabel, U. Pirfenidone safety and adverse event management in idiopathic pulmonary fibrosis. Eur. Respir. Rev. 2017, 26, 170057. [Google Scholar] [CrossRef]
- Glass, D.S.; Grossfeld, D.; Renna, H.A.; Agarwala, P.; Spiegler, P.; DeLeon, J.; Reiss, A.B. Idiopathic pulmonary fibrosis: Current and future treatment. Clin. Respir. J. 2022, 16, 84–96. [Google Scholar] [CrossRef]
- Habermann, A.C.; Gutierrez, A.J.; Bui, L.T.; Yahn, S.L.; Winters, N.I.; Calvi, C.L.; Peter, L.; Chung, M.I.; Taylor, C.J.; Jetter, C.; et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 2020, 6, eaba1972. [Google Scholar] [CrossRef]
- Adams, T.S.; Schupp, J.C.; Poli, S.; Ayaub, E.A.; Neumark, N.; Ahangari, F.; Chu, S.G.; Raby, B.A.; DeIuliis, G.; Januszyk, M.; et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 2020, 6, eaba1983. [Google Scholar] [CrossRef]
- Budi, E.H.; Schaub, J.R.; Decaris, M.; Turner, S.; Derynck, R. TGF-β as a driver of fibrosis: Physiological roles and therapeutic opportunities. J. Pathol. 2021, 254, 358–373. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Lyu, C.; Liao, H.; Du, Y. Collagen crosslinking: Effect on structure, mechanics and fibrosis progression. Biomed. Mater. 2021, 16, 062005. [Google Scholar] [CrossRef] [PubMed]
- Leask, A. Potential therapeutic targets for cardiac fibrosis: TGFbeta, angiotensin, endothelin, CCN2, and PDGF, partners in fibroblast activation. Circ. Res. 2010, 106, 1675–1680. [Google Scholar] [CrossRef] [PubMed]
- Morabito, S.; Reese, F.; Rahimzadeh, N.; Miyoshi, E.; Swarup, V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell. Rep. Methods 2023, 3, 100498. [Google Scholar] [CrossRef] [PubMed]
- Gervasi, M.; Bianchi-Smiraglia, A.; Cummings, M.; Zheng, Q.; Wang, D.; Liu, S.; Bakin, A.V. JunB contributes to Id2 repression and the epithelial-mesenchymal transition in response to transforming growth factor-β. J. Cell Biol. 2012, 196, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, Y.; Miller, M.; Cao, L.; Zhao, J.; Wu, J.; Wang, J.; Liu, L.; Li, S.; Zou, M.; et al. Autophagy plays a role in FSTL1-induced epithelial mesenchymal transition and airway remodeling in asthma. Am. J. Physiol. Lung Cell Mol. Physiol. 2017, 313, L27–L40. [Google Scholar] [CrossRef]
- Jana, S.; Aujla, P.; Hu, M.; Kilic, T.; Zhabyeyev, P.; McCulloch, C.A.; Oudit, G.Y.; Kassiri, Z. Gelsolin is an important mediator of Angiotensin II-induced activation of cardiac fibroblasts and fibrosis. FASEB J. 2021, 35, e21932. [Google Scholar] [CrossRef]
- Tsukui, T.; Sun, K.H.; Wetter, J.B.; Wilson-Kanamori, J.R.; Hazelwood, L.A.; Henderson, N.C.; Adams, T.S.; Schupp, J.C.; Poli, S.D.; Rosas, I.O.; et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat. Commun. 2020, 11, 1920. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, L.; Wang, L.; Wang, H.; Wu, J.; Cai, H.; Mo, C.; Yang, J. Machine learning identified MDK score has prognostic value for idiopathic pulmonary fibrosis based on integrated bulk and single cell expression data. Front. Genet. 2023, 14, 1246983. [Google Scholar] [CrossRef]
- Morse, C.; Tabib, T.; Sembrat, J.; Buschur, K.L.; Bittar, H.T.; Valenzi, E.; Jiang, Y.; Kass, D.J.; Gibson, K.; Chen, W.; et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 2019, 54, 1802441. [Google Scholar] [CrossRef]
- Martinez, F.J.; Collard, H.R.; Pardo, A.; Raghu, G.; Richeldi, L.; Selman, M.; Swigris, J.J.; Taniguchi, H.; Wells, A.U. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Primers 2017, 3, 17074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shen, H.; Yang, T.; Li, T.; Liu, X.; Wang, J.; Liao, Z.; Wei, J.; Lu, J.; Liu, H.; et al. A single-cell analysis reveals tumor heterogeneity and immune environment of acral melanoma. Nat. Commun. 2022, 13, 7250. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]

| Dataset | Year | Area | Species | Platform | Data Type | Number of Samples | |
|---|---|---|---|---|---|---|---|
| Normal | IPF | ||||||
| GSE128033 | 2019 | United States | Homo | GPL18573 | scRNA-seq | 10 | 8 |
| GSE32537 | 2011 | United States | Homo | GPL6244 | Bulk RNA-seq | 39 | 131 |
| GSE110147 | 2018 | Canada | Homo | GPL6244 | Bulk RNA-seq | 11 | 22 |
| GSE70866 | 2015 | Germany | Homo | GPL14550 | Bulk RNA-seq | 20 | 212 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, J.; Yang, Y.; Han, X. Machine Learning and Single-Cell Analysis Identify Molecular Features of IPF-Associated Fibroblast Subtypes and Their Implications on IPF Prognosis. Int. J. Mol. Sci. 2024, 25, 94. https://doi.org/10.3390/ijms25010094
Hou J, Yang Y, Han X. Machine Learning and Single-Cell Analysis Identify Molecular Features of IPF-Associated Fibroblast Subtypes and Their Implications on IPF Prognosis. International Journal of Molecular Sciences. 2024; 25(1):94. https://doi.org/10.3390/ijms25010094
Chicago/Turabian StyleHou, Jiwei, Yanru Yang, and Xin Han. 2024. "Machine Learning and Single-Cell Analysis Identify Molecular Features of IPF-Associated Fibroblast Subtypes and Their Implications on IPF Prognosis" International Journal of Molecular Sciences 25, no. 1: 94. https://doi.org/10.3390/ijms25010094
APA StyleHou, J., Yang, Y., & Han, X. (2024). Machine Learning and Single-Cell Analysis Identify Molecular Features of IPF-Associated Fibroblast Subtypes and Their Implications on IPF Prognosis. International Journal of Molecular Sciences, 25(1), 94. https://doi.org/10.3390/ijms25010094







